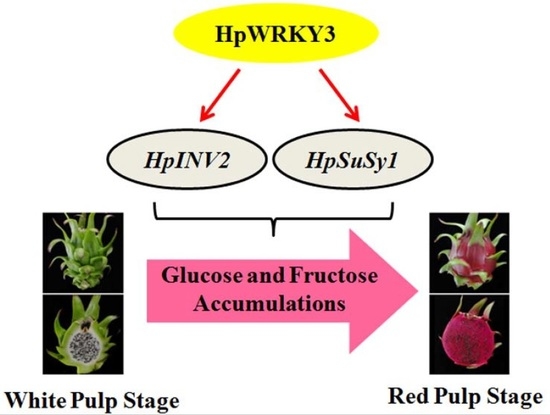
Pitaya HpWRKY3 Is Associated with Fruit Sugar Accumulation by Transcriptionally Modulating Sucrose Metabolic Genes HpINV2 and HpSuSy1
Abstract
:1. Introduction
2. Results and Discussion
2.1. Changes in Soluble Sugars during Pitaya Fruit Maturation
2.2. Expression Levels of HpINV2 and HpSuSy1 Were Positively Correlated with Accumulations of Glucose and Fructose
2.3. Identification of HpWRKY3 as A Putative Transcriptional Regulator of HpINV2 and HpSuSy1
2.4. Bioinformatics and Molecular Characteristics of HpWRKY3
2.5. HpWRKY3 Enhances the Transcriptions of HpINV2 and HpSuSy1
3. Materials and Methods
3.1. Fruit Samples
3.2. Soluble Sugar Quantification by HPLC
3.3. RNA Preparation, Gene Isolation, and Bioinformatic Analysis
3.4. Analysis of Gene Expression by qRT-PCR
3.5. Gene Promoter Analysis
3.6. Yeast One-Hybrid Assay
3.7. Recombinant Protein Induction, Purification, and EMSA Assay
3.8. Subcellular Localization Assay
3.9. Trans-Activation Activity Assay in Yeast
3.10. Transient Trans-Activation Assay in Tobacco Leaves
3.11. Statistics
3.12. Primers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, H.; Choi, H.K.; Moon, J.Y.; Kim, Y.S.; Mosaddik, A.; Cho, S.K. Comparative antioxidant and antiproliferative activities of red and white pitayas and their correlation with flavonoid and polyphenol content. J. Food Sci. 2011, 76, C38–C45. [Google Scholar] [CrossRef] [PubMed]
- Hua, Q.; Chen, C.; Tel Zur, N.; Wang, H.; Wu, J.; Chen, J.; Zhang, Z.; Zhao, J.; Hu, G.; Qin, Y. Metabolomic characterization of pitaya fruit from three red-skinned cultivars with different pulp colors. Plant Physiol. Biochem. 2018, 126, 117–125. [Google Scholar] [CrossRef]
- García-Cruz, L.; Dueñas, M.; Santos-Buelgas, C.; Valle-Guadarrama, S.; Salinas-Moreno, Y. Betalains and phenolic compounds profiling and antioxidant capacity of pitaya (Stenocereus spp.) fruit from two species (S. Pruinosus and S. stellatus). Food Chem. 2017, 234, 111–118. [Google Scholar] [CrossRef]
- Suh, D.H.; Lee, S.; Heo do, Y.; Kim, Y.S.; Cho, S.K.; Lee, S.; Lee, C.H. Metabolite profiling of red and white pitayas (Hylocereus polyrhizus and Hylocereus undatus) for comparing betalain biosynthesis and antioxidant activity. J. Agric. Food Chem. 2014, 62, 8764–8771. [Google Scholar] [CrossRef]
- Menezes Cordeiro, M.H.; Da Silva, J.M.; Mizobutsi, G.P.; Mizobutsi, E.H.; Da Mota, W.F. Physical, chemical and nutritional characterization of pink pitaya of red pulp. Rev. Bras. Frutic. 2015, 37, 20–26. [Google Scholar]
- Song, H.Z.; Chu, Q.; Xu, D.D.; Xu, Y.; Zheng, X.D. Purified betacyanins from Hylocereus undatus peel ameliorate obesity and insulin resistance in high-fat-diet-fed mice. J. Agric. Food Chem. 2016, 64, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Hua, Q.Z.; Chen, C.J.; Chen, Z.; Chen, P.K.; Ma, Y.W.; Wu, J.Y.; Zheng, J.; Hu, G.B.; Zhao, J.T.; Qin, Y.H. Transcriptomic analysis reveals key genes related to betalain biosynthesis in pulp coloration of Hylocereus polyrhizus. Front. Plant Sci. 2016, 6, 1179. [Google Scholar]
