Plant Cell Wall Proteomics: A Focus on Monocot Species, Brachypodium distachyon, Saccharum spp. and Oryza sativa
Abstract
1. Introduction
2. Methods of Monocots CWPs Extraction and Analysis
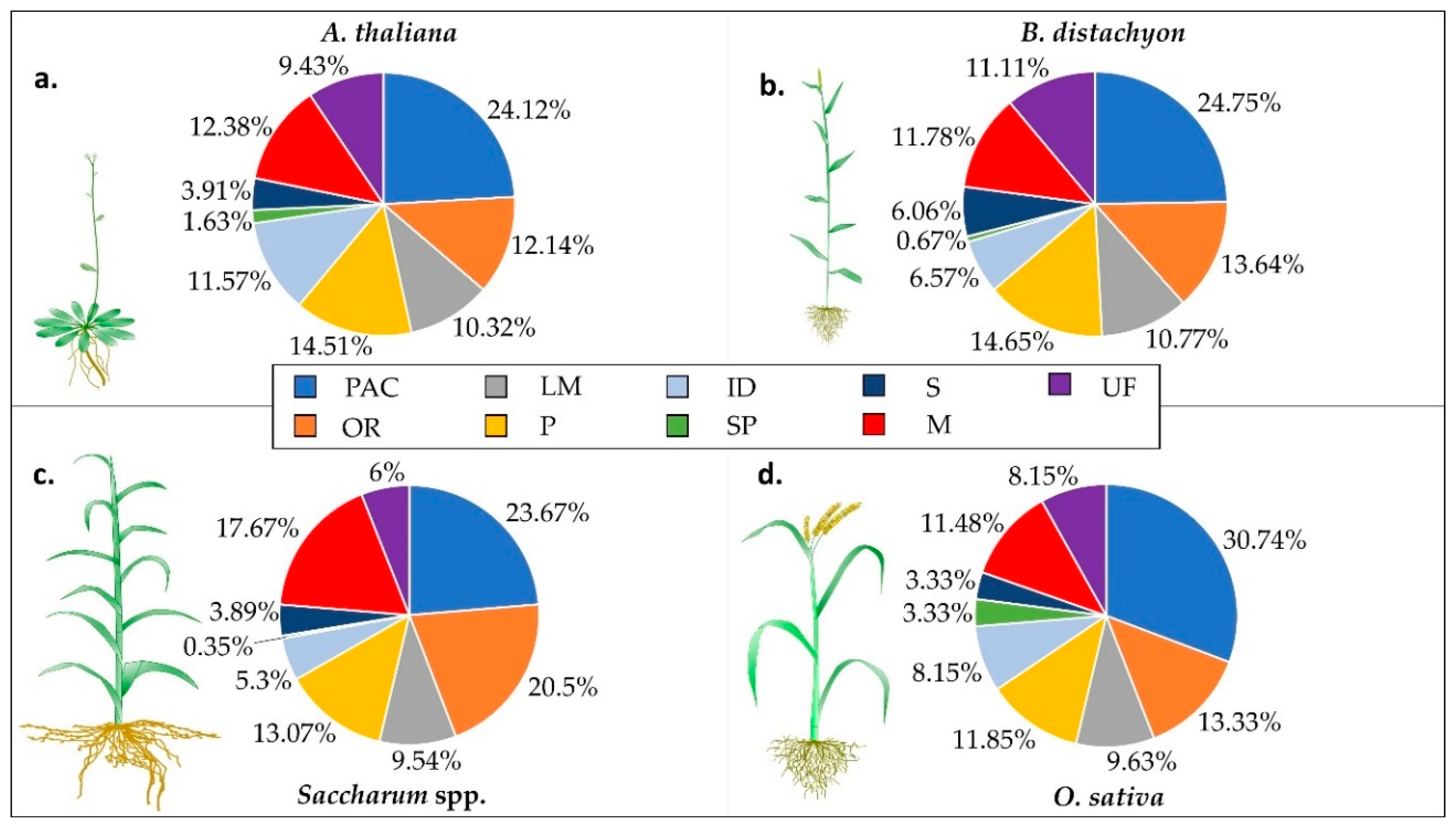
3. Functional Class Distribution in Monocots
3.1. Proteins Acting on Carbohydrates
3.2. Oxidoreductases
3.3. Proteins Related to Lipid Metabolism
3.4. Proteases
3.5. Proteins with Interacting Domains
3.6. Proteins Possibly Related to Signaling
3.7. Miscellaneous
3.8. Structural Proteins
3.9. Proteins of Unkown Function
4. Applicative Aspects of Research on CWPs
5. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
Abbreviations
| 1D-PAGE | one-dimensional polyacrylamide gel electrophoresis |
| 2D | two-dimensional |
| 2D-PAGE | two-dimensional polyacrylamide gel electrophoresis |
| ABA | abscisic acid |
| Asp | aspartyl |
| BBE | berberine-bridge enzyme |
| BBI | bowman-birk serine protease inhibitor |
| CaCl2 | calcium chloride |
| CWP | cell wall protein |
| COBL | COBRA-like protein |
| DAF | days after flowering |
| DUF | protein with domains of unknown function |
| ER | endoplasmic reticulum |
| EXT | extensin |
| FLA | fasciclin-like arabinogalactan |
| GDPD | glycerophosphodiester phosphodiesterase |
| GDPDL | glycerophosphodiester phosphodiesterase-like |
| GH | glycosyl hydrolase |
| ID | protein with predicted interaction domains |
| LiCl | lithium chloride |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| LM | protein related to lipid metabolism |
| LRR | leucine-rich repeat |
| LRR-RK | leucine-rich repeat receptor kinase |
| LRX | leucine-rich repeat extensin |
| LTP | lipid transfer protein |
| M | miscellaneous protein |
| MS | mass spectrometry |
| OR | oxidoreductase |
| P | protease |
| PAC | proteins acting on carbohydrate |
| PME | pectin methyl esterase |
| PMEI | pectin methyl esterase inhibitor |
| Prx | class III peroxidase |
| S | protein possibly involved in signaling |
| SP | structural protein |
| UF | proteins of unknown function |
References
- Roberts, K. The plant extracellular matrix: In a new expansive mood. Curr. Opin. Cell Biol. 1994, 6, 688–694. [Google Scholar] [CrossRef]
- Brownlee, C. Role of the extracellular matrix in cell-cell signalling: Paracrine paradigms. Curr. Opin. Plant Biol. 2002, 5, 396–401. [Google Scholar] [CrossRef]
- Franková, L.; Fry, S.C. Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. J. Exp. Bot. 2013, 64, 3519–3550. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Gruissem, W.; Jones, R.L. Biochemistry & Molecular Biology of Plants, 2nd ed.; John Wiley & Sons: Chichester, UK, 2015; pp. 1–1221. ISBN 9780470714225. [Google Scholar]
- Cassab, G.I.; Varner, J.E. Cell Wall Proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 321–353. [Google Scholar] [CrossRef]
- Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Jamet, E.; Albenne, C.; Boudart, G.; Irshad, M.; Canut, H.; Pont-Lezica, R. Recent advances in plant cell wall proteomics. Proteomics 2008, 8, 893–908. [Google Scholar] [CrossRef]
- Rapoport, T.A. Transport of proteins across the endoplasmic reticulum membrane. Science 1992, 258, 931–936. [Google Scholar] [CrossRef][Green Version]
- Vitale, A.; Denecke, J. The endoplasmic reticulum—Gateway of the secretory pathway. Plant Cell 1999, 11, 615–628. [Google Scholar] [CrossRef]
- Crofts, A.; Leborgne-Castel, N.; Hillmer, S.; Robinson, D.G.; Phillipson, B.; Carlsson, L.E.; Ashford, D.A.; Denecke, J. Saturation of the endoplasmic reticulum retention machinery reveals anterograde bulk flow. Plant Cell 1999, 11, 2233–2248. [Google Scholar] [CrossRef] [PubMed]
