Gene Copy Number and Post-Transductional Mechanisms Regulate TRAP1 Expression in Human Colorectal Carcinomas
Abstract
1. Introduction
2. Results
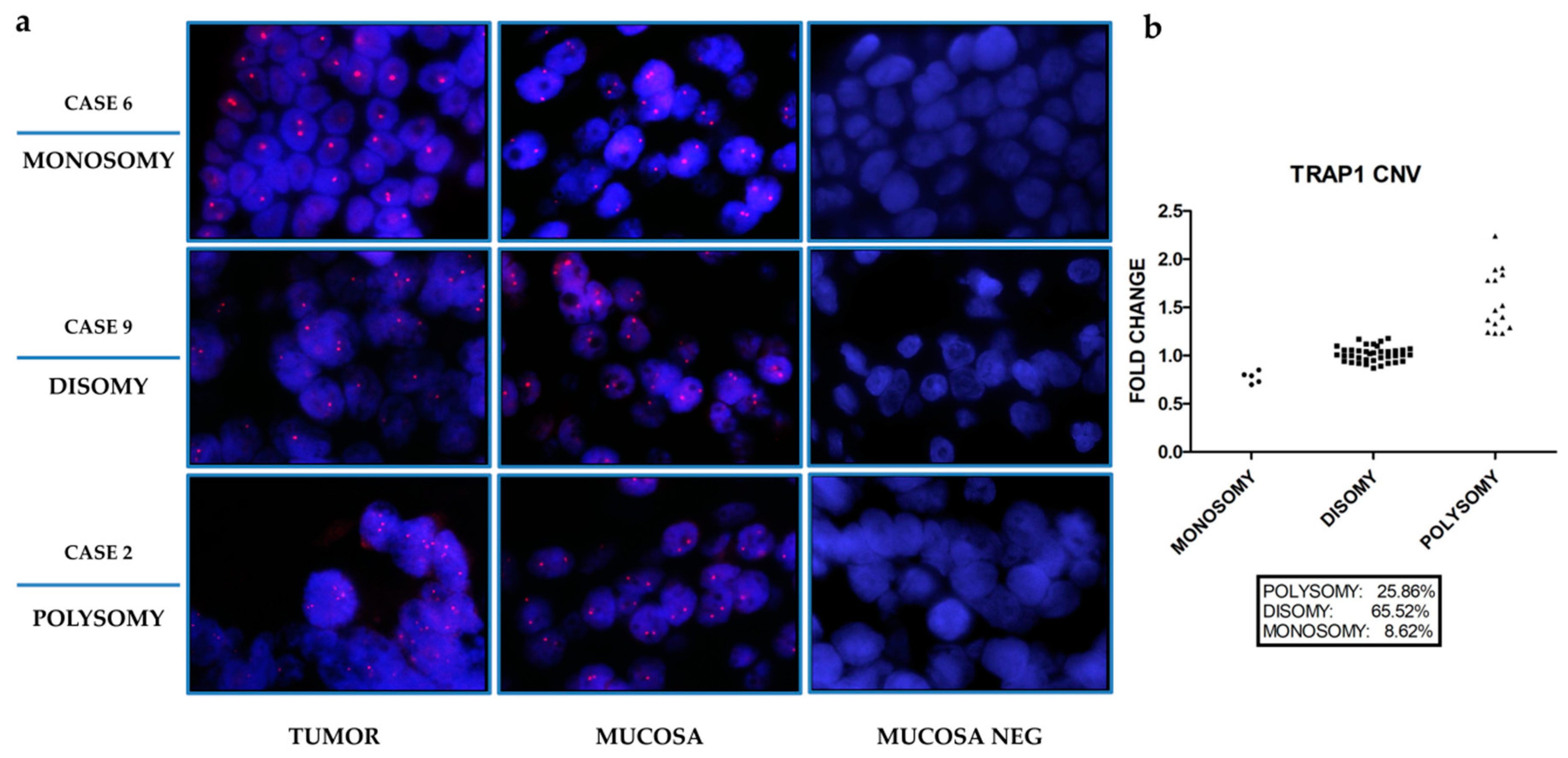
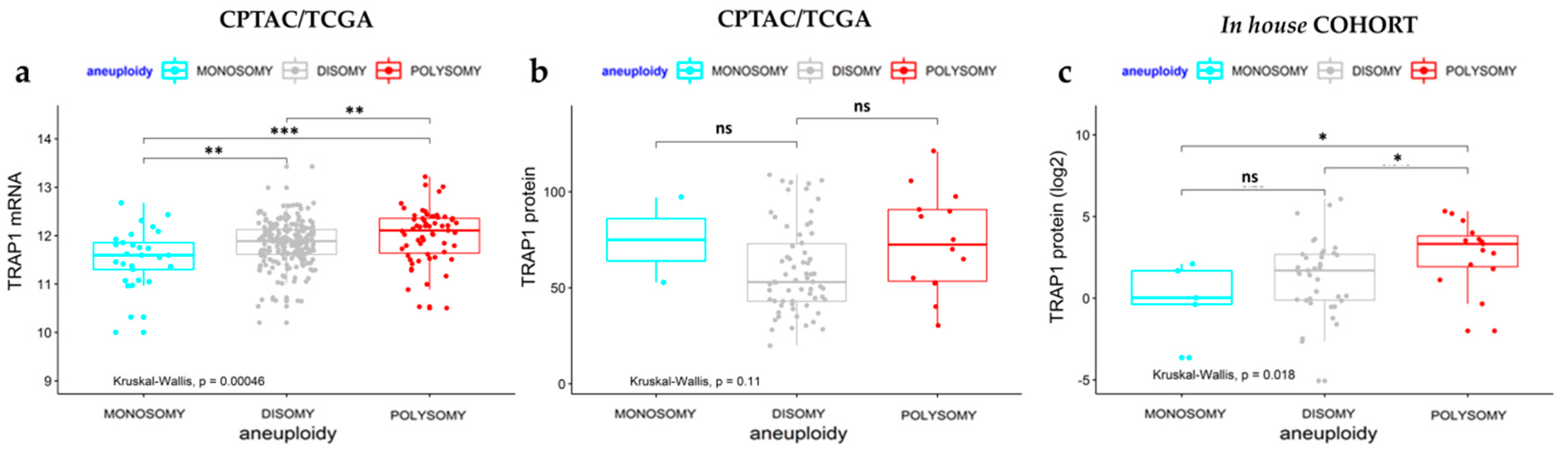
2.1. TRAP1 Expression Is Partially Dependent on Transcriptional Mechanisms
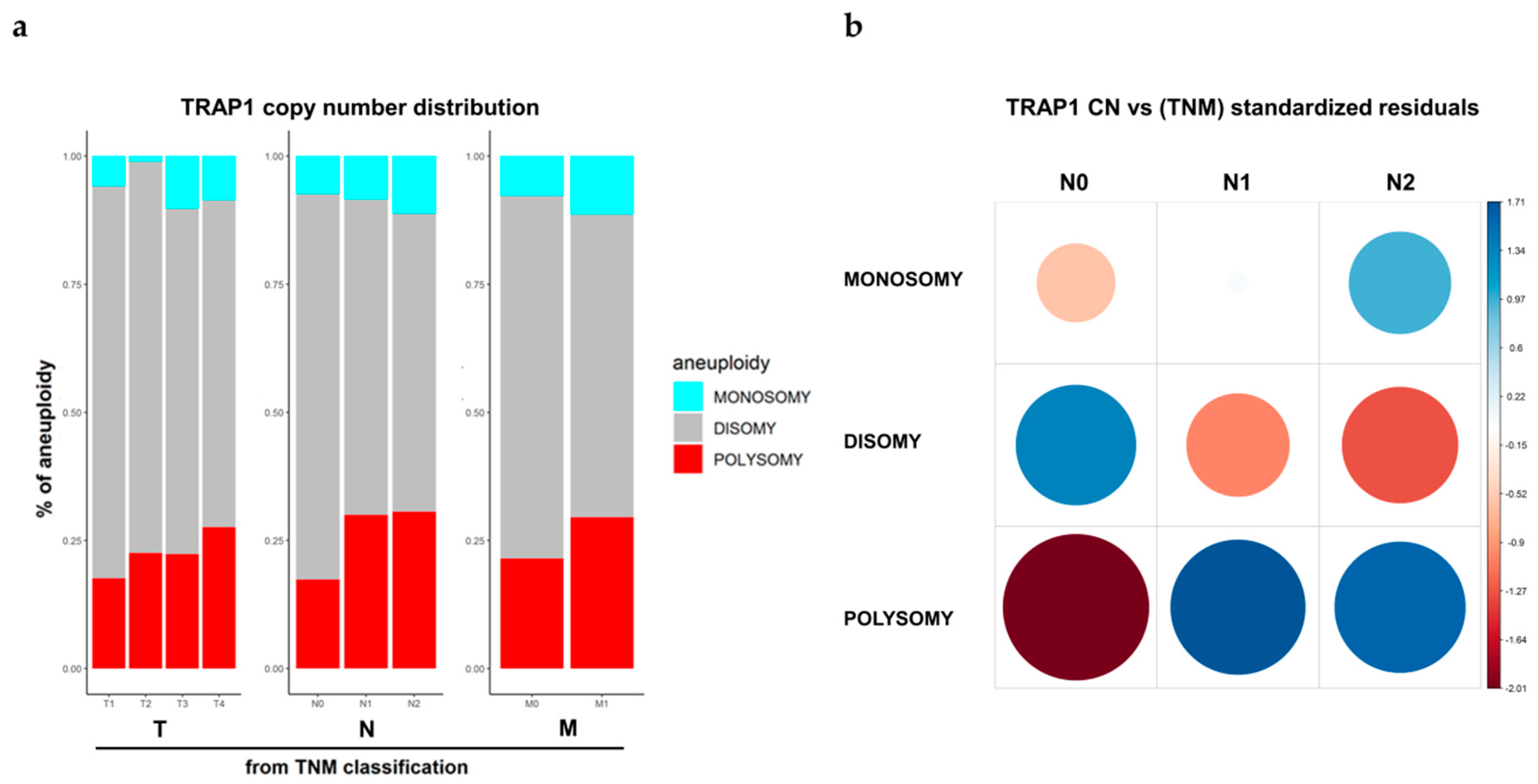
2.2. TRAP1 Polysomy Correlates with N Stage
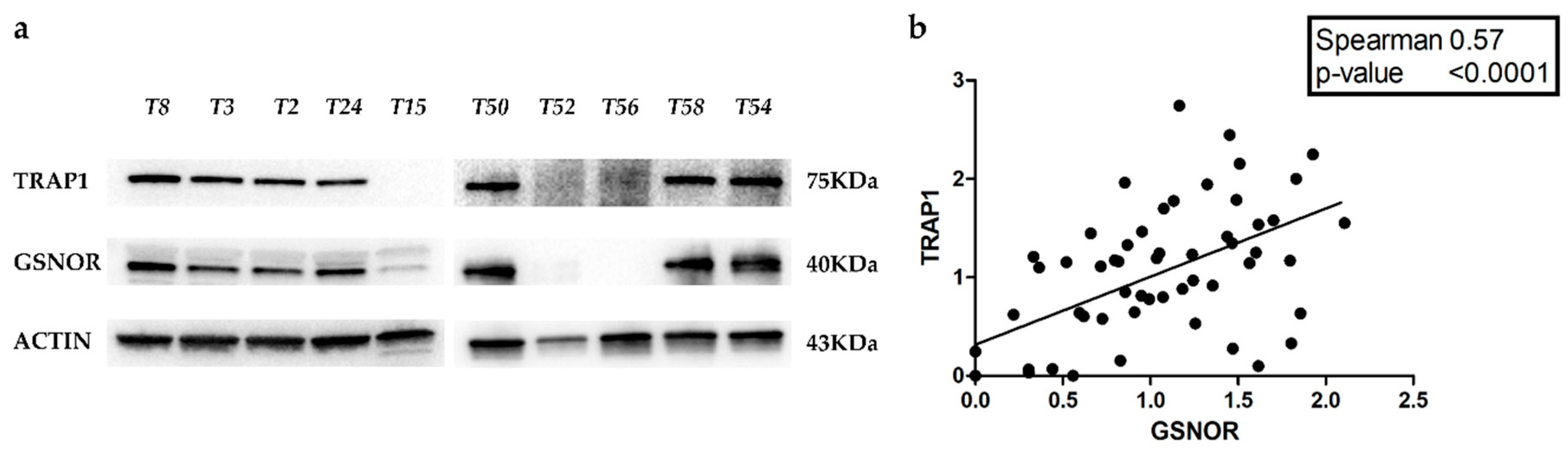
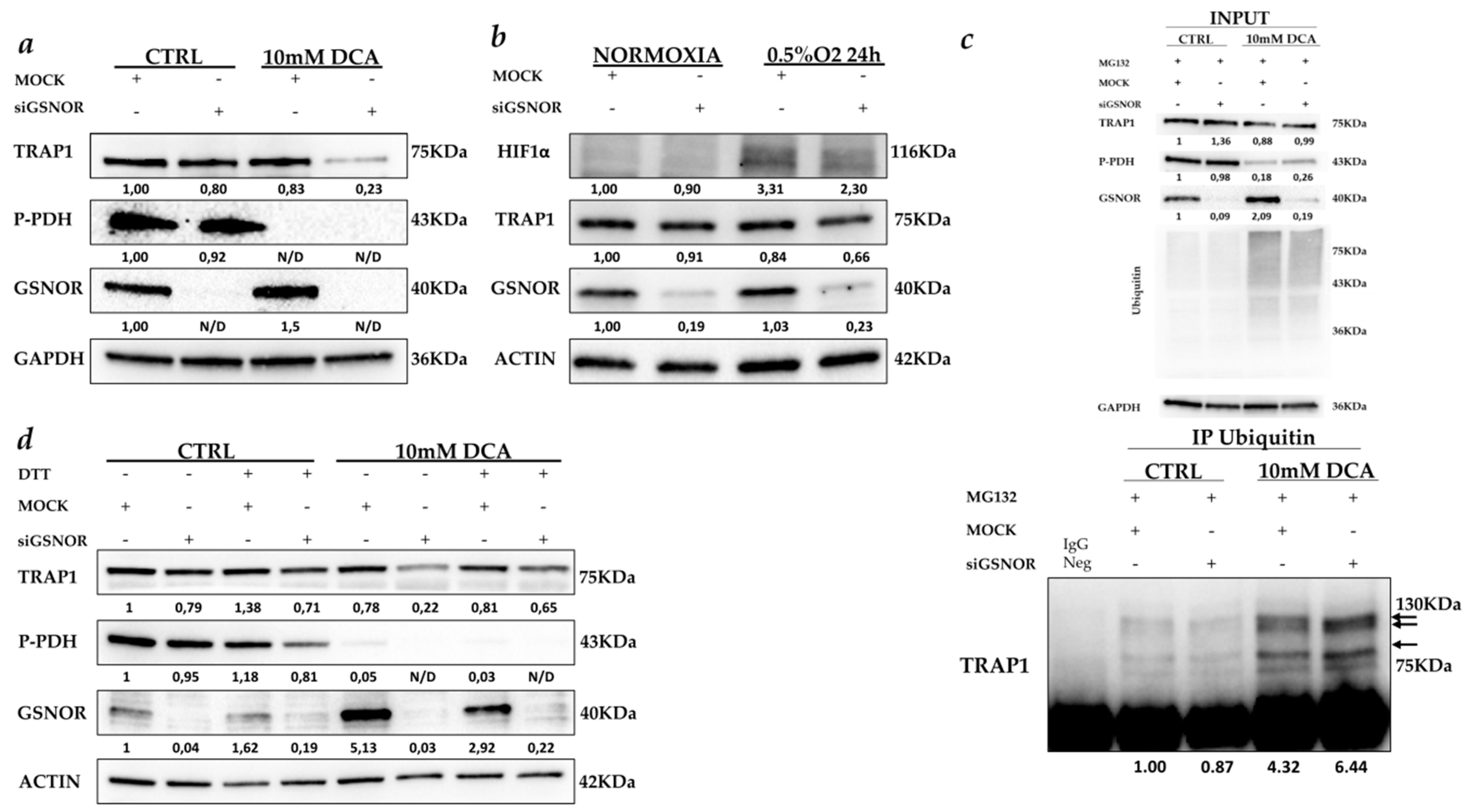
2.3. GSNOR Is Responsible for TRAP1 Post-Transductional Regulation
3. Discussion
4. Materials and Methods
4.1. Tumor Specimens, Clinical Data, Reagents and Cell Cultures
4.2. Cell Extract, Immunoblot Analysis, and Antibodies
4.3. RT-PCR Copy Number Variation Analysis
4.4. Fluorescent In Situ Hybridization (FISH)
4.5. High-Throughput Sequencing and Other Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| TRAP1 | Tumor Necrosis Factor Receptor-Associated Protein 1 |
| TCGA | The Cancer Genome Atlas |
| CPTAC | the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium |
| CRCs | Human Colorectal Carcinomas |
| CN | Copy number |
| COADREAD | Colon and rectum adenocarcinoma |
| GSNOR | S-nitrosoglutathione reductase |
| DCA | Dichloroacetate |
| PDH | pyruvate dehydrogenase |
| DTT | dithiothreitol |
References
- Kolligs, F.T. Diagnostics and Epidemiology of Colorectal Cancer. Visc. Med. 2016, 32, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Sadanandam, A.; Wang, X.; de Sousa, E.M.F.; Gray, J.W.; Vermeulen, L.; Hanahan, D.; Medema, J.P. Reconciliation of classification systems defining molecular subtypes of colorectal cancer: Interrelationships and clinical implications. Cell Cycle 2014, 13, 353–357. [Google Scholar] [CrossRef]