- Cheng, M.N.; Huang, Z.J.; Hua, Q.Z.; Shan, W.; Kuang, J.F.; Lu, W.J.; Qin, Y.H.; Chen, J.Y. The WRKY transcription factor HpWRKY44 regulates CytP450-like1 expression in red pitaya fruit (Hylocereus polyrhizus). Hortic. Res. 2017, 4, 17039. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.J.; Sun, M.H.; Lu, J.; Liu, Y.J.; Hu, D.G.; Hao, Y.J. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 2017, 174, 2348–2362. [Google Scholar] [CrossRef] [PubMed]
- Li, M.J.; Li, P.M.; Ma, F.W.; Abhaya, M.; Dandekar, A.M.; Cheng, L.L. Sugar metabolism and accumulation in the fruit of transgenic apple trees with decreased sorbitol synthesis. Hortic. Res. 2018, 5, 60. [Google Scholar] [CrossRef]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Zheng, H.; Peng, Q.; Jiang, Q.; Wang, H.; Fang, T.; Liao, L.; Wang, L.; He, H.; Han, Y. Assessment of sugar components and genes involved in the regulation of sucrose accumulation in peach fruit. J. Agric. Food Chem. 2016, 64, 6723–6729. [Google Scholar] [CrossRef]
- Cirilli, M.; Bassi, D.; Ciacciulli, A. Sugars in peach fruit: A breeding perspective. Hortic. Res. 2016, 3, 15067. [Google Scholar] [CrossRef]
- Zhu, Z.; Liu, R.; Li, B.; Tian, S. Characterisation of genes encoding key enzymes involved in sugar metabolism of apple fruit in controlled atmosphere storage. Food Chem. 2013, 141, 3323–3328. [Google Scholar] [CrossRef]
- Yativ, M.; Harary, I.; Wolf, S. Sucrose accumulation in watermelon fruits: Genetic variation and biochemical analysis. J. Plant Physiol. 2010, 167, 589–596. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Alseekh, S.; Tohge, T.; Rallapalli, G.; Luo, J.; Kawar, P.G.; Hill, L.; Santino, A.; Fernie, A.R.; et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015, 6, 8635. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yan, S.; Huang, W.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; Zhu, X.; et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 2018, 98, 289–302. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Viana, V.E.; Busanello, C.; da Maia, L.C.; Pegoraro, C.; Costa de Oliveira, A. Activation of rice WRKY transcription factors: An army of stress fighting soldiers? Curr. Opin. Plant Biol. 2018, 45, 268–275. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Schluttenhofer, C.; Yuan, L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, S.R.; Dwivedi, V.; Rai, A.; Pal, S.; Shasany, A.K.; Nagegowda, D.A. A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 2017, 215, 1115–1131. [Google Scholar] [CrossRef]
- Luo, Y.; Yu, S.; Li, J.; Li, Q.; Wang, K.; Huang, J.; Liu, Z. Molecular characterization of WRKY transcription factors that act as negative regulators of O-methylated catechin biosynthesis in tea plants (Camellia sinensis L.). J. Agric. Food Chem. 2018, 66, 11234–11243. [Google Scholar] [CrossRef]
- Vannozzi, A.; Wong, D.C.J.; Höll, J.; Hmmam, I.; Matus, J.T.; Bogs, J.; Ziegler, T.; Dry, I.; Barcaccia, G.; Lucchin, M. Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol. 2018, 59, 1043–1059. [Google Scholar] [CrossRef]