- Chivasa, S.; Ndimba, B.K.; Simon, W.J.; Robertson, D.; Yu, X.L.; Knox, J.P.; Bolwell, P.; Slabas, A.R. Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis 2002, 23, 1754–1765. [Google Scholar] [CrossRef]
- Carpita, N.; Tierney, M.; Campbell, M. Molecular biology of the plant cell wall: Searching for the genes that define structure, architecture and dynamics. Plant Mol. Biol. 2001, 47, 1–5. [Google Scholar] [CrossRef]
- Jamet, E.; Canut, H.; Boudart, G.; Pont-Lezica, R.F. Cell wall proteins: A new insight through proteomics. Trends Plant Sci. 2006, 11, 33–39. [Google Scholar] [CrossRef]
- Tan, L.; Eberhard, S.; Pattathil, S.; Warder, C.; Glushka, J.; Yuan, C.; Hao, Z.; Zhu, X.; Avci, U.; Miller, J.S.; et al. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 2013, 25, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Saravanan, R.S.; Damasceno, C.M.B.; Yamane, H.; Kim, B.-D.; Rose, J.K.C. Digging deeper into the plant cell wall proteome. Plant Physiol. Biochem. 2004, 42, 979–988. [Google Scholar] [CrossRef]
- Borderies, G.; Jamet, E.; Lafitte, C.; Rossignol, M.; Jauneau, A.; Boudart, G.; Monsarrat, B.; Esquerré-Tugayé, M.-T.; Boudet, A.; Pont-Lezica, R. Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: A critical analysis. Electrophoresis 2003, 24, 3421–3432. [Google Scholar] [CrossRef] [PubMed]
- Boudart, G.; Jamet, E.; Rossignol, M.; Lafitte, C.; Borderies, G.; Jauneau, A.; Esquerré-Tugayé, M.-T.; Pont-Lezica, R. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: Identification by mass spectrometry and bioinformatics. Proteomics 2005, 5, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.M.; Bottrill, A.R.; Walshaw, J.; Vigouroux, M.; Naldrett, M.J.; Thomas, C.L.; Maule, A.J. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 2006, 6, 301–311. [Google Scholar] [CrossRef]
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008, 8, 94. [Google Scholar] [CrossRef]
- Nguyen-Kim, H.; San Clamente, H.; Balliau, T.; Zivy, M.; Dunand, C.; Albenne, C.; Jamet, E. Arabidopsis thaliana root cell wall proteomics: Increasing the proteome coverage using a combinatorial peptide ligand library and description of unexpected Hyp in peroxidase amino acid sequences. Proteomics 2016, 16, 491–503. [Google Scholar] [CrossRef]
- Hervé, V.; Duruflé, H.; San Clemente, H.; Albenne, C.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 2016, 16, 3183–3187. [Google Scholar] [CrossRef] [PubMed]
- Duruflé, H.; San Clemente, H.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. Proline hydroxylation in cell wall proteins: Is it yet possible to define rules? Front. Plant Sci. 2017, 8, 1802. [Google Scholar] [CrossRef]
- Kehr, J.; Buhtz, A.; Giavalisco, P. Analysis of xylem sap proteins from Brassica napus. BMC Plant Biol. 2005, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Ligat, L.; Lauber, E.; Albenne, C.; San Clemente, H.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Arlat, M.; Jamet, E. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 2011, 11, 1798–1813. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Alvarez, S.; Marsh, E.L.; LeNoble, M.E.; Cho, I.-J.; Sivaguru, M.; Chen, S.; Nguyen, H.T.; Wu, Y.; Schachtman, D.P.; et al. Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol. 2007, 145, 1533–1548. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, D.; Pandey, A.; Chattopadhyay, A.; Choudhary, M.K.; Chakraborty, S.; Datta, A.; Chakraborty, N. Extracellular matrix proteome of chickpea (Cicer arietinum L.) illustrates pathway abundance, novel protein functions and evolutionary perspect. J. Proteome Res. 2006, 5, 1711–1720. [Google Scholar] [CrossRef]
