Map-Based Cloning, Phylogenetic, and Microsynteny Analyses of ZmMs20 Gene Regulating Male Fertility in Maize
Abstract
1. Introduction
2. Results
2.1. Genetic and Phenotypic Analyses of ms20 Mutant
2.2. Defective Anther Cuticle and Abortion Pollen Grain in ms20 Mutant
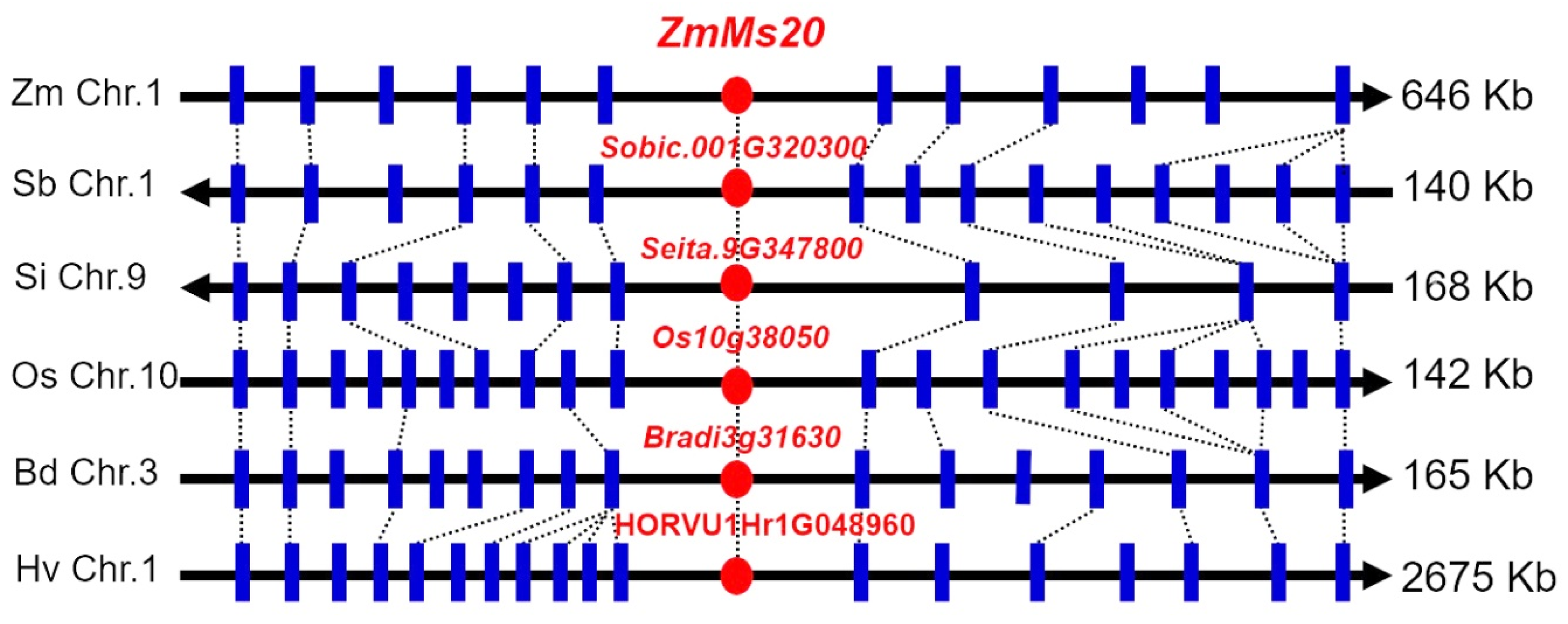
2.3. Isolation of ZmMs20
2.4. Phylogenetic Evolution of ZmMs20
2.5. The Rigorous Stage-specificity Expression Patterns of GMS Genes in Maize
2.6. Two Co-segregating Functional Markers Developed for Creating ms20 Male-sterility Lines by MAS Strategy under Different Genetic Backgrounds
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Condition, and Phenotypic Characterization
4.2. SEM Analysis
4.3. Genetic Analysis and Map-Based Cloning of ZmMs20
4.4. Sequence Alignment and Phylogenetic Analysis
4.5. Expression Analysis and Clustering of the Maize Cloned GMS Genes Using RNA-seq
4.6. Microsynteny Analysis of ZmMs20
4.7. Development of the Co-segregating Markers of ZmMs20/ms20 for MAS Breeding
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CMS | Cytoplasmic Male Sterility |
| GMC | Glucose Methanol Choline |
| GMS | Genic Male Sterility |
| MAS | Marker Assisted Selection |
| MCS, | Multi-Control Sterility |
| RNA-seq | RNA-sequencing |
| SEM | Scanning Electron Microscope |
| SSR | Simple Sequence Repeat |
| WT | Wild Type |
References
- Wan, X.Y.; Wu, S.W.; Li, Z.W.; Dong, Z.Y.; An, X.L.; Ma, M.; Tian, Y.H.; Li, J.P. Maize genic male-sterility genes and their applications in hybrid breeding: Progress and Perspectives. Mol. Plant 2019, 12, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K. The formation and function of plant cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Heredia, A. Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim. Biophys. Acta 2003, 1620, 1–7. [Google Scholar] [CrossRef]
- Walton, T.I.; Harwood, J.L.; Bowyer, J.R. Methods in Plant Biochemistry: Lipids, Membranes and Aspects of Photobiology; Academic Press: San Diego, CA, USA, 1990; Volume 4, pp. 105–158. [Google Scholar]
- Chen, W.; Yu, X.H.; Zhang, K.; Shi, J.; De Oliveira, S.; Schreiber, L.; Shanklin, J.; Zhang, D.B. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 2011, 157, 842–853. [Google Scholar] [CrossRef]
- Shi, J.; Tan, H.; Yu, X.H.; Liu, Y.; Liang, W.; Ranathunge, K.; Franke, R.B.; Schreiber, L.; Wang, Y.; Kai, G.; et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 2011, 23, 2225–2246. [Google Scholar] [CrossRef]
- Tian, Y.; Xiao, S.; Liu, J.; Somaratne, Y.; Zhang, H.; Wang, M.; Zhang, H.R.; Zhao, L.; Chen, H.B. MALE STERILE6021 (MS6021) is required for the development of anther cuticle and pollen exine in maize. Sci. Rep. 2017, 7, 16736. [Google Scholar] [CrossRef]