- Herzig, D.O.; Tsikitis, V.L. Molecular markers for colon diagnosis, prognosis and targeted therapy. J. Surg. Oncol. 2015, 111, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Maddalena, F.; Simeon, V.; Vita, G.; Bochicchio, A.; Possidente, L.; Sisinni, L.; Lettini, G.; Condelli, V.; Matassa, D.S.; Li Bergolis, V.; et al. TRAP1 protein signature predicts outcome in human metastatic colorectal carcinoma. Oncotarget 2017, 8, 21229–21240. [Google Scholar] [CrossRef] [PubMed]
- Costantino, E.; Maddalena, F.; Calise, S.; Piscazzi, A.; Tirino, V.; Fersini, A.; Ambrosi, A.; Neri, V.; Esposito, F.; Landriscina, M. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009, 279, 39–46. [Google Scholar] [CrossRef]
- Landriscina, M.; Laudiero, G.; Maddalena, F.; Amoroso, M.R.; Piscazzi, A.; Cozzolino, F.; Monti, M.; Garbi, C.; Fersini, A.; Pucci, P.; et al. Mitochondrial chaperone Trap1 and the calcium binding protein Sorcin interact and protect cells against apoptosis induced by antiblastic agents. Cancer Res. 2010, 70, 6577–6586. [Google Scholar] [CrossRef]
- Sisinni, L.; Maddalena, F.; Condelli, V.; Pannone, G.; Simeon, V.; Li Bergolis, V.; Lopes, E.; Piscazzi, A.; Matassa, D.S.; Mazzoccoli, C.; et al. TRAP1 controls cell cycle G2-M transition through the regulation of CDK1 and MAD2 expression/ubiquitination. J. Pathol. 2017, 243, 123–134. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Guzzo, G.; Morello, V.; Frezza, C.; Zheng, L.; Nannini, N.; Calabrese, F.; Laudiero, G.; Esposito, F.; Landriscina, M.; et al. The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. Cell Metab. 2013, 17, 988–999. [Google Scholar] [CrossRef]