- Negri, A.S.; Prinsi, B.; Scienza, A.; Morgutti, S.; Cocucci, M.; Espen, L. Analysis of grape berry cell wall proteome: A comparative evaluation of extraction methods. J. Plant Physiol. 2008, 165, 1379–1389. [Google Scholar] [CrossRef]
- Jung, Y.-H.; Jeong, S.-H.; Kim, S.H.; Singh, R.; Lee, J.; Cho, Y.-S.; Agrawal, G.K.; Rakwal, R.; Jwa, N.-S. Systematic secretome analyses of rice leaf and seed callus suspension-cultured cells: Workflow development and establishment of high-density two-dimensional gel reference maps. J. Proteome Res. 2008, 7, 5187–5210. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Kim, S.T.; Cho, W.K.; Rim, Y.; Kim, S.; Kim, S.W.; Kang, K.Y.; Park, Z.Y.; Kim, J.Y. Proteomics of weakly bound cell wall proteins in rice calli. J. Plant Physiol. 2009, 166, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.K.; Chen, X.Y.; Chu, H.; Rim, Y.; Kim, S.; Kim, S.T.; Kim, S.-W.; Park, Z.-Y.; Kim, J.-Y. Proteomic analysis of the secretome of rice calli. Physiol. Plant. 2009, 135, 331–341. [Google Scholar] [CrossRef]
- Zhou, L.; Bokhari, S.A.; Dong, C.-J.; Liu, J.-Y. Comparative proteomics analysis of the root apoplasts of rice seedlings in response to hydrogen peroxide. PLoS ONE 2011, 6, e16723. [Google Scholar] [CrossRef]
- Millar, D.J.; Whitelegge, J.P.; Bindschedler, L.V.; Rayon, C.; Boudet, A.-M.; Rossignol, M.; Borderies, G.; Bolwell, G.P. The cell wall and secretory proteome of a tobacco cell line synthesising secondary wall. Proteomics 2009, 9, 2355–2372. [Google Scholar] [CrossRef] [PubMed]
- Douché, T.; San Clemente, H.; Burlat, V.; Roujol, D.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Jamet, E. Brachypodium distachyon as a model plant toward improved biofuel crops: Search for secreted proteins involved in biogenesis and disassembly of cell wall polymers. Proteomics 2013, 13, 2438–2454. [Google Scholar] [CrossRef] [PubMed]
- Francin-Allami, M.; Lollier, V.; Pavlovic, M.; San Clemente, H.; Rogniaux, H.; Jamet, E.; Guillon, F.; Larré, C. Understanding the remodelling of cell walls during Brachypodium distachyon grain development through a sub-cellular quantitative proteomic approach. Proteomes 2016, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, G.-X.; Lv, D.-W.; Li, X.-H.; Yan, Y.-M. N-Linked Glycoproteome profiling of seedling leaf in Brachypodium distachyon L. J. Proteome Res. 2015, 14, 1727–1738. [Google Scholar] [CrossRef]
- Francin-Allami, M.; Merah, K.; Albenne, C.; Rogniaux, H.; Pavlovic, M.; Lollier, V.; Sibout, R.; Guillon, F.; Jamet, E.; Larré, C. Cell wall proteomic of Brachypodium distachyon grains: A focus on cell wall remodeling proteins. Proteomics 2015, 15, 2296–2306. [Google Scholar] [CrossRef]
- Calderan-Rodrigues, M.J.; Jamet, E.; Bonassi, M.B.C.R.; Guidetti-Gonzalez, S.; Begossi, A.C.; Setem, L.V.; Franceschini, L.M.; Fonseca, J.G.; Labate, C.A. Cell wall proteomics of sugarcane cell suspension cultures. Proteomics 2014, 14, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Calderan-Rodrigues, M.J.; Jamet, E.; Douché, T.; Bonassi, M.B.R.; Cataldi, T.R.; Fonseca, J.G.; San Clemente, H.; Pont-Lezica, R.; Labate, C.A. Cell wall proteome of sugarcane stems: Comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol. 2016, 16, 14. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, J.G.; Calderan-Rodrigues, M.J.; de Moraes, F.E.; Cataldi, T.R.; Jamet, E.; Labate, C.A. Cell wall proteome of sugarcane young and mature leaves and stems. Proteomics 2018, 18, 9853. [Google Scholar] [CrossRef] [PubMed]
- Labate, C.A.; Calderan-Rodrigues, M.J.; Cataldi, T.R. Cell Wall Proteome of Seven Month-Old Sugarcane Leaves and Internodes. 2019. Available online: http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD012761 (accessed on 11 February 2019).
