RNA-Binding Proteins in Acute Leukemias
Abstract
1. Introduction
2. IGF2BP Family
3. MSI2
4. HnRNP K
5. Nucleolin
6. WT1
7. ZFP36L1/2
8. RBM15-MKL1
9. CPSF6
10. Tyrosine Kinase Fusions Involving RBPs
11. Discussion
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AA | Amino acid |
| AICD | Activation induced cell death |
| ALDH1A1 | Aldehyde dehydrogenase 1 member A1 |
| AKT | RAC-alpha serine/threonine-protein kinase |
| ALL | Acute lymphoblastic leukemia |
| AMKL | Acute megakaryoblastic leukemia |
| AML | Acute myeloid leukemia |
| APL | Acute promyelocytic leukemia |
| ARE | Adenine uridine rich element |
| ATRA | All-trans retinoid acid |
| AU | Adenine uridine |
| BAG3 | BCL2 associated athanogene 3 |
| Bax | BCL2 associated X protein |
| BCL2 | B-cell lymphoma 2 |
| BSAC | Basic SAP and coiled-coil domain containing protein |
| β-TRCP1 | Beta transducin repeat containing E3 Ubiquitin protein ligase 1 |
| BTYNB | 2-[[(5-bromo-2-thienyl)methylene]amino]-benzamide |
| CALM1 | Calmodulin 1 |
| CCCH | Cys-Cys-Cys-His |
| CDC34 | Cell Division Cycle Cycle 34 |
| CDK2 | Cyclin dependent kinase 2 |
| CDK6 | Cyclin dependent kinase 6 |
| CEBPA | CCAAT Enhancer binding protein alpha |
| ChIP | Chromatin precipitation assay |
| CML | Chronic myeloid leukemia |
| Col5A1 | Collagen Type V Alpha 1 Chain |
| CpG | CpG islands |
| CPSF6 | Cleavage and polyadenylation specifity factor 6 |
| CSF1R | Colony-stimulating factor 1 receptor |
| DBD | DNA-binding domain |
| DICE | Differentiation control element |
| eEF2 | Eukaryotic elongation factor 2 |
| ERK | Extracellular signal-regulated kinase |
| ETV6 | Translocation-Ets-leukemia virus 6 |
| GATA-2 | GATA-binding factor 2 |
| hnRNP | Heterogeneous nuclear ribonucleoproteins |
| HOXB4 | Homeobox B4 |
| HSP70 | 70kD heat shock protein |
| I-BET151 | 7,3,5-dimethyl-4-isoxazolyl-1,3-dihydro-8-methoxy-1-[1R-1-(2-pyridinyl)ethyl]-2H-imidazo[4,5-c]quinolin-2-one |
| iCLIP | Individual-nucleotide resolution crolllinking and immunoprecipitation |
| IDH1 | Isocitrate dehydrogenase 1 |
| IDH2 | Isocitrate dehydrogenase 2 |
| IGF2BP | Insulin-like growth factor 2 mRNA binding protein |
| IGH | Immunoglobulin Heavy Locus |
| IRES | Internal ribosomal entry site |
| KH | HnRNP-K homology |
| KI | K protein interactive |
| KNS | hnRNP K-specific nuclear shuttling signal |
| LBD | Ligand-binding domain |
| LSC | Leukemic stem cell |
| LSK | Lin-Sca1+Kit- |
| LT-HSC | Long-term hematopoietic stem cell |
| LOX | Lipoxygenase |
| MAL | Megakaryocytic acute leukemia |
| MAPK | Mitogen-activated protein kinase |
| MEF | Mouse embryo fibroblast |
| MKL1 | Megakaryocytic leukemia-1 |
| MLL | Mixed lineage leukemia |
| MPP | Multipotent progenitors |
| MRTF-A | Myocardin-related transcription factor A |
| MSI | Musashi |
| MSI2 | Musashi-2 Protein |
| mRNA | Messenger RNA |
| MiRNAs | microRNAs |
| MYB | MYB proto-oncogene |
| MYC | MYC proto-oncogene protein |
| NCL | Nucleolin |
| NLS | Nuclear localization signal |
| NPM | Nucleophosmin |
| NUMB | NUMB endocytic adaptor protein |
| numb5 | NUMB endocytic adaptor protein 5 |
| PH | Pleckstrin homology domain |
| PI3K | Phosphoinositide 3-kinases |
| PIAS3 | Protein inhibitor of activated STAT3 |
| PU.1 | Transcription factor PU.1 |
| p53 | Tumor protein p53 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 |
| RARG | Retinoic acid receptor gamma |
| RBM6 | RNA-binding motif 6 |
| RBM15 | RNA-binding motif protein-15 |
| RBP | RNA-binding protein |
| RNP | Ribonucleoprotein |
| RRM | RNA-recognition motif |
| RUNX1 | Runt related transcription factor 1 |
| SENP2 | SUMO-specific protease 2 |
| SF3B1 | Splicing Factor 3b Subunit 1 |
| SUMO | Small ubiquitin-related modifier |