- Chae, Y.C.; Angelin, A.; Lisanti, S.; Kossenkov, A.V.; Speicher, K.D.; Wang, H.; Powers, J.F.; Tischler, A.S.; Pacak, K.; Fliedner, S.; et al. Landscape of the mitochondrial Hsp90 metabolome in tumours. Nat. Commun. 2013, 4, 2139. [Google Scholar] [CrossRef]
- Yoshida, S.; Tsutsumi, S.; Muhlebach, G.; Sourbier, C.; Lee, M.J.; Lee, S.; Vartholomaiou, E.; Tatokoro, M.; Beebe, K.; Miyajima, N.; et al. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc. Natl. Acad. Sci. USA 2013, 110, E1604–E1612. [Google Scholar] [CrossRef]
- Lettini, G.; Sisinni, L.; Condelli, V.; Matassa, D.S.; Simeon, V.; Maddalena, F.; Gemei, M.; Lopes, E.; Vita, G.; Del Vecchio, L.; et al. TRAP1 regulates stemness through Wnt/beta-catenin pathway in human colorectal carcinoma. Cell Death Differ. 2016, 23, 1792–1803. [Google Scholar] [CrossRef] [PubMed]
- Pak, M.G.; Koh, H.J.; Roh, M.S. Clinicopathologic significance of TRAP1 expression in colorectal cancer: A large scale study of human colorectal adenocarcinoma tissues. Diagn. Pathol. 2017, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Y.; Song, B.R.; Peng, J.J.; Lu, Y.M. Correlation between mitochondrial TRAP-1 expression and lymph node metastasis in colorectal cancer. World J. Gastroenterol. 2012, 18, 5965–5971. [Google Scholar] [CrossRef] [PubMed]
- Han, J.J.; Baek, S.K.; Lee, J.J.; Kim, G.Y.; Kim, S.Y.; Lee, S.H. Combination of TRAP1 and ERCC1 Expression Predicts Clinical Outcomes in Metastatic Colorectal Cancer Treated with Oxaliplatin/5-Fluorouracil. Cancer Res. Treat. 2014, 46, 55–64. [Google Scholar] [CrossRef]
- Rizza, S.; Montagna, C.; Cardaci, S.; Maiani, E.; Di Giacomo, G.; Sanchez-Quiles, V.; Blagoev, B.; Rasola, A.; De Zio, D.; Stamler, J.S.; et al. S-nitrosylation of the Mitochondrial Chaperone TRAP1 Sensitizes Hepatocellular Carcinoma Cells to Inhibitors of Succinate Dehydrogenase. Cancer Res. 2016, 76, 4170–4182. [Google Scholar] [CrossRef]
