Preeclampsia is Associated with Sex-Specific Transcriptional and Proteomic Changes in Fetal Erythroid Cells
Abstract
:1. Introduction
2. Results
2.1. Preeclampsia Does Not Alter Migration/Homing or Differentiation Capacity of UCB HSPCs
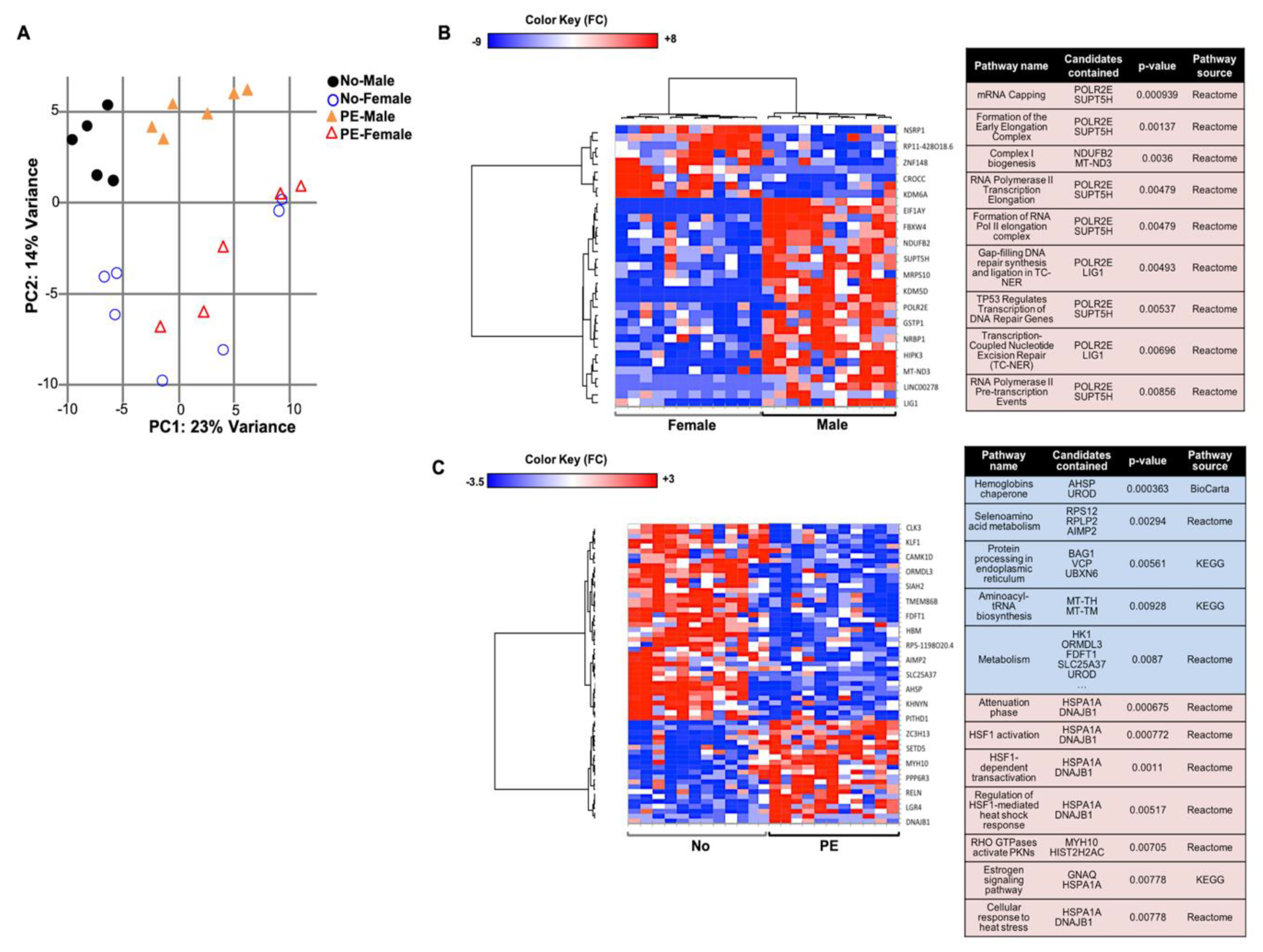
2.2. Preeclampsia Affects the Gene Expression in UCB HSPCs
2.3. Preeclampsia Is Associated with Changes in Metabolic and Protein Synthesis Pathways of In Vitro Differentiated Erythroid Cells
2.4. Preeclampsia Does Not Alter the UCB Profile of Terminally Differentiating Erythroblasts
2.5. Gene Expression Differences between Male vs. Female Samples Are Irrespective of Pregnancy Outcome
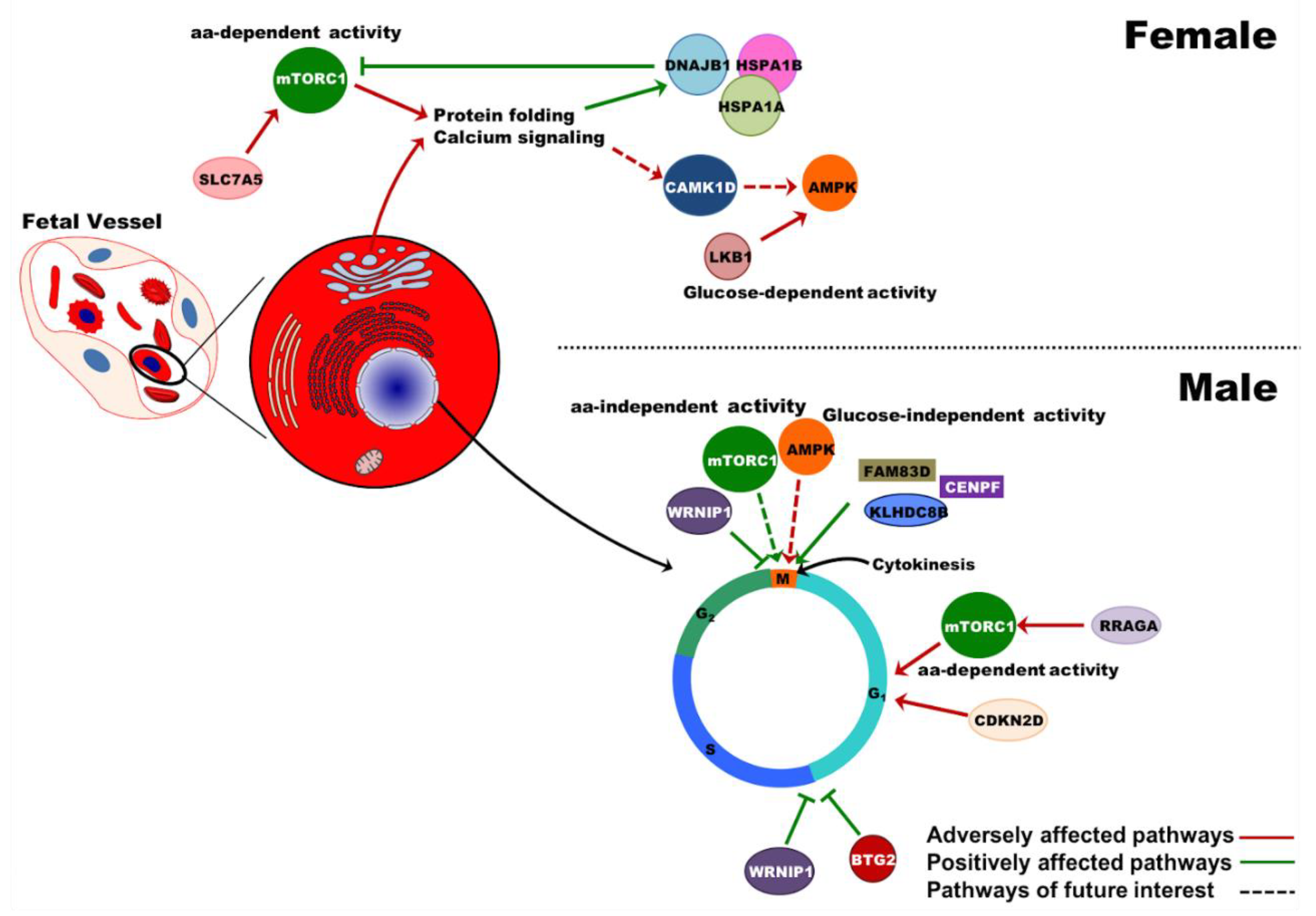
2.6. Effects of PE on Gene Expression in UCB Erythroid Cells Are Sex-Specific
2.7. Data Archiving
3. Discussion
4. Materials and Methods
4.1. Ethical Approval and Sample Collection
4.2. Mononuclear Cell Isolation from the UCB
4.3. Isolation of UCB CD34+ Cells
4.4. Flow Cytometric Analysis of SAM Expression on UCB HSPCs
4.5. Flow Cytometric Analysis of UCB Stem Cells and Colony Formation Assay
4.6. RNA Extraction and cDNA Subtractive Hybridization
4.7. Quantitative Proteomic Analysis
4.8. Fluorescent-Activated Sorting of Erythroblasts from the UCB
4.9. RNA Extraction, Library Preparation and Quality Check
4.10. Bioinformatics Analysis
4.11. Pathway Analysis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PE | Preeclampsia/preeclamptic |
| UCB | Umbilical cord blood |
| HS(P)Cs | Hematopoietic stem (progenitor) cells |
| SAM | Surface adhesion molecule |
| mTORC1 | mammalian target of rapamycin complex 1 |
| AMPK | AMP-activated protein kinase |
| FDR | False discovery rate |
| DE | Differentially expressed |
References
- WHO. WHO Recommendations for Prevention and Treatment of Pre-Eclampsia and Eclampsia; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Davey, D.A.; MacGillivray, I. The classification and definition of the hypertensive disorders of pregnancy. Am. J. Obstet. Gynecol. 1988, 158, 892–898. [Google Scholar] [CrossRef]
- Redman, C.W.; Sargent, I.L. Latest advances in understanding preeclampsia. Science 2005, 308, 1592–1594. [Google Scholar] [CrossRef] [PubMed]
- Kingdom, J.C.; Kaufmann, P. Oxygen and placental villous development: Origins of fetal hypoxia. Placenta 1997, 18, 613–621. [Google Scholar] [CrossRef]
- Redman, C.W.; Sargent, I.L. Placental stress and pre-eclampsia: A revised view. Placenta 2009, 30, S38–S42. [Google Scholar] [CrossRef] [PubMed]
- Browne, V.A.; Julian, C.G.; Toledo-Jaldin, L.; Cioffi-Ragan, D.; Vargas, E.; Moore, L.G. Uterine artery blood flow, fetal hypoxia and fetal growth. Philos. Trans R Soc. Lond. B Biol. Sci. 2015, 370, 20140068. [Google Scholar] [CrossRef] [PubMed]
- Soleymanlou, N.; Jurisica, I.; Nevo, O.; Ietta, F.; Zhang, X.; Zamudio, S.; Post, M.; Caniggia, I. Molecular evidence of placental hypoxia in preeclampsia. J. Clin. Endocrinol. Metab. 2005, 90, 4299–4308. [Google Scholar] [CrossRef]
- Sultana, Z.; Maiti, K.; Aitken, J.; Morris, J.; Dedman, L.; Smith, R. Oxidative stress, placental ageing-related pathologies and adverse pregnancy outcomes. Am. J. Reprod. Immunol. 2017, 77, e1265. [Google Scholar] [CrossRef]