| SPEN | Split end |
| SPOC | SPEN paralogue and orthologue C-terminal |
| SRSF2 | Serine and arginine rich splicing factor 2 |
| STAT3 | Signal transducer and activator of transcription 3 |
| ST-HSCs | short-term hematopoietic stem cells |
| TA | Transactivation |
| TET2 | Ten-eleven translocation methylcytosine dioxygenase 2 |
| TSS | Transcription start sites |
| TTC40 | Tetratricopeptide repeat domain 40 |
| U2AF1 | U2 small nuclear RNA auxiliary factor 1 |
| VEGF | Vascular endothelial growth factor |
| WT1 | Wilms Tumor 1 |
| YBX1 | Y-box binding protein 1 |
| ZF | Zinc finger domain |
| ZFP36 | Zinc finger protein homolog 36 |
References
- Lindsley, R.C.; Ebert, B.L.; Lindsley, R.C. The biology and clinical impact of genetic lesions in myeloid malignancies. Blood 2013, 122, 3741–3748. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.-H.; Yang, J.J.; Hunger, S.P.; Pieters, R.; Schrappe, M.; Biondi, A.; Vora, A.; Baruchel, A.; Silverman, L.B.; Schmiegelow, K.; et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J. Clin. Oncol. 2015, 33, 2938–2948. [Google Scholar] [CrossRef] [PubMed]
- Maynadié, M.; Girodon, F.; Manivet-Janoray, I.; Mounier, M.; Mugneret, F.; Bailly, F.; Favre, B.; Caillot, D.; Petrella, T.; Flesch, M.; et al. Twenty-five years of epidemiological recording on myeloid malignancies: Data from the specialized registry of hematologic malignancies of Côte d’Or (Burgundy, France). Haematologica 2010, 96, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Kuhlen, M.; Klusmann, J.-H.; Hoell, J.I. Molecular Approaches to Treating Pediatric Leukemias. Front. Pediatr. 2019, 7, 368. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Hope, K.J.; Jin, L.; Dick, J. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 2004, 5, 738–743. [Google Scholar] [CrossRef]
- Silverman, L.B. Balancing cure and long-term risks in acute lymphoblastic leukemia. Hematologica 2014, 2014, 190–197. [Google Scholar] [CrossRef][Green Version]
- El Marabti, E.; Younis, I. The Cancer Spliceome: Reprograming of Alternative Splicing in Cancer. Front. Mol. Biosci. 2018, 5, 80. [Google Scholar] [CrossRef]
- Ciafrè, S.A.; Galardi, S. microRNAs and RNA-binding proteins. RNA Boil. 2013, 10, 935–942. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Boil. 2007, 8, 479–490. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.; Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science 1994, 265, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Maris, C.; Dominguez, C.; Allain, F.H.-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Nagata, T.; Tsuda, K.; Kobayashi, N.; Imai, T.; Okano, H.; Yamazaki, T.; Katahira, M. Structure of Musashi1 in a complex with target RNA: The role of aromatic stacking interactions. Nucleic Acids Res. 2011, 40, 3218–3231. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.; Edwards, L.; Regan, L. Structure and function of KH domains. FEBS J. 2008, 275, 2712–2726. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.M.T. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Boil. 2005, 15, 367–373. [Google Scholar] [CrossRef]
- Lukong, K.E.; Chang, K.-W.; Khandjian, E.W.; Richard, S. RNA-binding proteins in human genetic disease. Trends Genet. 2008, 24, 416–425. [Google Scholar] [CrossRef]
- Wurth, L. Versatility of RNA-Binding Proteins in Cancer. Comp. Funct. Genom. 2012, 2012, 178525. [Google Scholar] [CrossRef]
- Hidalgo-Curtis, C.; Chase, A.; Drachenberg, M.; Roberts, M.W.; Finkelstein, J.Z.; Mould, S.; Oscier, D.; Cross, N.C.P.; Grand, F.H. The t(1;9)(p34;q34) and t(8;12)(p11;q15) fuse pre-mRNA processing proteinsSFPQ (PSF) andCPSF6 toABL andFGFR1. Genes, Chromosom. Cancer 2008, 47, 379–385. [Google Scholar] [CrossRef]