- Albenne, C.; Canut, H.; Hoffmann, L.; Jamet, E. Plant cell wall Proteins: A large body of data, but what about runaways? Proteomes 2014, 2, 224–242. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Sarkar, P.; Bosneaga, E.; Auer, M. Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 2009, 60, 3615–3635. [Google Scholar] [CrossRef]
- Nogueira, L.A.H.; Leal, M.R.L.V.; Fernandes, E.; Chum, H.L.; Diaz-Chavez, R.; Endres, J.; Mahakhant, A.; Otto, M.; Seebaluck, V.; van der Wielen, L. Sustainable development and innovation. In SCOPE Bioenergy & Sustainability: Bridging the Gaps, 1st ed.; Souza, G.M., Victoria, R.I., Joly, C.A., Verdade, L.M., Eds.; Scientific Committee on Problems of the Environment (SCOPE): Paris, France, 2015; Volume 1, pp. 184–217. ISBN 978-2-9545557-0-6. [Google Scholar]
- Yong, W.; Link, B.; O’Malley, R.; Tewari, J.; Hunter, C.T.; Lu, C.A.; Li, X.; Bleecker, A.B.; Koch, K.E.; McCann, M.C.; et al. Genomics of plant cell wall biogenesis. Planta 2005, 221, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Feiz, L.; Irshad, M.; Pont-Lezica, R.F.; Canut, H.; Jamet, E. Evaluation of cell wall preparations for proteomics: A new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2006, 2, 10. [Google Scholar] [CrossRef]
- Jamet, E.; Boudart, G.; Borderies, G.; Charmont, S.; Lafitte, C.; Rossignol, M.; Canut, H.; Pont-Lezica, R. Isolation of plant cell wall proteins. In Methods in Molecular Biology, Applications and Protocols in Expression Proteomics, 1st ed.; Posch, A., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2007; Volume 2, pp. 187–201. [Google Scholar]
- WallProtDB. Available online: http://www.polebio.lrsv.ups-tlse.fr/WallProtDB/ (accessed on 29 October 2018).
- Calderan-Rodrigues, M.J.; Fonseca, J.G.; Clemente, H.S.; Labate, C.A.; Jamet, E. Glycoside hydrolases in plant cell wall proteomes: Predicting functions that could be relevant for improving biomass transformation processes. In Advances in Biofuels and Bioenergy, 1st ed.; Nageswara-Rao, M., Ed.; IntechOpen: London, UK, 2018; Volume 1, pp. 165–182. [Google Scholar]
- Tyler, L.; Bragg, J.N.; Wu, J.; Yang, X.; Tuskan, G.A.; Vogel, J.P. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC Genom. 2010, 11, 600. [Google Scholar] [CrossRef]
- Chapelle, A.; Morreel, K.; Vanholme, R.; Le-Bris, P.; Morin, H.; Lapierre, C.; Boerjan, W.; Jouanin, L.; Demont-Caulet, N. Impact of absence of stem-specific β-glucosidases on lignin and monolignols. Plant Physiol. 2012, 160, 1204–1217. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.N.; Hrmova, M.; Fincher, G.B. Three-dimensional structure of a barley beta-d-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 1999, 7, 179–190. [Google Scholar] [CrossRef]
- Minic, Z. Physiological roles of plant glycoside hydrolases. Planta 2008, 227, 723–740. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Jouanin, L. Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol. Biochem. 2006, 44, 435–449. [Google Scholar] [CrossRef]
- Jordan, D.B.; Wagschal, K. Properties and applications of microbial β-d-xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Appl. Microbiol. Biotechnol. 2010, 86, 1647–1658. [Google Scholar] [CrossRef] [PubMed]
- Gebruers, K.; Brijs, K.; Courtin, C.; Fierens, K.; Goesaert, H.; Rabijns, A.; Raedschelders, G.; Robben, J.; Sansen, S.; Sørensen, J.F.; et al. Properties of TAXI-type endoxylanase inhibitors. Biochim. Biophys. Acta Proteins Proteomics 2004, 1696, 213–221. [Google Scholar] [CrossRef]
- Brunner, F.; Stintzi, A.; Fritig, B.; Legrand, M. Substrate specificities of tobacco chitinases. Plant J. 1998, 14, 225–234. [Google Scholar] [CrossRef]
- Sasaki, C.; Vårum, K.M.; Itoh, Y.; Tamoi, M.; Fukamizo, T. Rice chitinases: Sugar recognition specificities of the individual subsites. Glycobiology 2006, 16, 1242–1250. [Google Scholar] [CrossRef]
- Veillet, F.; Gaillard, C.; Coutos-Thévenot, P.; La Camera, S. Targeting the AtCWIN1 gene to explore the role of invertases in sucrose transport in roots and during Botrytis cinerea infection. Front. Plant Sci. 2016, 7, 1899. [Google Scholar] [CrossRef] [PubMed]
- Moneo-Sánchez, M.; Alonso-Chico, A.; Knox, J.P.; Dopico, B.; Labrador, E.; Martín, I. β-(1,4)-Galactan remodelling in Arabidopsis cell walls affects the xyloglucan structure during elongation. Planta 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, K.; Elashry, A.; Quentin, M.; Grundler, F.M.; Favery, B.; Seifert, J.G.; Bohlmann, H. A distinct role of pectate lyases in the formation of feeding structures induced by cyst and root-knot nematodes. Mol. Plant-Microbe Interact. 2014, 27, 901–912. [Google Scholar] [CrossRef]