- Tannetta, D.; Sargent, I. Placental disease and the maternal syndrome of preeclampsia: Missing links? Curr. Hypertens. Rep. 2013, 15, 590–599. [Google Scholar] [CrossRef]
- Anderson, U.D.; Olsson, M.G.; Rutardottir, S.; Centlow, M.; Kristensen, K.H.; Isberg, P.E.; Thilaganathan, B.; Akerstrom, B.; Hansson, S.R. Fetal hemoglobin and alpha1-microglobulin as first- and early second-trimester predictive biomarkers for preeclampsia. Am. J. Obstet. Gynecol. 2011, 204, e521–e525. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Oxidative reactions of hemoglobin. Methods Enzymol. 1990, 186, 265–272. [Google Scholar]
- May, K.; Rosenlof, L.; Olsson, M.G.; Centlow, M.; Morgelin, M.; Larsson, I.; Cederlund, M.; Rutardottir, S.; Siegmund, W.; Schneider, H.; et al. Perfusion of human placenta with hemoglobin introduces preeclampsia-like injuries that are prevented by alpha1-microglobulin. Placenta 2011, 32, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.W.; Chou, H.C.; Tsou, K.I.; Fang, L.J.; Tsao, P.N. Delivery before 32 weeks of gestation for maternal pre-eclampsia: Neonatal outcome and 2-year developmental outcome. Early Hum. Dev. 2004, 76, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.H.; Markham, K.; Moorehead, P.; Cordero, L.; Nankervis, C.A.; Giannone, P.J. Maternal preeclampsia and neonatal outcomes. J. Pregnancy 2011, 2011, 214365. [Google Scholar] [CrossRef]
- Williams, D. Long-term complications of preeclampsia. Semin. Nephrol. 2011, 31, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Ratsep, M.T.; Paolozza, A.; Hickman, A.F.; Maser, B.; Kay, V.R.; Mohammad, S.; Pudwell, J.; Smith, G.N.; Brien, D.; Stroman, P.W.; et al. Brain Structural and Vascular Anatomy Is Altered in Offspring of Pre-Eclamptic Pregnancies: A Pilot Study. AJNR Am. J. Neuroradiol. 2016, 37, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Masoumi, Z.; Familari, M.; Kallen, K.; Ranstam, J.; Olofsson, P.; Hansson, S.R. Fetal hemoglobin in umbilical cord blood in preeclamptic and normotensive pregnancies: A cross-sectional comparative study. PLoS ONE 2017, 12, e0176697. [Google Scholar] [CrossRef] [PubMed]
- Akercan, F.; Cirpan, T.; Saydam, G. Nucleated red blood cells in infants of women with preterm labor and pre-eclampsia. Int. J. Gynaecol. Obstet. 2005, 90, 138–139. [Google Scholar] [CrossRef]
- Catarino, C.; Rebelo, I.; Belo, L.; Rocha-Pereira, P.; Rocha, S.; Bayer Castro, E.; Patricio, B.; Quintanilha, A.; Santos-Silva, A. Erythrocyte changes in preeclampsia: Relationship between maternal and cord blood erythrocyte damage. J. Perinat. Med. 2009, 37, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Hebbar, S.; Misha, M.; Rai, L. Significance of maternal and cord blood nucleated red blood cell count in pregnancies complicated by preeclampsia. J. Pregnancy 2014, 2014, 496416. [Google Scholar] [CrossRef]
- Aali, B.S.; Malekpour, R.; Sedig, F.; Safa, A. Comparison of maternal and cord blood nucleated red blood cell count between pre-eclamptic and healthy women. J. Obstet. Gynaecol. Res. 2007, 33, 274–278. [Google Scholar] [CrossRef]
- Hermansen, M.C. Nucleated red blood cells in the fetus and newborn. Arch. Dis. Child Fetal Neonatal. Ed. 2001, 84, F211–F215. [Google Scholar] [CrossRef] [Green Version]
- Teramo, K.A.; Widness, J.A. Increased fetal plasma and amniotic fluid erythropoietin concentrations: Markers of intrauterine hypoxia. Neonatology 2009, 95, 105–116. [Google Scholar] [CrossRef]