- Duhoux, F.P.; Auger, N.; De Wilde, S.; Wittnebel, S.; Ameye, G.; Bahloula, K.; Berg, C.V.D.; Libouton, J.-M.; Saussoy, P.; Grand, F.H.; et al. The t(1;9)(p34;q34) fusing ABL1 with SFPQ, a pre-mRNA processing gene, is recurrent in acute lymphoblastic leukemias. Leuk. Res. 2011, 35, e114–e117. [Google Scholar] [CrossRef]
- Saleki, R.; Christensen, T.; Liu, W.; Wang, X.; Chen, Q.C.; Aakre, M.; Gomes, N.M.V.; Alexiev, B.A.; Schappert, J.; Baer, M.R.; et al. A novel TTC40-MSI2 fusion in de novo acute myeloid leukemia with an unbalanced 10;17 translocation. Leuk. Lymphoma 2014, 56, 1137–1139. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.-L.; Mercher, T.; Tyner, J.W.; Goss, V.L.; Walters, D.K.; Cornejo, M.G.; Reeves, C.; Popova, L.; Lee, K.; Heinrich, M.C.; et al. A novel fusion of RBM6 to CSF1R in acute megakaryoblastic leukemia. Blood 2007, 110, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Huang, X.-J.; Zhu, H.-H. Identification of a novel CPSF6-RARG fusion transcript in acute myeloid leukemia resembling acute promyelocytic leukemia. Leukemia 2018, 32, 2285–2287. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wen, L.; Yuan, H.; Wang, Y.; Yao, L.; Xu, Y.; Cen, J.; Ruan, C.; Wu, D.; Chen, S. Identification of novel recurrent CPSF6-RARG fusions in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood 2018, 131, 1870–1873. [Google Scholar] [CrossRef]
- Narla, A.; Ebert, B.L. Translational medicine: Ribosomopathies. Blood 2011, 118, 4300–4301. [Google Scholar] [CrossRef]
- Calado, R.; Regal, J.A.; Hills, M.; Yewdell, W.T.; Dalmazzo, L.F.; Zago, M.A.; Lansdorp, P.M.; Hogge, N.; Chanock, S.J.; Estey, E.H.; et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2009, 106, 1187–1192. [Google Scholar] [CrossRef]
- Devlin, E.E.; Dacosta, L.; Mohandas, N.; Elliott, G.; Bodine, D.M. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood 2010, 116, 2826–2835. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.G.; Hoadley, K.A.; Triche, T.J.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Dvinge, H.; Kim, E.; Abdel-Wahab, O.; Bradley, R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 2016, 16, 413–430. [Google Scholar] [CrossRef]
- Wong, A.; Rasko, J.E.J.; Wong, J.J.-L. We skip to work: Alternative splicing in normal and malignant myelopoiesis. Leukemia 2018, 32, 1081–1093. [Google Scholar] [CrossRef]
- Lee, S.C.-W.; Abdel-Wahab, O. Therapeutic targeting of splicing in cancer. Nat. Med. 2016, 22, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.-M.; Ashi, M.O.; Srour, N.; Delpy, L.; Saulière, J. Mechanisms and Regulation of Nonsense-Mediated mRNA Decay and Nonsense-Associated Altered Splicing in Lymphocytes. Int. J. Mol. Sci. 2020, 21, 1335. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Chen, R. Understanding aberrant RNA splicing to facilitate cancer diagnosis and therapy. Oncogene 2019, 39, 2231–2242. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Lee, S.C. Mutations in spliceosome genes and therapeutic opportunities in myeloid malignancies. Genes, Chromosom. Cancer 2019, 58, 889–902. [Google Scholar] [CrossRef]
- Prieto, C.; Kharas, M.G. RNA Regulators in Leukemia and Lymphoma. Cold Spring Harb. Perspect. Med. 2019, 10, a034967. [Google Scholar] [CrossRef]
- Ianniello, Z.; Fatica, A. N6-Methyladenosine Role in Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2018, 19, 2345. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Weng, H.; Huang, H.; Li, Z.; Chen, J. RNA N6-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 2018, 28, 507–517. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Feng, X.; Wei, M.; Chen, J. Role of N6-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 2017, 48, 1–7. [Google Scholar] [CrossRef]