- Park, H.K.; Hong, J.H.; Oh, Y.T.; Kim, S.S.; Yin, J.; Lee, A.J.; Chae, Y.C.; Kim, J.H.; Park, S.H.; Park, C.K.; et al. Interplay between TRAP1 and Sirtuin-3 Modulates Mitochondrial Respiration and Oxidative Stress to Maintain Stemness of Glioma Stem Cells. Cancer Res. 2019, 79, 1369–1382. [Google Scholar] [CrossRef]
- Piantadosi, C.A. Regulation of mitochondrial processes by protein S-nitrosylation. Biochim. Biophys. Acta 2012, 1820, 712–721. [Google Scholar] [CrossRef]
- Burrows, N.; Cane, G.; Robson, M.; Gaude, E.; Howat, W.J.; Szlosarek, P.W.; Pedley, R.B.; Frezza, C.; Ashcroft, M.; Maxwell, P.H. Hypoxia-induced nitric oxide production and tumour perfusion is inhibited by pegylated arginine deiminase (ADI-PEG20). Sci. Rep. 2016, 6, 22950. [Google Scholar] [CrossRef]
- Pietrafesa, M. Rionero in Vulture, TRAP1 Modulation upon Hypoxic and DCA Treatments; Laboratory of Preclinical and Translational Research, IRCCS, Referral Cancer Center of Basilicata: Foggia, Italy, 2019. [Google Scholar]
- Radtke, F.; Raj, K. The role of Notch in tumorigenesis: Oncogene or tumour suppressor? Nat. Rev. Cancer 2003, 3, 756–767. [Google Scholar] [CrossRef]
- Otalora-Otalora, B.A.; Henriquez, B.; Lopez-Kleine, L.; Rojas, A. RUNX family: Oncogenes or tumor suppressors (Review). Oncol. Rep. 2019, 42, 3–19. [Google Scholar] [CrossRef]
- De Carcer, G. The Mitotic Cancer Target Polo-Like Kinase 1: Oncogene or Tumor Suppressor? Genes 2019, 10, 208. [Google Scholar] [CrossRef]