- Thilaganathan, B.; Athanasiou, S.; Ozmen, S.; Creighton, S.; Watson, N.R.; Nicolaides, K.H. Umbilical cord blood erythroblast count as an index of intrauterine hypoxia. Arch. Dis. Child Fetal Neonatal. Ed. 1994, 70, F192–F194. [Google Scholar] [CrossRef] [PubMed]
- Korst, L.M.; Phelan, J.P.; Ahn, M.O.; Martin, G.I. Nucleated red blood cells: An update on the marker for fetal asphyxia. Am. J. Obstet. Gynecol. 1996, 175, 843–846. [Google Scholar] [CrossRef]
- Santillan, D.A.; Hamilton, W.; Christensen, A.; Talcott, K.; Gravatt, L.; Santillan, M.K.; Hunter, S.K. The effects of preeclampsia on signaling to hematopoietic progenitor cells. Proc. Obstet. Gynecol. 2013, 3, 11. [Google Scholar] [CrossRef]
- Surbek, D.V.; Danzer, E.; Steinmann, C.; Tichelli, A.; Wodnar-Filipowicz, A.; Hahn, S.; Holzgreve, W. Effect of preeclampsia on umbilical cord blood hematopoietic progenitor-stem cells. Am. J. Obstet. Gynecol. 2001, 185, 725–729. [Google Scholar] [CrossRef]
- Keele, D.K.; Kay, J.L. Plasma free fatty acid and blood sugar levels in newborn infants and their mothers. Pediatrics 1966, 37, 597–604. [Google Scholar]
- Sahasrabuddhe, A.; Pitale, S.; Raje, D.; Sagdeo, M.M. Cord blood levels of insulin and glucose in full-term pregnancies. J. Assoc. Phys. India 2013, 61, 378–382. [Google Scholar]
- Yang, J.M.; Wang, K.G. Relationship between acute fetal distress and maternal-placental-fetal circulations in severe preeclampsia. Acta Obstet. Gynecol. Scand. 1995, 74, 419–424. [Google Scholar] [CrossRef]
- Guillemette, L.; Lacroix, M.; Allard, C.; Patenaude, J.; Battista, M.C.; Doyon, M.; Moreau, J.; Menard, J.; Ardilouze, J.L.; Perron, P.; et al. Preeclampsia is associated with an increased pro-inflammatory profile in newborns. J. Reprod. Immunol. 2015, 112, 111–114. [Google Scholar] [CrossRef]
- Luo, S.T.; Zhang, D.M.; Qin, Q.; Lu, L.; Luo, M.; Guo, F.C.; Shi, H.S.; Jiang, L.; Shao, B.; Li, M.; et al. The Promotion of Erythropoiesis via the Regulation of Reactive Oxygen Species by Lactic Acid. Sci. Rep. 2017, 7, 38105. [Google Scholar] [CrossRef] [Green Version]
- Morceau, F.; Dicato, M.; Diederich, M. Pro-inflammatory cytokine-mediated anemia: Regarding molecular mechanisms of erythropoiesis. Mediators Inflamm. 2009, 2009, 405016. [Google Scholar] [CrossRef]
- Oburoglu, L.; Romano, M.; Taylor, N.; Kinet, S. Metabolic regulation of hematopoietic stem cell commitment and erythroid differentiation. Curr. Opin. Hematol. 2016, 23, 198–205. [Google Scholar] [CrossRef]
- Oburoglu, L.; Tardito, S.; Fritz, V.; de Barros, S.C.; Merida, P.; Craveiro, M.; Mamede, J.; Cretenet, G.; Mongellaz, C.; An, X.; et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell 2014, 15, 169–184. [Google Scholar] [CrossRef]
- Prince, O.D.; Langdon, J.M.; Layman, A.J.; Prince, I.C.; Sabogal, M.; Mak, H.H.; Berger, A.E.; Cheadle, C.; Chrest, F.J.; Yu, Q.; et al. Late stage erythroid precursor production is impaired in mice with chronic inflammation. Haematologica 2012, 97, 1648–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galacteros, F.; Guilloud-Bataille, M.; Feingold, J. Sex, gestational age, and weight dependancy of adult hemoglobin concentration in normal newborns. Blood 1991, 78, 1121–1124. [Google Scholar]