- Ianniello, Z.; Paiardini, A.; Fatica, A. N6-Methyladenosine (m6A): A Promising New Molecular Target in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 251. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Wu, J.; Liu, J.; Qin, Z.; Fan, H. N6-Methyladenosine: A Potential Breakthrough for Human Cancer. Mol. Ther. - Nucleic Acids 2020, 19, 804–813. [Google Scholar] [CrossRef]
- Hong, K. Emerging function of N6-methyladenosine in cancer. Oncol. Lett. 2018, 16, 5519–5524. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Searles, M.A.; Klug, A. Crystal structure of a zinc-finger–RNA complex reveals two modes of molecular recognition. Nature 2003, 426, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Feracci, M.; Foot, J.N.; Grellscheid, S.N.; Danilenko, M.; Stehle, R.; Gonchar, O.; Kang, H.-S.; Dalgliesh, C.; Meyer, N.H.; Liu, Y.; et al. Structural basis of RNA recognition and dimerization by the STAR proteins T-STAR and Sam68. Nat. Commun. 2016, 7, 10355. [Google Scholar] [CrossRef] [PubMed]
- Pontén, F.; Jirström, K.; Uhlén, M. The Human Protein Atlas—a tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 2012, 70, 2657–2675. [Google Scholar] [CrossRef]
- Wächter, K.; Köhn, M.; Stöhr, N.; Hüttelmaier, S. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Boil. Chem. 2013, 394, 1077–1090. [Google Scholar] [CrossRef]
- Nielsen, J.; Kristensen, M.A.; Willemoës, M.; Nielsen, F.C.; Christiansen, J. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: A cooperative mechanism providing RNP stability. Nucleic Acids Res. 2004, 32, 4368–4376. [Google Scholar] [CrossRef]
- Bell, J.; Turlapati, R.; Liu, T.; Schulte, J.H.; Hüttelmaier, S. IGF2BP1 Harbors Prognostic Significance by Gene Gain and Diverse Expression in Neuroblastoma. J. Clin. Oncol. 2015, 33, 1285–1293. [Google Scholar] [CrossRef]
- Stoskus, M.; Gineikiene, E.; Valceckiene, V.; Valatkaite, B.; Pileckyte, R.; Griskevicius, L. Identification of characteristic IGF2BP expression patterns in distinct B-ALL entities. Blood Cells, Mol. Dis. 2011, 46, 321–326. [Google Scholar] [CrossRef]
- Degrauwe, N.; Suvà, M.-L.; Janiszewska, M.; Riggi, N.; Stamenkovic, I. IMPs: An RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016, 30, 2459–2474. [Google Scholar] [CrossRef]
- Hattori, A.; Buac, K.; Ito, T. Regulation of Stem Cell Self-Renewal and Oncogenesis by RNA-Binding Proteins. Adv. Exp. Med. Biol. 2016, 907, 153–188. [Google Scholar] [CrossRef] [PubMed]
- Elcheva, I.; Wood, T.; Chiarolanzio, K.; Chim, B.; Wong, M.; Singh, V.; Gowda, C.P.; Lu, Q.; Hafner, M.; Dovat, S.; et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia 2019, 34, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Umeda, S.; Yamamoto, K.; Murayama, T.; Hidaka, M.; Kurata, M.; Ohshima, T.; Suzuki, S.; Sugawara, E.; Kawano, F.; Kitagawa, M. Prognostic significance of HOXB4 in de novo acute myeloid leukemia. Hematology 2012, 17, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Frech, M.; Teichler, S.; Feld, C.; Bouchard, C.; Berberich, H.; Sorg, K.; Mernberger, M.; Bullinger, L.; Bauer, U.-M.; Neubauer, A. MYB induces the expression of the oncogenic corepressor SKI in acute myeloid leukemia. Oncotarget 2018, 9, 22423–22435. [Google Scholar] [CrossRef]
- Gasparetto, M.; Smith, C.A. ALDHs in normal and malignant hematopoietic cells: Potential new avenues for treatment of AML and other blood cancers. Chem. Interactions 2017, 276, 46–51. [Google Scholar] [CrossRef]
- Mahapatra, L.; Andruska, N.; Mao, C.; Le, J.; Shapiro, D.J. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl. Oncol. 2017, 10, 818–827. [Google Scholar] [CrossRef]