- Jiang, J.; Xi, H.; Dai, Z.; Lecourieux, F.; Yuan, L.; Liu, X.; Patra, B.; Wei, Y.; Li, S.; Wang, L. VvWRKY8 represses stilbene synthase gene through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J. Exp. Bot. 2018. [Google Scholar] [CrossRef]
- Nomura, K.; Ide, M.; Yonemoto, Y. Changes in sugars and acids in pitaya (Hylocereus undatus) fruit during development. J. Hortic. Sci. Biotech. 2005, 80, 711–715. [Google Scholar] [CrossRef]
- Le Bellec, F.; Vaillant, F.; Imbert, E. Pitahaya (Hylocereus spp.): A new fruit crop, a market with future. Fruits 2006, 61, 237–250. [Google Scholar] [CrossRef]
- Nguyen-Quoc, B.; Foyer, C.H. A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J. Exp. Bot. 2001, 52, 881–889. [Google Scholar] [CrossRef] [Green Version]
- Beauvoit, B.P.; Colombié, S.; Monier, A.; Andrieu, M.H.; Biais, B.; Bénard, C.; Chéniclet, C.; Dieuaide-Noubhani, M.; Nazaret, C.; Mazat, J.P.; et al. Model-assisted analysis of sugar metabolism throughout tomato fruit development reveals enzyme and carrier properties in relation to vacuole expansion. Plant Cell 2014, 26, 3224–3242. [Google Scholar] [CrossRef]
- Zhang, X.M.; Wang, W.; Du, L.Q.; Xie, J.H.; Yao, Y.L.; Sun, G.M. Expression patterns, activities and carbohydrate-metabolizing regulation of sucrose phosphate synthase, sucrose synthase and neutral invertase in pineapple fruit during development and ripening. Int. J. Mol. Sci. 2012, 13, 9460–9477. [Google Scholar] [CrossRef]
- Ruan, Y.L.; Jin, Y.; Yang, Y.J.; Li, G.J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Fan, Z.Q.; Tan, X.L.; Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Characterization of a transcriptional regulator, BrWRKY6, associated with gibberellin-suppressed leaf senescence of Chinese flowering cabbage. J. Agric. Food Chem. 2018, 66, 1791–1799. [Google Scholar] [CrossRef]
- Islam, M.Z.; Jin, L.F.; Shi, C.Y.; Liu, Y.Z.; Peng, S.A. Citrus sucrose transporter genes: Genome-wide identification and transcript analysis in ripening and ABA-injected fruits. Tree Genet. Genomes 2015, 11, 1–9. [Google Scholar] [CrossRef]
- Ren, Y.; Guo, S.; Zhang, J.; He, H.; Sun, H.; Tian, S.; Gong, G.; Zhang, H.; Levi, A.; Tadmor, Y.; et al. A tonoplast sugar transporter underlies a sugar accumulation QTL in watermelon. Plant Physiol. 2018, 176, 836–850. [Google Scholar] [CrossRef]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.Q.; Chen, Z. Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef]
- Shan, W.; Chen, J.Y.; Kuang, J.F.; Lu, W.J. Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against Colletotrichum musae. Mol. Plant Pathol. 2016, 17, 330–338. [Google Scholar] [CrossRef]
- Ye, Y.J.; Xiao, Y.Y.; Han, Y.C.; Shan, W.; Fan, Z.Q.; Xu, Q.G.; Kuang, J.F.; Lu, W.J.; Lakshmanan, P.; Chen, J.Y. Banana fruit VQ motif-containing protein5 represses cold-responsive transcription factor MaWRKY26 involved in the regulation of JA biosynthetic genes. Sci. Rep. 2016, 6, 23632. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Mao, W.; Jia, M.; Xing, S.; Ali, U.; Zhao, Y.; Chen, Y.; Cao, M.; Dai, Z.; Zhang, K.; et al. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J. Exp. Bot. 2018, 69, 4805–4820. [Google Scholar] [CrossRef]