- Shen, L.; Shi, Q.; Wang, W. Double agents: Genes with both oncogenic and tumor-suppressor functions. Oncogenesis 2018, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Lettini, G.; Maddalena, F.; Sisinni, L.; Condelli, V.; Matassa, D.S.; Costi, M.P.; Simoni, D.; Esposito, F.; Landriscina, M. TRAP1: A viable therapeutic target for future cancer treatments? Expert Opin. Ther. Targets 2017, 21, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Matassa, D.S.; Agliarulo, I.; Avolio, R.; Landriscina, M.; Esposito, F. TRAP1 Regulation of Cancer Metabolism: Dual Role as Oncogene or Tumor Suppressor. Genes 2018, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Sisinni, L.; Maddalena, F.; Lettini, G.; Condelli, V.; Matassa, D.S.; Esposito, F.; Landriscina, M. TRAP1 role in endoplasmic reticulum stress protection favors resistance to anthracyclins in breast carcinoma cells. Int. J. Oncol. 2014, 44, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Matassa, D.S.; Amoroso, M.R.; Lu, H.; Avolio, R.; Arzeni, D.; Procaccini, C.; Faicchia, D.; Maddalena, F.; Simeon, V.; Agliarulo, I.; et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ. 2016, 23, 1542–1554. [Google Scholar] [CrossRef]
- Condelli, V.; Piscazzi, A.; Sisinni, L.; Matassa, D.S.; Maddalena, F.; Lettini, G.; Simeon, V.; Palladino, G.; Amoroso, M.R.; Trino, S.; et al. TRAP1 is involved in BRAF regulation and downstream attenuation of ERK phosphorylation and cell-cycle progression: A novel target for BRAF-mutated colorectal tumors. Cancer Res. 2014, 74, 6693–6704. [Google Scholar] [CrossRef]
- Condelli, V.; Maddalena, F.; Sisinni, L.; Lettini, G.; Matassa, D.S.; Piscazzi, A.; Palladino, G.; Amoroso, M.R.; Esposito, F.; Landriscina, M. Targeting TRAP1 as a downstream effector of BRAF cytoprotective pathway: A novel strategy for human BRAF-driven colorectal carcinoma. Oncotarget 2015, 6, 22298–22309. [Google Scholar] [CrossRef]