- Guan, Y.; Nothnagel, E.A. Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiol. 2004, 135, 1346–1366. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef]
- Welinder, K.G.; Justesen, A.F.; Kjærsgård, I.V.; Jensen, R.B.; Rasmussen, S.K.; Jespersen, H.M.; Duroux, L. Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 2002, 269, 6063–6081. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.A.; Anoni, C.O.; Mancini, M.C.; Santos, F.R.C.; Marconi, T.G.; Gazaffi, R.; Pastina, M.M.; Perecin, D.; Mollinari, M.; Xavier, M.A.; et al. QTL mapping including codominant SNP markers with ploidy level information in a sugarcane progeny. Euphytica 2016, 211, 1–16. [Google Scholar] [CrossRef]
- Shah, K.; Penel, C.; Gagnon, J.; Dunand, C. Purification and identification of a Ca2+-pectate binding peroxidase from Arabidopsis leaves. Phytochemistry 2004, 65, 307–312. [Google Scholar] [CrossRef]
- Kunieda, T.; Shimada, T.; Kondo, M.; Nishimura, M.; Nishitani, K.; Hara-Nishimura, I. Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 2013, 25, 1355–1367. [Google Scholar] [CrossRef]
- Fernández-Pérez, F.; Pomar, F.; Pedreño, M.A.; Novo-Uzal, E. The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol. Plant. 2015, 154, 395–406. [Google Scholar] [CrossRef]
- Herrero, J.; Fernández-Pérez, F.; Yebra, T.; Novo-Uzal, E.; Pomar, F.; Pedreño, M.Á.; Cuello, J.; Guéra, A.; Esteban-Carrasco, A.; Zapata, J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 2013, 237, 1599–1612. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rubio, M.C.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Linkies, A.; Schuster-Sherpa, U.; Tintelnot, S.; Leubner-Metzger, G.; Müller, K. Peroxidases identified in a subtractive cDNA library approach show tissue-specific transcript abundance and enzyme activity during seed germination of Lepidium sativum. J. Exp. Bot. 2010, 61, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Hewezi, T.; Baum, T.J. Arabidopsis peroxidase AtPRX53 influences cell elongation and susceptibility to Heterodera schachtii. Plant Signal. Behav. 2011, 6, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Ranocha, P.; Francoz, E.; Burlat, V.; Zheng, Y.; Perry, S.E.; Ripoll, J.-J.; Yanofsky, M.; Dunand, C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2017, 213, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Kim, S.Y.; Nam, K.H. Genes encoding plant-specific class III peroxidases are responsible for increased cold tolerance of the brassinosteroid-insensitive 1 mutant. Mol. Cells 2012, 34, 539–548. [Google Scholar] [CrossRef]
- Daniel, B.; Pavkov-Keller, T.; Steiner, B.; Dordic, A.; Gutmann, A.; Nidetzky, B.; Sensen, C.W.; van der Graaff, E.; Wallner, S.; Gruber, K.; et al. Oxidation of monolignols by members of the berberine bridge enzyme family suggests a role in plant cell wall metabolism. J. Biol. Chem. 2015, 290, 18770–18781. [Google Scholar] [CrossRef]
- Benedetti, M.; Verrascina, I.; Pontiggia, D.; Locci, F.; Mattei, B.; De Lorenzo, G.; Cervone, F. Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 2018, 94, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Daniel, B.; Konrad, B.; Toplak, M.; Lahham, M.; Messenlehner, J.; Winkler, A.; Macheroux, P. The family of berberine bridge enzyme-like enzymes: A treasure-trove of oxidative reactions. Arch. Biochem. Biophys. 2017, 632, 88–103. [Google Scholar] [CrossRef]
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 2018, 59, 1374–1382. [Google Scholar] [CrossRef]
- Bosch, M.; Mayer, C.D.; Cookson, A.; Donnison, I.S. Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J. Exp. Bot. 2011, 62, 3545–3561. [Google Scholar] [CrossRef]
- Ariizumi, T.; Amagai, M.; Shibata, D.; Hatakeyama, K.; Watanabe, M.; Toriyama, K. Comparative study of promoter activity of three anther-specific genes encoding lipid transfer protein, xyloglucan endotransglucosylase/hydrolase and polygalacturonase in transgenic Arabidopsis thaliana. Plant Cell Rep. 2002, 21, 90–96. [Google Scholar] [CrossRef]
- Gao, S.; Guo, W.; Feng, W.; Liu, L.; Song, X.; Chen, J.; Hou, W.; Zhu, H.; Tang, S.; Hu, J. LTP3 contributes to disease susceptibility in Arabidopsis by enhancing abscisic acid (ABA) biosynthesis. Mol. Plant Pathol. 2016, 17, 412–426. [Google Scholar] [CrossRef]