- Burman, D. Haemoglobin levels in normal infants aged 3 to 24 months, and the effect of iron. Arch. Dis. Child 1972, 47, 261–271. [Google Scholar] [CrossRef]
- Elsmen, E.; Kallen, K.; Marsal, K.; Hellstrom-Westas, L. Fetal gender and gestational-age-related incidence of pre-eclampsia. Acta Obstet. Gynecol. Scand. 2006, 85, 1285–1291. [Google Scholar] [CrossRef] [PubMed]
- Global Pregnancy, C.; Schalekamp-Timmermans, S.; Arends, L.R.; Alsaker, E.; Chappell, L.; Hansson, S.; Harsem, N.K.; Jalmby, M.; Jeyabalan, A.; Laivuori, H.; et al. Fetal sex-specific differences in gestational age at delivery in pre-eclampsia: A meta-analysis. Int. J. Epidemiol. 2016, 46, 632–642. [Google Scholar] [CrossRef]
- Brown, R.N. Maternal adaptation to pregnancy is at least in part influenced by fetal gender. BJOG 2015, 123, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Bytautiene, E.; Tamayo, E.; Gamble, P.; Anderson, G.D.; Hankins, G.D.; Longo, M.; Saade, G.R. Gender-specific effect of overexpression of sFlt-1 in pregnant mice on fetal programming of blood pressure in the offspring later in life. Am. J. Obstet. Gynecol. 2007, 197, e411–e415. [Google Scholar] [CrossRef]
- Higgins, M.; Keller, J.; Moore, F.; Ostrander, L.; Metzner, H.; Stock, L. Studies of blood pressure in Tecumseh, Michigan. I. Blood pressure in young people and its relationship to personal and familial characteristics and complications of pregnancy in mothers. Am. J. Epidemiol. 1980, 111, 142–155. [Google Scholar] [CrossRef]
- Langford, H.G.; Watson, R.L. Prepregnant blood pressure, hypertension during pregnancy, and later blood pressure of mothers and offspring. Hypertension 1980, 2, 130–133. [Google Scholar] [CrossRef]
- Palti, H.; Rothschild, E. Blood pressure and growth at 6 years of age among offsprings of mothers with hypertension of pregnancy. Early Hum. Dev. 1989, 19, 263–269. [Google Scholar] [CrossRef]
- Spinillo, A.; Montanari, L.; Gardella, B.; Roccio, M.; Stronati, M.; Fazzi, E. Infant sex, obstetric risk factors, and 2-year neurodevelopmental outcome among preterm infants. Dev. Med. Child Neurol. 2009, 51, 518–525. [Google Scholar] [CrossRef]
- Hu, J.; Liu, J.; Xue, F.; Halverson, G.; Reid, M.; Guo, A.; Chen, L.; Raza, A.; Galili, N.; Jaffray, J.; et al. Isolation and functional characterization of human erythroblasts at distinct stages: Implications for understanding of normal and disordered erythropoiesis in vivo. Blood 2013, 121, 3246–3253. [Google Scholar] [CrossRef]
- Wisgrill, L.; Schuller, S.; Bammer, M.; Berger, A.; Pollak, A.; Radke, T.F.; Kogler, G.; Spittler, A.; Helmer, H.; Husslein, P.; et al. Hematopoietic stem cells in neonates: Any differences between very preterm and term neonates? PLoS ONE 2014, 9, e106717. [Google Scholar] [CrossRef]
- Timeus, F.; Crescenzio, N.; Basso, G.; Ramenghi, U.; Saracco, P.; Gabutti, V. Cell adhesion molecule expression in cord blood CD34+ cells. Stem Cells 1998, 16, 120–126. [Google Scholar] [CrossRef]
- Roy, V.; Verfaillie, C.M. Expression and function of cell adhesion molecules on fetal liver, cord blood and bone marrow hematopoietic progenitors: Implications for anatomical localization and developmental stage specific regulation of hematopoiesis. Exp. Hematol. 1999, 27, 302–312. [Google Scholar] [CrossRef]