- Agliarulo, I.; Matassa, D.S.; Amoroso, M.R.; Maddalena, F.; Sisinni, L.; Sepe, L.; Ferrari, M.C.; Arzeni, D.; Avolio, R.; Paolella, G.; et al. TRAP1 controls cell migration of cancer cells in metabolic stress conditions: Correlations with AKT/p70S6K pathways. Biochim. Biophys. Acta 2015, 1853, 2570–2579. [Google Scholar] [CrossRef]
- Matassa, D.S.; Agliarulo, I.; Amoroso, M.R.; Maddalena, F.; Sepe, L.; Ferrari, M.C.; Sagar, V.; D’Amico, S.; Loreni, F.; Paolella, G.; et al. TRAP1-dependent regulation of p70S6K is involved in the attenuation of protein synthesis and cell migration: Relevance in human colorectal tumors. Mol. Oncol. 2014, 8, 1482–1494. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, M.R.; Matassa, D.S.; Agliarulo, I.; Avolio, R.; Lu, H.; Sisinni, L.; Lettini, G.; Gabra, H.; Landriscina, M.; Esposito, F. TRAP1 downregulation in human ovarian cancer enhances invasion and epithelial-mesenchymal transition. Cell Death Dis. 2016, 7, e2522. [Google Scholar] [CrossRef] [PubMed]
- Montesano Gesualdi, N.; Chirico, G.; Pirozzi, G.; Costantino, E.; Landriscina, M.; Esposito, F. Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress 2007, 10, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, M.R.; Matassa, D.S.; Laudiero, G.; Egorova, A.V.; Polishchuk, R.S.; Maddalena, F.; Piscazzi, A.; Paladino, S.; Sarnataro, D.; Garbi, C.; et al. TRAP1 and the proteasome regulatory particle TBP7/Rpt3 interact in the endoplasmic reticulum and control cellular ubiquitination of specific mitochondrial proteins. Cell Death Differ. 2012, 19, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Matassa, D.S.; Amoroso, M.R.; Agliarulo, I.; Maddalena, F.; Sisinni, L.; Paladino, S.; Romano, S.; Romano, M.F.; Sagar, V.; Loreni, F.; et al. Translational control in the stress adaptive response of cancer cells: A novel role for the heat shock protein TRAP1. Cell Death Dis. 2013, 4, e851. [Google Scholar] [CrossRef] [PubMed]