- Jones, M.A.; Raymond, M.J.; Smirnoff, N. Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 2006, 45, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Beers, E.P.; Jones, A.M.; Dickerman, A.W. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 2004, 65, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.E.; Borniego, M.L.; Battchikova, N.; Aro, E.-M.; Tyystjärvi, E.; Guiamét, J.J. SASP, a senescence-associated subtilisin protease, is involved in reproductive development and determination of silique number in Arabidopsis. J. Exp. Bot. 2015, 66, 161–174. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Q.; Guo, Y.; Yang, J.; Wang, M.; Duan, X.; Niu, J.; Liu, S.; Zhang, J.; Lu, Y.; et al. Arabidopsis subtilase SASP is involved in the regulation of ABA signaling and drought tolerance by interacting with OPEN STOMATA 1. J. Exp. Bot. 2018, 69, 4403–4417. [Google Scholar] [CrossRef]
- Pathirana, R.; West, P.; Hedderley, D.; Eason, J. Cell death patterns in Arabidopsis cells subjected to four physiological stressors indicate multiple signalling pathways and cell cycle phase specificity. Protoplasma 2017, 254, 635–647. [Google Scholar] [CrossRef]
- Hocq, L.; Sénéchal, F.; Lefebvre, V.; Lehner, A.; Domon, J.-M.; Mollet, J.-C.; Dehors, J.; Pageau, J.; Marcelo, P.; Guérineau, F.; et al. Combined experimental and computational approaches reveal distinct pH dependence of pectin methylesterase inhibitors. Plant Physiol. 2017, 173, 1075–1093. [Google Scholar] [CrossRef]
- Carotenuto, G.; Chabaud, M.; Miyata, K.; Capozzi, M.; Takeda, N.; Kaku, H.; Shibuya, N.; Nakagawa, T.; Barker, D.G.; Genre, A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017, 214, 1440–1446. [Google Scholar] [CrossRef]
- Qi, R.-F.; Song, Z.-W.; Chi, C.-W. Structural features and molecular evolution of Bowman-Birk protease inhibitors and their potential application. Acta Biochim. Biophys. Sin. 2005, 37, 283–292. [Google Scholar] [CrossRef]
- Guo, K.; Bu, Y.; Takano, T.; Liu, S.; Zhang, X. Arabidopsis cysteine proteinase inhibitor AtCYSb interacts with a Ca2+-dependent nuclease, AtCaN2. FEBS Lett. 2013, 587, 3417–3421. [Google Scholar] [CrossRef]
- Helft, L.; Reddy, V.; Chen, X.; Koller, T.; Federici, L.; Fernández-Recio, J.; Gupta, R.; Bent, A. LRR conservation mapping to predict functional sites within protein leucine-rich repeat domains. PLoS ONE 2011, 6, e21614. [Google Scholar] [CrossRef]
- Shah, S.J.; Anjam, M.S.; Mendy, B.; Anwer, M.A.; Habash, S.S.; Lozano-Torres, J.L.; Grundler, F.M.W.; Siddique, S. Damage-associated responses of the host contribute to defence against cyst nematodes but not root-knot nematodes. J. Exp. Bot. 2017, 68, 5949–5960. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Szewińska, J.; Sobczak, M.; Dzik, J.M. Protease activity and phytocystatin expression in Arabidopsis thaliana upon Heterodera schachtii infection. Plant Physiol. Biochem. 2016, 109, 416–429. [Google Scholar] [CrossRef]
- Song, C.; Kim, T.; Chung, W.S.; Lim, C.O. The Arabidopsis phytoscystatin AtCYS5 enhances seed germination and seedling growth under heat stress conditions. Mol. Cells 2017, 40, 577–586. [Google Scholar] [CrossRef]
- Johnson, K.L.; Jones, B.J.; Bacic, A.; Schultz, C.J. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003, 133, 1911–1925. [Google Scholar] [CrossRef]
- Kotake, T.; Hirata, N.; Kitazawa, K.; Soga, K.; Tsumuraya, Y. Arabinogalactan-proteins in the evolution of gravity resistance in land plants. Soc. Biol. Sci. Space 2009, 23, 143–149. [Google Scholar] [CrossRef]
- MacMillan, C.P.; Mansfield, S.D.; Stachurski, Z.H.; Evans, R.; Southerton, S.G. Fasciclin-like arabinogalactan proteins: Specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 2010, 62, 689–703. [Google Scholar] [CrossRef]
- Johnson, K.L.; Kibble, N.A.; Bacic, A.; Schultz, C.J. A Fasciclin-Like Arabinogalactan-Protein (FLA) Mutant of Arabidopsis thaliana, fla1, Shows Defects in Shoot Regeneration. PLoS ONE 2011, 6, e25154. [Google Scholar] [CrossRef]
- Schindelman, G.; Morikami, A.; Jung, J.; Baskin, T.I.; Carpita, N.C.; Derbyshire, P.; McCann, M.C.; Benfey, P.N. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001, 15, 1115–1127. [Google Scholar] [CrossRef]
- Niu, E.; Shang, X.; Cheng, C.; Bao, Z.; Zeng, Y.; Cai, C.; Du, X.; Guo, W. Comprehensive analysis of the COBRA-Like (COBL) gene family in Gossypium identifies two COBLs potentially associated with fiber quality. PLoS ONE 2015, 10, e0145725. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, Y.; Tan, W.; Chen, J.; Zhu, M.; Lv, Y.; Liu, Y.; Yu, S.; Zhang, W.; Cai, H. Brittle Culm 1 encodes a COBRA-like protein involved in secondary cell wall cellulose biosynthesis in Sorghum. Plant Cell Physiol. 2018. [Google Scholar] [CrossRef]
- Holton, N.; Nekrasov, V.; Ronald, P.C.; Zipfel, C. The phylogenetically-related pattern recognition receptors EFR and XA21 recruit similar immune signaling components in monocots and dicots. PLoS Pathog. 2015, 11, e1004602. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Song, L.; Zhang, Y.; Zheng, Z.; Liu, D. Arabidopsis PHL2 and PHL1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol. 2016, 170, 499–514. [Google Scholar] [CrossRef]