- Surbek, D.V.; Steinmann, C.; Burk, M.; Hahn, S.; Tichelli, A.; Holzgreve, W. Developmental changes in adhesion molecule expressions in umbilical cord blood CD34 hematopoietic progenitor and stem cells. Am. J. Obstet. Gynecol. 2000, 183, 1152–1157. [Google Scholar] [CrossRef]
- Kohn, L.A.; Hao, Q.L.; Sasidharan, R.; Parekh, C.; Ge, S.; Zhu, Y.; Mikkola, H.K.; Crooks, G.M. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat. Immunol. 2012, 13, 963–971. [Google Scholar] [CrossRef]
- Gunji, Y.; Nakamura, M.; Hagiwara, T.; Hayakawa, K.; Matsushita, H.; Osawa, H.; Nagayoshi, K.; Nakauchi, H.; Yanagisawa, M.; Miura, Y.; et al. Expression and function of adhesion molecules on human hematopoietic stem cells: CD34+ LFA-1- cells are more primitive than CD34+ LFA-1+ cells. Blood 1992, 80, 429–436. [Google Scholar]
- Chen, F.W.; Ioannou, Y.A. Ribosomal proteins in cell proliferation and apoptosis. Int. Rev. Immunol. 1999, 18, 429–448. [Google Scholar] [CrossRef]
- Shama, S.; Avni, D.; Frederickson, R.M.; Sonenberg, N.; Meyuhas, O. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr. Patt. 1995, 4, 241–252. [Google Scholar]
- Robledo, S.; Idol, R.A.; Crimmins, D.L.; Ladenson, J.H.; Mason, P.J.; Bessler, M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 2008, 14, 1918–1929. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef] [Green Version]
- Blagosklonny, M.V. Geroconversion: Irreversible step to cellular senescence. Cell Cycle 2014, 13, 3628–3635. [Google Scholar] [CrossRef]
- Green, D.R.; Kroemer, G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009, 458, 1127–1130. [Google Scholar] [CrossRef]
- Levine, B.; Abrams, J. p53: The Janus of autophagy? Nat. Cell Biol. 2008, 10, 637–639. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Weidberg, H.; Gonen, C.; Wilder, S.; Elazar, Z.; Oren, M. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc. Natl. Acad. Sci. USA 2010, 107, 18511–18516. [Google Scholar] [CrossRef] [Green Version]
- Raiser, D.M.; Narla, A.; Ebert, B.L. The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leuk. Lymphoma 2014, 55, 491–500. [Google Scholar] [CrossRef]
- Flygare, J.; Karlsson, S. Diamond-Blackfan anemia: Erythropoiesis lost in translation. Blood 2007, 109, 3152–3154. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, K.; Xiao, X.; Liao, J.; Hu, Q.; Chen, H.; Liu, J.; An, X. Autophagy as a regulatory component of erythropoiesis. Int. J. Mol. Sci. 2015, 16, 4083–4094. [Google Scholar] [CrossRef]
- Steelman, L.S.; Abrams, S.L.; Whelan, J.; Bertrand, F.E.; Ludwig, D.E.; Basecke, J.; Libra, M.; Stivala, F.; Milella, M.; Tafuri, A.; et al. Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways to leukemia. Leukemia 2008, 22, 686–707. [Google Scholar] [CrossRef] [Green Version]
- Constantino, B.T.; Cogionis, b. Nucleated RBCs—Significance in the Peripheral Blood Film. Lab. Med. 2000, 31, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Roussel, M.F. The INK4 family of cell cycle inhibitors in cancer. Oncogene 1999, 18, 5311–5317. [Google Scholar] [CrossRef] [Green Version]
- Jin, G.; Lee, S.W.; Zhang, X.; Cai, Z.; Gao, Y.; Chou, P.C.; Rezaeian, A.H.; Han, F.; Wang, C.Y.; Yao, J.C.; et al. Skp2-Mediated RagA Ubiquitination Elicits a Negative Feedback to Prevent Amino-Acid-Dependent mTORC1 Hyperactivation by Recruiting GATOR1. Mol. Cell 2015, 58, 989–1000. [Google Scholar] [CrossRef]