- Gunderson, L.L.; Jessup, J.M.; Sargent, D.J.; Greene, F.L.; Stewart, A. Revised tumor and node categorization for rectal cancer based on surveillance, epidemiology, and end results and rectal pooled analysis outcomes. J. Clin. Oncol. 2010, 28, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Piscazzi, A.; Costantino, E.; Maddalena, F.; Natalicchio, M.I.; Gerardi, A.M.; Antonetti, R.; Cignarelli, M.; Landriscina, M. Activation of the RAS/RAF/ERK signaling pathway contributes to resistance to sunitinib in thyroid carcinoma cell lines. J. Clin. Endocrinol. Metab. 2012, 97, E898–E906. [Google Scholar] [CrossRef][Green Version]
- Pannone, G.; Santoro, A.; Pasquali, D.; Zamparese, R.; Mattoni, M.; Russo, G.; Landriscina, M.; Piscazzi, A.; Toti, P.; Cignarelli, M.; et al. The role of survivin in thyroid tumors: Differences of expression in well-differentiated, non-well-differentiated, and anaplastic thyroid cancers. Thyroid 2014, 24, 511–519. [Google Scholar] [CrossRef]
- Cappuzzo, F.; Hirsch, F.R.; Rossi, E.; Bartolini, S.; Ceresoli, G.L.; Bemis, L.; Haney, J.; Witta, S.; Danenberg, K.; Domenichini, I.; et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J. Natl. Cancer Inst. 2005, 97, 643–655. [Google Scholar] [CrossRef]
- Alvarez, G.; Perry, A.; Tan, B.R.; Wang, H.L. Expression of epidermal growth factor receptor in squamous cell carcinomas of the anal canal is independent of gene amplification. Mod. Pathol. 2006, 19, 942–949. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available online: https://github.com/taiyun/corrplot; 2017 (accessed on 1 December 2019).