- Morant, M.; Jorgensen, K.; Schaller, H.; Pinot, F.; Moller, B.L.; Werck-Reichhart, D.; Bak, S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 2007, 19, 1473–1487. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, D.; Shi, J.; He, Y.; Pinot, F.; Grausem, B.; Yin, C.; Zhu, L.; Chen, M.; Luo, Z. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 2014, 56, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Somaratne, Y.; Tian, Y.; Zhang, H.; Wang, M.; Huo, Y.; Cao, F.; Zhao, L.; Chen, H.B. ABNORMAL POLLEN VACUOLATION1 (APV1) is required for male fertility by contributing to anther cuticle and pollen exine formation in maize. Plant J. 2017, 90, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.A.; Shrestha, J.; Morant, M.; Pinot, F.; Matsuno, M.; Swanson, R.; Moller, B.L.; Preuss, D. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 2009, 151, 574–589. [Google Scholar] [CrossRef]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef]
- Men, X.; Shi, J.; Liang, W.; Zhang, Q.; Lian, G.; Quan, S.; Zhu, L.; Luo, Z.; Chen, M.; Zhang, D. Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J. Exp. Bot. 2017, 68, 513–526. [Google Scholar] [CrossRef]
- Xie, K.; Wu, S.; Li, Z.; Zhou, Y.; Zhang, D.; Dong, Z.; An, X.; Zhu, T.; Zhang, S.; Liu, S.; et al. Map-based cloning and characterization of Zea mays male sterility33 (ZmMs33) gene, encoding a glycerol-3-phosphate acyltransferase. Theor. Appl. Genet. 2018, 131, 1363–1378. [Google Scholar] [CrossRef]
- Zhu, T.T.; Wu, S.W.; Zhang, D.F.; Li, Z.W.; Xie, K.; An, X.L.; Ma, M.; Hou, Q.C.; Dong, Z.Y.; Tian, Y.H.; et al. Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theor. Appl. Genet. 2019, TAAG-D-18-00637R1. (Accepted). [Google Scholar]
- An, X.L.; Dong, Z.Y.; Tian, Y.H.; Xie, K.; Wu, S.W.; Zhu, T.T.; Zhang, D.F.; Zhou, Y.; Niu, C.F.; Ma, B.; et al. ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol. Plant 2019, 12, 343–359. [Google Scholar] [CrossRef]
- Yang, C.; Vizcay-Barrena, G.; Conner, K.; Wilson, Z.A. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 2007, 19, 3530–3548. [Google Scholar] [CrossRef]
- Li, H.; Yuan, Z.; Vizcay-Barrena, G.; Yang, C.; Liang, W.; Zong, J.; Wilson, Z.A.; Zhang, D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011, 156, 615–630. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Dong, Z.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S.; et al. Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef]
- Cavener, D.R. GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J. Mol. Biol. 1992, 223, 811–814. [Google Scholar] [CrossRef]
- Wongnate, T.; Chaiyen, P. The substrate oxidation mechanism of pyranose 2-oxidase and other related enzymes in the glucose-methanol-choline superfamily. FEBS J. 2013, 280, 3009–3027. [Google Scholar] [CrossRef] [PubMed]