- Kalaitzidis, D.; Lee, D.; Efeyan, A.; Kfoury, Y.; Nayyar, N.; Sykes, D.B.; Mercier, F.E.; Papazian, A.; Baryawno, N.; Victora, G.D.; et al. Amino acid-insensitive mTORC1 regulation enables nutritional stress resilience in hematopoietic stem cells. J. Clin. Investig. 2017, 127, 1405–1413. [Google Scholar] [CrossRef]
- Lim, I.K. TIS21 (/BTG2/PC3) as a link between ageing and cancer: Cell cycle regulator and endogenous cell death molecule. J. Cancer Res. Clin. Oncol. 2006, 132, 417–426. [Google Scholar] [CrossRef]
- Kanu, N.; Zhang, T.; Burrell, R.A.; Chakraborty, A.; Cronshaw, J.; DaCosta, C.; Gronroos, E.; Pemberton, H.N.; Anderton, E.; Gonzalez, L.; et al. RAD18, WRNIP1 and ATMIN promote ATM signalling in response to replication stress. Oncogene 2016, 35, 4020. [Google Scholar] [CrossRef]
- Krem, M.M.; Luo, P.; Ing, B.I.; Horwitz, M.S. The kelch protein KLHDC8B guards against mitotic errors, centrosomal amplification, and chromosomal instability. J. Biol. Chem. 2012, 287, 39083–39093. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Wang, Y.; Qu, X.; Li, J.; Hale, J.; Huang, Y.; An, C.; Papoin, J.; Guo, X.; Chen, L.; et al. Distinct roles for TET family proteins in regulating human erythropoiesis. Blood 2017, 129, 2002–2012. [Google Scholar] [CrossRef]
- An, X.; Schulz, V.P.; Li, J.; Wu, K.; Liu, J.; Xue, F.; Hu, J.; Mohandas, N.; Gallagher, P.G. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014, 123, 3466–3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuyas, E.; Corominas-Faja, B.; Joven, J.; Menendez, J.A. Cell cycle regulation by the nutrient-sensing mammalian target of rapamycin (mTOR) pathway. Methods Mol. Biol. 2014, 1170, 113–144. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diekmann, F.; Rovira, J.; Diaz-Ricart, M.; Arellano, E.M.; Vodenik, B.; Jou, J.M.; Vives-Corrons, J.L.; Escolar, G.; Campistol, J.M. mTOR inhibition and erythropoiesis: Microcytosis or anaemia? Nephrol. Dial Transpl. 2012, 27, 537–541. [Google Scholar] [CrossRef]
- Knight, Z.A.; Schmidt, S.F.; Birsoy, K.; Tan, K.; Friedman, J.M. A critical role for mTORC1 in erythropoiesis and anemia. Elife 2014, 3, e01913. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Camprecios, G.; Rimmele, P.; Liang, R.; Yalcin, S.; Mungamuri, S.K.; Barminko, J.; D’Escamard, V.; Baron, M.H.; Brugnara, C.; et al. FOXO3-mTOR metabolic cooperation in the regulation of erythroid cell maturation and homeostasis. Am. J. Hematol. 2014, 89, 954–963. [Google Scholar] [CrossRef]
- Mortensen, M.; Ferguson, D.J.; Edelmann, M.; Kessler, B.; Morten, K.J.; Komatsu, M.; Simon, A.K. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Suzuki, M.; Satoh, H.; Kihara-Negishi, F.; Nakano, H.; Oikawa, T. Effects of PU.1-induced mouse calcium-calmodulin-dependent kinase I-like kinase (CKLiK) on apoptosis of murine erythroleukemia cells. Exp. Cell Res. 2004, 294, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Bauer, D.E.; Ghamari, A.; Nizzi, C.P.; Deck, K.M.; Kingsley, P.D.; Yien, Y.Y.; Huston, N.C.; Chen, C.; Schultz, I.J.; et al. The mTORC1/4E-BP pathway coordinates hemoglobin production with L-leucine availability. Sci. Sig. 2015, 8, ra34. [Google Scholar] [CrossRef] [PubMed]