- Yin, K.; Han, X.; Xu, Z.; Xue, H. Arabidopsis GLP4 is localized to the Golgi and binds auxin in vitro. Acta Biochim. Biophys. Sin. 2009, 41, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Pickel, B.; Pfannstiel, J.; Steudle, A.; Lehmann, A.; Gerken, U.; Pleiss, J.; Schaller, A. A model of dirigent proteins derived from structural and functional similarities with allene oxide cyclase and lipocalins. FEBS J. 2012, 279, 1980–1993. [Google Scholar] [CrossRef] [PubMed]
- Papini-Terzi, F.S.; Rocha, F.R.; Vêncio, R.Z.N.; Felix, J.M.; Branco, D.S.; Waclawovsky, A.J.; Del Bem, L.E.V.; Lembke, C.G.; Costa, M.D.L.; Nishiyama, M., Jr.; et al. Sugarcane genes associated with sucrose content. BMC Genomics 2009, 10, 1–21. [Google Scholar] [CrossRef]
- Lamport, D.T.A.; Kieliszewski, M.J.; Chen, Y.; Cannon, M.C. Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 2011, 156, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.C.; Terneus, K.; Hall, Q.; Tan, L.; Wang, W.; Wegenhart, B.L.; Chen, L.; Lamport, D.T.A.; Chen, Y.; Kieliszewski, M.J. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 2008, 105, 2226–2231. [Google Scholar] [CrossRef]
- Betekhtin, A.; Milewska-Hendel, A.; Lusinska, J.; Chajec, L.; Kurczynska, E.; Hasterok, R. Organ and Tissue-Specific Localisation of selected cell wall epitopes in the zygotic embryo of Brachypodium distachyon. Int. J. Mol. Sci. 2018, 19, 725. [Google Scholar] [CrossRef]
- Liu, X.; Wolfe, R.; Welch, L.R.; Domozych, D.S.; Popper, Z.A.; Showalter, A.M. Bioinformatic identification and analysis of extensins in the plant kingdom. PLoS ONE 2016, 11, e0150177. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Doesseger, B.; Guyot, R.; Diet, A.; Parsons, R.L.; Clark, M.A.; Simmons, M.P.; Bedinger, P.; Goff, S.A.; Ringli, C.; et al. Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 2003, 131, 1313–1326. [Google Scholar] [CrossRef]
- Vázquez-Lobo, A.; Roujol, D.; Zuñiga-Sánchez, E.; Albenne, C.; Piñero, D.; Gamboa de Buen, A.; Jamet, E. The highly conserved spermatophyte cell wall DUF642 protein family: Phylogeny and first evidence of interaction with cell wall polysaccharides in vitro. Mol. Phylogenet. Evol. 2012, 63, 510–520. [Google Scholar] [CrossRef]
- Zúñiga-Sanchez, E.; Soriano, D.; Martínez-Barajas, E.; Orozco-Segovia, A.; Gamboa-deBuen, A. BIIDXI, the At4g32460 DUF642 gene, is involved in pectin methyl esterase regulation during Arabidopsis thaliana seed germination and plant development. BMC Plant Biol. 2014, 14, 338. [Google Scholar] [CrossRef]
- Salazar-Iribe, A.; Cruz-Valderrama, J.E.; Jímenez-Durán, K.; Gómez-Maqueo, X.; Gamboa-deBuen, A. BIIDXI, a DUF642 cell wall protein, is involved in hypocotyl growth via auxin efflux. J. Plant Physiol. 2018, 231, 105–109. [Google Scholar] [CrossRef]
- Garza-Caligaris, L.E.; Avendaño-Vázquez, A.O.; Alvarado-López, S.; Zúñiga-Sánchez, E.; Orozco-Segovia, A.; Pérez-Ruíz, R.V.; Gamboa-DeBuen, A. At3g08030 transcript: A molecular marker of seed ageing. Ann. Bot. 2012, 110, 1253–1260. [Google Scholar] [CrossRef]
- Xu, B.; Gou, J.-Y.; Li, F.-G.; Shangguan, X.X.; Zhao, B.; Yang, C.-Q.; Wang, L.-J.; Yuan, S.; Liu, C.-J.; Chen, X.-Y. A cotton BURP domain protein interacts with α-expansin and their co-expression promotes plant growth and fruit production. Mol. Plant 2013, 6, 945–958. [Google Scholar] [CrossRef]
- Donohoe, B.S.; Wei, H.; Mittal, A.; Shollenberger, T.; Lunin, V.V.; Himmel, M.E.; Brunecky, R. Towards an understanding of enhanced biomass digestibility by In Planta expression of a family 5 glycoside hydrolase. Sci. Rep. 2017, 7, 4389. [Google Scholar] [CrossRef]
- Kidwai, M.; Dhar, Y.V.; Gautam, N.; Tiwari, M.; Ahmad, I.Z.; Asif, M.H.; Chakrabarty, D. Oryza sativa class III peroxidase (OsPRX38) overexpression in Arabidopsis thaliana reduces arsenic accumulation due to apoplastic lignification. J. Hazard. Mater 2019, 362, 383–393. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, L.; Wang, Y.; Li, H.; Li, S.; Zhao, H.; Zhang, H. Constitutive expression of miR408 improves biomass and seed yield in Arabidopsis. Front. Plant Sci. 2018, 8, 2114. [Google Scholar] [CrossRef]
- Endo, S.; Iwamoto, K.; Fukuda, H. Overexpression and cosuppression of xylem-related genes in an early xylem differentiation stage-specific manner by the AtTED4 promoter. Plant Biotechnol. J. 2018, 16, 451–458. [Google Scholar] [CrossRef]
- Popper, Z. Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 2008, 11, 286–292. [Google Scholar] [CrossRef]
- Phytozome. Available online: https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 23 January 2019).