- Sorigue, D.; Legeret, B.; Cuine, S.; Blangy, S.; Moulin, S.; Billon, E.; Richaud, P.; Brugiere, S.; Coute, Y.; Nurizzo, D. An algal photoenzyme converts fatty acids to hydrocarbons. Science 2017, 357, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Kurdyukov, S.; Faust, A.; Trenkamp, S.; Bar, S.; Franke, R.; Efremova, N.; Tietjen, K.; Schreiber, L.; Saedler, H.; Yephremov, A. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta 2006, 224, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Akiba, T.; Hibara, K.; Kimura, F.; Tsuda, K.; Shibata, K.; Ishibashi, M.; Moriya, C.; Nakagawa, K.; Kurata, N.; Itoh, J. Organ fusion and defective shoot development in oni3 mutants of rice. Plant Cell Physiol. 2014, 55, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Hu, J.; Xu, J.; Yu, H.; Shi, Z.; Xiong, G.; Zhu, L.; Zeng, D.; Zhang, G.; Gao, Z. Identification and characterization of Mini1, a gene regulating rice shoot development. J. Integr. Plant Biol. 2015, 57, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, S.; Liu, Y.; Ling, S.; Chen, C.; Yao, J. HOTHEAD-Like HTH1 is Involved in anther cutin biosynthesis and is required for pollen fertility in rice. Plant Cell Physiol. 2017, 58, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Chen, Z.; Wang, N.; Xie, G.; Lu, J.; Yan, W.; Zhou, J.; Tang, X.; Deng, X.W. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. USA 2016, 113, 14145–14150. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lin, S.; Shi, J.; Yu, J.; Zhu, L.; Yang, X.; Zhang, D.; Liang, W. Rice No Pollen 1 (NP1) is required for anther cuticle formation and pollen exine patterning. Plant J. 2017, 91, 263–277. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, H.; Sun, H.; Luo, H.; Zhao, L.; Dong, Z.; Yan, S.; Zhao, C.; Liu, R.; Xu, C. IRREGULAR POLLEN EXINE1 is a novel factor in anther cuticle and pollen exine formation. Plant Physiol. 2017, 173, 307–325. [Google Scholar] [CrossRef]
- Evans, M.M. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Vernoud, V.; Laigle, G.; Rozier, F.; Meeley, R.B.; Perez, P.; Rogowsky, P.M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009, 59, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, M.; Fox, T.; Trimnell, M.; Wu, Y.; Lowe, L.; Li, B.; Faller, M. Msca1 Nucleotide Sequences Impacting Plant Male Fertility and Method of Using Same. U.S. Patent US20090038027A1, 5 Feburary 2009. [Google Scholar]
- Wang, C.J.; Nan, G.L.; Kelliher, T.; Timofejeva, L.; Vernoud, V.; Golubovskaya, I.N.; Harper, L.; Egger, R.; Walbot, V.; Cande, W.Z. Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 2012, 139, 2594–2603. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Skibbe, D.; Timofejeva, L.; Wang, C.J.; Kelliher, T.; Kremling, K.; Walbot, V.; Cande, W.Z. Regulation of cell divisions and differentiation by MALE STERILITY32 is required for anther development in maize. Plant J. 2013, 76, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Nan, G.L.; Zhai, J.; Arikit, S.; Morrow, D.; Fernandes, J.; Mai, L.; Nguyen, N.; Meyers, B.C.; Walbot, V. MS23, a master basic helix-loop-helix factor, regulates the specification and development of the tapetum in maize. Development 2017, 144, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, M.; Fox, T.; Leonard, A.; Li, B.; Loveland, B.; Trimnell, M. Cloning and Use of the ms9 Gene from Maize. U.S. Patent US20160024520A1, 8 January 2016. [Google Scholar]