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000; ISBN 0-387-98784-3. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plot. 2018. Available online: https://rpkgs.datanovia.com/ggpubr (accessed on 1 December 2019).
| Patients | 58 | |
|---|---|---|
| Age | ||
| Median (years) | 70 | |
| Range | 35–89 | |
| Sex | No of patients | |
| Female | 26 | |
| Male | 32 | |
| Tumor stage | No of patients | (%) |
| Tumor | ||
| T1 | 2 | 4 |
| T2 | 4 | 7 |
| T3 | 38 | 65 |
| T4 | 14 | 24 |
| Nodes | ||
| N0 | 19 | 33 |
| N1 | 19 | 33 |
| N2 | 20 | 34 |
| Metastasis | ||
| M0 | 38 | 65 |
| M1 | 20 | 35 |
| Cases (n) | TRAP1 Copy Number for Cells (%) | Pattern | Fold Change | ||
|---|---|---|---|---|---|
| 1 Signals/Cell | 2 Signals/Cell | >2 Signals/Cell | |||
| 1 | 100 | Polisomy | 1.84 | ||
| 2 | 3 | 97 | Polisomy | 1.24 | |
| 3 | 4 | 96 | Polisomy | 1.23 | |
| 4 | 63 | 31 | 6 | Monosomy | 0.73 |
| 5 | 16 | 80 | 4 | Disomy | 1.05 |
| 6 | 63 | 37 | Monosomy | 0.85 | |
| 7 | 17 | 29 | 54 | Polisomy | 1.52 |
| 8 | 21 | 72 | 7 | Disomy | 1.12 |
| 9 | 41 | 56 | 3 | Disomy | 0.91 |
| 10 | 12 | 82 | 6 | Disomy | 1.17 |
| 11 | 15 | 80 | 5 | Disomy | 1.06 |
| 12 | 10 | 82 | 8 | Disomy | 1.03 |
| 13 | 17 | 75 | 8 | Disomy | 1.1 |
| 14 | 15 | 79 | 6 | Disomy | 1.04 |
| 15 | 38 | 60 | 2 | Disomy | 0.94 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietrafesa, M.; Maddalena, F.; Possidente, L.; Condelli, V.; Zoppoli, P.; Li Bergolis, V.; Rodriquenz, M.G.; Aieta, M.; Vita, G.; Esposito, F.; et al. Gene Copy Number and Post-Transductional Mechanisms Regulate TRAP1 Expression in Human Colorectal Carcinomas. Int. J. Mol. Sci. 2020, 21, 145. https://doi.org/10.3390/ijms21010145
Pietrafesa M, Maddalena F, Possidente L, Condelli V, Zoppoli P, Li Bergolis V, Rodriquenz MG, Aieta M, Vita G, Esposito F, et al. Gene Copy Number and Post-Transductional Mechanisms Regulate TRAP1 Expression in Human Colorectal Carcinomas. International Journal of Molecular Sciences. 2020; 21(1):145. https://doi.org/10.3390/ijms21010145
Chicago/Turabian StylePietrafesa, Michele, Francesca Maddalena, Luciana Possidente, Valentina Condelli, Pietro Zoppoli, Valeria Li Bergolis, Maria Grazia Rodriquenz, Michele Aieta, Giulia Vita, Franca Esposito, and et al. 2020. "Gene Copy Number and Post-Transductional Mechanisms Regulate TRAP1 Expression in Human Colorectal Carcinomas" International Journal of Molecular Sciences 21, no. 1: 145. https://doi.org/10.3390/ijms21010145
APA StylePietrafesa, M., Maddalena, F., Possidente, L., Condelli, V., Zoppoli, P., Li Bergolis, V., Rodriquenz, M. G., Aieta, M., Vita, G., Esposito, F., & Landriscina, M. (2020). Gene Copy Number and Post-Transductional Mechanisms Regulate TRAP1 Expression in Human Colorectal Carcinomas. International Journal of Molecular Sciences, 21(1), 145. https://doi.org/10.3390/ijms21010145