- Daniels, G. Xg blood group system. In Human Blood Groups, 2nd ed.; Blackwell Science Ltd.: Oxford, UK, 2002. [Google Scholar]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Goffin, S.M.; Derraik, J.G.B.; Groom, K.M.; Cutfield, W.S. Maternal pre-eclampsia and long-term offspring health: Is there a shadow cast? Pregnancy Hypertens. 2018, 12, 11–15. [Google Scholar] [CrossRef]
- Figueiro-Filho, E.A.; Mak, L.E.; Reynolds, J.N.; Stroman, P.W.; Smith, G.N.; Forkert, N.D.; Paolozza, A.; Ratsep, M.T.; Croy, B.A. Neurological function in children born to preeclamptic and hypertensive mothers—A systematic review. Pregnancy Hypertens. 2017, 10, 1–6. [Google Scholar] [CrossRef]
- Sood, R.; Zehnder, J.L.; Druzin, M.L.; Brown, P.O. Gene expression patterns in human placenta. Proc. Natl. Acad. Sci. USA 2006, 103, 5478–5483. [Google Scholar] [CrossRef] [Green Version]
- Sedlmeier, E.M.; Brunner, S.; Much, D.; Pagel, P.; Ulbrich, S.E.; Meyer, H.H.; Amann-Gassner, U.; Hauner, H.; Bader, B.L. Human placental transcriptome shows sexually dimorphic gene expression and responsiveness to maternal dietary n-3 long-chain polyunsaturated fatty acid intervention during pregnancy. BMC Genom. 2014, 15, 941. [Google Scholar] [CrossRef] [PubMed]
- Dearden, L.; Bouret, S.G.; Ozanne, S.E. Sex and gender differences in developmental programming of metabolism. Mol. Metab. 2018, 15, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, C.S. Sex-Specific Placental Responses in Fetal Development. Endocrinology 2015, 156, 3422–3434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, S.A.; Roberts, J.M.; Bodnar, L.M.; Haggerty, C.L.; Youk, A.O.; Catov, J.M. Newborns of preeclamptic women show evidence of sex-specific disparity in fetal growth. Gend. Med. 2012, 9, 424–435. [Google Scholar] [CrossRef]
- Wu, C.S.; Nohr, E.A.; Bech, B.H.; Vestergaard, M.; Catov, J.M.; Olsen, J. Health of children born to mothers who had preeclampsia: A population-based cohort study. Am. J. Obstet. Gynecol. 2009, 201, e261–e269. [Google Scholar] [CrossRef]
- Turner, M.L.; McIlwaine, K.; Anthony, R.S.; Parker, A.C. Differential expression of cell adhesion molecules by human hematopoietic progenitor cells from bone marrow and mobilized adult peripheral blood. Stem Cells 1995, 13, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Voermans, C.; van Hennik, P.B.; van der Schoot, C.E. Homing of human hematopoietic stem and progenitor cells: New insights, new challenges? J. Hematother. Stem Cell Res. 2001, 10, 725–738. [Google Scholar] [CrossRef]
- Deguchi, T.; Komada, Y.; Sugiyama, K.; Zhang, X.L.; Azuma, E.; Yamamoto, H.; Sakurai, M. Expression of homing-associated cell adhesion molecule (H-CAM/CD44) on human CD34+ hematopoietic progenitor cells. Exp. Hematol. 1999, 27, 542–552. [Google Scholar] [CrossRef]
- Yanai, N.; Sekine, C.; Yagita, H.; Obinata, M. Roles for integrin very late activation antigen-4 in stroma-dependent erythropoiesis. Blood 1994, 83, 2844–2850. [Google Scholar] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Bushnell, B. BBTools. Available online: http://jgi.doe.gov/data-and-tools/bbtools/ (accessed on 1 May 2017).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Herzeel, C.; Costanza, P.; Decap, D.; Fostier, J.; Reumers, J. elPrep: High-Performance Preparation of Sequence Alignment/Map Files for Variant Calling. PLoS ONE 2015, 10, e0132868. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB—A database for integrating human functional interaction networks. Nucleic Acids Res. 2009, 37, D623–D628. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [Green Version]