- Chen, M.; Jiang, Q.; Yin, X.R.; Lin, Q.; Chen, J.Y.; Allan, A.C.; Xu, C.J.; Chen, K.S. Effect of hot air treatment on organic acid- and sugar metabolism in Ponkan (Citrus reticulata) fruits. Sci. Hortic. 2012, 147, 118–125. [Google Scholar] [CrossRef]
- Fan, Z.Q.; Ba, L.J.; Shan, W.; Xiao, Y.Y.; Lu, W.J.; Kuang, J.F.; Chen, J.Y. A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J. 2018, 96, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.Q.; Tan, X.L.; Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. BrWRKY65, a WRKY transcription factor, is involved in regulating three leaf senescence-associated genes in Chinese flowering cabbage. Int. J. Mol. Sci. 2017, 18, 1228. [Google Scholar]
- Tan, X.L.; Fan, Z.Q.; Shan, W.; Yin, X.R.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Association of BrERF72 with methyl jasmonate-induced leaf senescence of Chinese flowering cabbage through activating JA biosynthesis-related genes. Hortic. Res. 2018, 5, 22. [Google Scholar] [CrossRef]
- Fan, Z.Q.; Tan, X.L.; Chen, J.W.; Liu, Z.L.; Kuang, J.F.; Lu, W.J.; Shan, W.; Chen, J.Y. BrNAC055, a novel transcriptional activator, regulates leaf senescence in Chinese flowering cabbage by modulating reactive oxygen species production and chlorophyll degradation. J. Agric. Food Chem. 2018, 66, 9399–9408. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, W.; Cheng, M.-n.; Ba, L.-j.; Zeng, R.-x.; Luo, D.-l.; Qin, Y.-h.; Liu, Z.-l.; Kuang, J.-f.; Lu, W.-j.; Chen, J.-y.; et al. Pitaya HpWRKY3 Is Associated with Fruit Sugar Accumulation by Transcriptionally Modulating Sucrose Metabolic Genes HpINV2 and HpSuSy1. Int. J. Mol. Sci. 2019, 20, 1890. https://doi.org/10.3390/ijms20081890
Wei W, Cheng M-n, Ba L-j, Zeng R-x, Luo D-l, Qin Y-h, Liu Z-l, Kuang J-f, Lu W-j, Chen J-y, et al. Pitaya HpWRKY3 Is Associated with Fruit Sugar Accumulation by Transcriptionally Modulating Sucrose Metabolic Genes HpINV2 and HpSuSy1. International Journal of Molecular Sciences. 2019; 20(8):1890. https://doi.org/10.3390/ijms20081890
Chicago/Turabian StyleWei, Wei, Mei-nv Cheng, Liang-jie Ba, Run-xi Zeng, Dong-lan Luo, Yong-hua Qin, Zong-li Liu, Jian-fei Kuang, Wang-jin Lu, Jian-ye Chen, and et al. 2019. "Pitaya HpWRKY3 Is Associated with Fruit Sugar Accumulation by Transcriptionally Modulating Sucrose Metabolic Genes HpINV2 and HpSuSy1" International Journal of Molecular Sciences 20, no. 8: 1890. https://doi.org/10.3390/ijms20081890
APA StyleWei, W., Cheng, M.-n., Ba, L.-j., Zeng, R.-x., Luo, D.-l., Qin, Y.-h., Liu, Z.-l., Kuang, J.-f., Lu, W.-j., Chen, J.-y., Su, X.-g., & Shan, W. (2019). Pitaya HpWRKY3 Is Associated with Fruit Sugar Accumulation by Transcriptionally Modulating Sucrose Metabolic Genes HpINV2 and HpSuSy1. International Journal of Molecular Sciences, 20(8), 1890. https://doi.org/10.3390/ijms20081890