- Wang, D.X.; Skibbe, D.S.; Walbot, V. Maize Male sterile 8 (Ms8), a putative β-1,3galactosyltransferase, modulates cell division, expansion, and differentiation during early maize anther development. Plant Reprod. 2013, 26, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Fox, T.; DeBruin, J.; Haug, C.K.; Trimnell, M.; Clapp, J.; Leonard, A.; Li, B.; Scolaro, E.; Collinson, S.; Glassman, K. A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 2017, 15, 942–952. [Google Scholar] [CrossRef]
- Cigan, A.M.; Unger, E.; Xu, R.J.; Kendall, T.; Fox, T.W. Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex. Plant Reprod. 2001, 14, 135–142. [Google Scholar] [CrossRef]
- Kannangara, R.; Branigan, C.; Liu, Y.; Penfield, T.; Rao, V.; Mouille, G.; Hofte, H.; Pauly, M.; Riechmann, J.L.; Broun, P. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 2007, 19, 1278–1294. [Google Scholar] [CrossRef]
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef]
- Zhou, X.Y.; Li, L.Z.; Xiang, J.H.; Gao, G.F.; Xu, F.X.; Liu, A.L.; Zhang, X.W.; Peng, Y.; Chen, X.B.; Wan, X.Y. OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS ONE 2015, 10, e116676. [Google Scholar] [CrossRef]
- Higgins, D.G.; Thompson, J.D.; Gibson, T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996, 266, 383–402. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]





| F2 Population Combination | Total Plants | Fertile Plants (F) | Sterile Plants (S) | F/S Ratio | χ2 | P | Significant Test p > 0.05 |
|---|---|---|---|---|---|---|---|
| ms20×Chang7-2 | 1365 | 1065 | 300 | 3.55:1 | 3.47 | 0.06 | ns |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, D.; Tian, Y.; Wu, S.; An, X.; Dong, Z.; Zhang, S.; Bao, J.; Li, Z.; Li, J.; et al. Map-Based Cloning, Phylogenetic, and Microsynteny Analyses of ZmMs20 Gene Regulating Male Fertility in Maize. Int. J. Mol. Sci. 2019, 20, 1411. https://doi.org/10.3390/ijms20061411
Wang Y, Liu D, Tian Y, Wu S, An X, Dong Z, Zhang S, Bao J, Li Z, Li J, et al. Map-Based Cloning, Phylogenetic, and Microsynteny Analyses of ZmMs20 Gene Regulating Male Fertility in Maize. International Journal of Molecular Sciences. 2019; 20(6):1411. https://doi.org/10.3390/ijms20061411
Chicago/Turabian StyleWang, Yanbo, Dongcheng Liu, Youhui Tian, Suowei Wu, Xueli An, Zhenying Dong, Simiao Zhang, Jianxi Bao, Ziwen Li, Jinping Li, and et al. 2019. "Map-Based Cloning, Phylogenetic, and Microsynteny Analyses of ZmMs20 Gene Regulating Male Fertility in Maize" International Journal of Molecular Sciences 20, no. 6: 1411. https://doi.org/10.3390/ijms20061411
APA StyleWang, Y., Liu, D., Tian, Y., Wu, S., An, X., Dong, Z., Zhang, S., Bao, J., Li, Z., Li, J., & Wan, X. (2019). Map-Based Cloning, Phylogenetic, and Microsynteny Analyses of ZmMs20 Gene Regulating Male Fertility in Maize. International Journal of Molecular Sciences, 20(6), 1411. https://doi.org/10.3390/ijms20061411