- Niklasson, A.; Albertsson-Wikland, K. Continuous growth reference from 24th week of gestation to 24 months by gender. BMC Pediatr. 2008, 8, 8. [Google Scholar] [CrossRef]
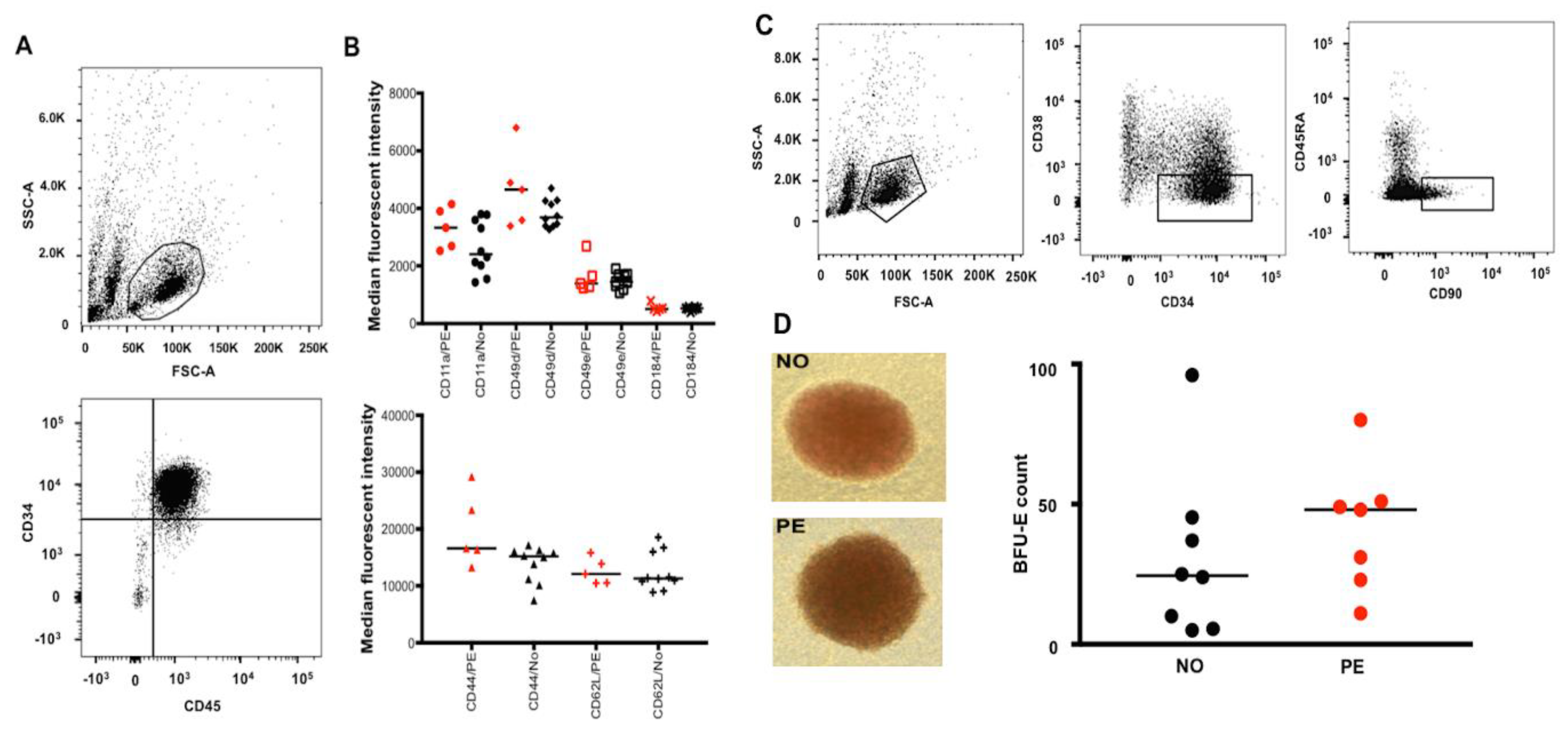




| To Recognize | Markers/Profile |
|---|---|
| Hematopoietic stem/progenitor cells (HSPCs) | CD34+ (clone 581) CD45+ (clone HI30) |
| Surface adhesion molecules (SAMs) | CD44 (clone 515) CD49d (clone 9F10) CD49e (clone IIA1) CD184 (clone 12G5) CD11a (clone HI111) CD62L (polyclonal) |
| Hematopoietic stem cells (HSCs) | CD34+ (clone 581) CD38lo (clone HIT2) CD45RA− (clone HI100) CD90+ (clone 5E10) |
| Erythroid cells (Flow cytometry) from proerythroblasts to mature erythrocytes | CD45− (clone HI30) GPA+ (clone HIR2) Band 3 (clone BRIC6) CD49d (clone 9F10) |
| Analysis | N | Pregnancy Condition | Gestational Age (Weeks) | Comments |
|---|---|---|---|---|
| SAM expression on UCB HSPCs | 10 | Normotensive | 36–42 | These samples were also used for cDNA subtractive hybridization. |
| 5 | PE | 36–39 | ||
| Colony formation assay | 8 | Normotensive | 38–40 | Performed in two sets of individual experiments, with three technical replicates in each set. |
| 7 | PE | 30–41 | ||
| Quantitative proteomic analysis | 5 | Normotensive | 38–40 | The colonies were obtained from the colony formation assay. |
| 5 | PE | 37–41 | ||
| UCB erythroid profile and transcriptome analysis | 7 | Normotensive | 36–40 | |
| 6 | PE | 36–40 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masoumi, Z.; Maes, G.E.; Herten, K.; Cortés-Calabuig, Á.; Alattar, A.G.; Hanson, E.; Erlandsson, L.; Mezey, E.; Magnusson, M.; Vermeesch, J.R.; et al. Preeclampsia is Associated with Sex-Specific Transcriptional and Proteomic Changes in Fetal Erythroid Cells. Int. J. Mol. Sci. 2019, 20, 2038. https://doi.org/10.3390/ijms20082038
Masoumi Z, Maes GE, Herten K, Cortés-Calabuig Á, Alattar AG, Hanson E, Erlandsson L, Mezey E, Magnusson M, Vermeesch JR, et al. Preeclampsia is Associated with Sex-Specific Transcriptional and Proteomic Changes in Fetal Erythroid Cells. International Journal of Molecular Sciences. 2019; 20(8):2038. https://doi.org/10.3390/ijms20082038
Chicago/Turabian StyleMasoumi, Zahra, Gregory E. Maes, Koen Herten, Álvaro Cortés-Calabuig, Abdul Ghani Alattar, Eva Hanson, Lena Erlandsson, Eva Mezey, Mattias Magnusson, Joris R Vermeesch, and et al. 2019. "Preeclampsia is Associated with Sex-Specific Transcriptional and Proteomic Changes in Fetal Erythroid Cells" International Journal of Molecular Sciences 20, no. 8: 2038. https://doi.org/10.3390/ijms20082038
APA StyleMasoumi, Z., Maes, G. E., Herten, K., Cortés-Calabuig, Á., Alattar, A. G., Hanson, E., Erlandsson, L., Mezey, E., Magnusson, M., Vermeesch, J. R., Familari, M., & Hansson, S. R. (2019). Preeclampsia is Associated with Sex-Specific Transcriptional and Proteomic Changes in Fetal Erythroid Cells. International Journal of Molecular Sciences, 20(8), 2038. https://doi.org/10.3390/ijms20082038