| Species | Plant Sample | Extraction Method | Pipeline of MS Analyses | Total of CWPs Identified |
|---|---|---|---|---|
| B. distachyon | young and mature leaves, apical and basal internodes [33] | destructive technique followed by salt-based extraction | separation through 1D-PAGE and LC-MS/MS | 594 |
| young seedlings [35] | destructive technique followed by SDS and phenol-based extraction, and enrichment by ConA affinity chromatography | 2D-LC-MS/MS | ||
| seeds [36] | destructive technique followed by salt-based extraction | separation through 1D-PAGE and LC-MS/MS | ||
| seeds (9, 13 and 19 days after flowering (DAF)) [34] | destructive technique followed by salt-based extraction | separation through 1D-PAGE and LC-MS/MS | ||
| Saccharum spp. | 7 days cells suspension cultures [37] | destructive technique followed by salt-based extraction | 2D-LC-MS/MS | 283 |
| 2 month-old internodes [38] | destructive and non-destructive techniques followed by salt-based extractions | 2D-LC-MS/MS | ||
| 4 month-old young and mature leaves and apical and basal internodes [39] | destructive technique followed by salt-based extraction | 2D-LC-MS/MS | ||
| 7 month-old young and mature leaves [40,48] | destructive and non-destructive techniques followed by salt-based extractions | 2D-LC-MS/MS | ||
| 7 month-old apical and basal internodes [40,48] | non-destructive technique followed by salt-based extraction | 2D-LC-MS/MS | ||
| O. sativa | 5 days cultures medium of cell suspension cultures [28] | non-destructive technique followed by TCAAEB-based extraction | separation through 2D-PAGE and LC-MS/MS | 270 |
| 2–3 weeks cell suspension cultures [29] | non-destructive technique followed by salt-based extraction | separation through 1D-PAGE and 2D-LC-MS/MS | ||
| 3 week-old 4th leaf [28] | non-destructive technique followed by Tween-20, CTAB and TCAAEB-based extraction | separation through 2D-PAGE and LC-MS/MS | ||
| 2–3 weeks culture medium of cell suspension cultures [30] | non-destructive technique followed by salt-based extraction | separation through 1D-PAGE and 2D-LC-MS/MS | ||
| roots [31] | non-destructive technique followed by salt-based extraction | separation through 2D-PAGE and MALDI-TOF/TOF MS |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calderan-Rodrigues, M.J.; Guimarães Fonseca, J.; de Moraes, F.E.; Vaz Setem, L.; Carmanhanis Begossi, A.; Labate, C.A. Plant Cell Wall Proteomics: A Focus on Monocot Species, Brachypodium distachyon, Saccharum spp. and Oryza sativa. Int. J. Mol. Sci. 2019, 20, 1975. https://doi.org/10.3390/ijms20081975
Calderan-Rodrigues MJ, Guimarães Fonseca J, de Moraes FE, Vaz Setem L, Carmanhanis Begossi A, Labate CA. Plant Cell Wall Proteomics: A Focus on Monocot Species, Brachypodium distachyon, Saccharum spp. and Oryza sativa. International Journal of Molecular Sciences. 2019; 20(8):1975. https://doi.org/10.3390/ijms20081975
Chicago/Turabian StyleCalderan-Rodrigues, Maria Juliana, Juliana Guimarães Fonseca, Fabrício Edgar de Moraes, Laís Vaz Setem, Amanda Carmanhanis Begossi, and Carlos Alberto Labate. 2019. "Plant Cell Wall Proteomics: A Focus on Monocot Species, Brachypodium distachyon, Saccharum spp. and Oryza sativa" International Journal of Molecular Sciences 20, no. 8: 1975. https://doi.org/10.3390/ijms20081975
APA StyleCalderan-Rodrigues, M. J., Guimarães Fonseca, J., de Moraes, F. E., Vaz Setem, L., Carmanhanis Begossi, A., & Labate, C. A. (2019). Plant Cell Wall Proteomics: A Focus on Monocot Species, Brachypodium distachyon, Saccharum spp. and Oryza sativa. International Journal of Molecular Sciences, 20(8), 1975. https://doi.org/10.3390/ijms20081975