- Stoskus, M.; Vaitkevičienė, G.; Eidukaite, A.; Griskevicius, L. ETV6/RUNX1 transcript is a target of RNA-binding protein IGF2BP1 in t(12;21)(p13;q22)-positive acute lymphoblastic leukemia. Blood Cells, Mol. Dis. 2016, 57, 30–34. [Google Scholar] [CrossRef]
- Stoskus, M.; Eidukaite, A.; Griskevicius, L. Defining the significance of IGF2BP1 overexpression in t(12;21)(p13;q22)-positive leukemia REH cells. Leuk. Res. 2016, 47, 16–21. [Google Scholar] [CrossRef]
- Guanizo, A.C.; Fernando, C.D.; Garama, D.J.; Gough, D.J. STAT3: A multifaceted oncoprotein. Growth Factors 2018, 36, 1–14. [Google Scholar] [CrossRef]
- Gu, G.; Sederberg, M.; Drachenberg, M.R.; South, S. IGF2BP1: A novel IGH translocation partner in B acute lymphoblastic leukemia. Cancer Genet. 2014, 207, 332–334. [Google Scholar] [CrossRef]
- He, X.; Li, W.; Liang, X.; Zhu, X.; Zhang, L.; Huang, Y.; Yu, T.; Li, S.; Chen, Z. IGF2BP2 Overexpression Indicates Poor Survival in Patients with Acute Myelocytic Leukemia. Cell. Physiol. Biochem. 2018, 51, 1945–1956. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.I.; Robson, S.C.; Chung, C.W.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Palanichamy, J.K.; Tran, T.M.; Howard, J.M.; Contreras, J.R.; Fernando, T.R.; Sterne-Weiler, T.; Katzman, S.; Toloue, M.; Yan, W.; Basso, G.; et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J. Clin. Investig. 2016, 126, 1495–1511. [Google Scholar] [CrossRef] [PubMed]
- Weidensdorfer, D.; Stöhr, N.; Baude, A.; Lederer, M.; Köhn, M.; Schierhorn, A.; Buchmeier, S.; Wahle, E.; Hüttelmaier, S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 2008, 15, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xue, X.; Zheng, L.; Bi, J.; Zhou, Y.; Zhi, K.; Gu, Y.; Fang, G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014, 281, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ouyang, M.; Rao, J.N.; Zou, T.; Xiao, L.; Chung, H.K.; Wu, J.; Donahue, J.M.; Gorospe, M.; Wang, J.Y. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol. Boil. Cell 2015, 26, 1797–1810. [Google Scholar] [CrossRef]
- Placke, T.; Faber, K.; Nonami, A.; Putwain, S.L.; Salih, H.R.; Heidel, F.H.; Krämer, A.; Root, D.E.; Barbie, D.A.; Krivtsov, A.V.; et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood 2014, 124, 13–23. [Google Scholar] [CrossRef]
- Chen, M.-T.; Dong, L.; Zhang, X.-H.; Yin, X.-L.; Ning, H.-M.; Shen, C.; Su, R.; Li, F.; Song, L.; Ma, Y.-N.; et al. ZFP36L1 promotes monocyte/macrophage differentiation by repressing CDK6. Sci. Rep. 2015, 5, 16229. [Google Scholar] [CrossRef]
- The UniProt Consortium; UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [CrossRef]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Nakamura, M.; Okano, H.; Blendy, J.A.; Montell, C. Musashi, a neural RNA-binding protein required for drosophila adult external sensory organ development. Neuron 1994, 13, 67–81. [Google Scholar] [CrossRef]
- Okano, H.; Imai, T.; Okabe, M. Musashi: A translational regulator of cell fate. J. Cell Sci. 2002, 115, 1355–1359. [Google Scholar] [PubMed]
- Kawahara, H.; Imai, T.; Imataka, H.; Tsujimoto, M.; Matsumoto, K.; Okano, H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Boil. 2008, 181, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Tokunaga, A.; Yoshida, T.; Hashimoto, M.; Mikoshiba, K.; Weinmaster, G.; Nakafuku, M.; Okano, H. The Neural RNA-Binding Protein Musashi1 Translationally Regulates Mammalian numb Gene Expression by Interacting with Its mRNA. Mol. Cell. Boil. 2001, 21, 3888–3900. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Yin, Y.; Yuan, H.; Sakamaki, T.; Okano, H.; Glazer, R. Musashi1 Modulates Mammary Progenitor Cell Expansion through Proliferin-Mediated Activation of the Wnt and Notch Pathways. Mol. Cell. Boil. 2008, 28, 3589–3599. [Google Scholar] [CrossRef]
- Nahas, G.R.; Murthy, R.G.; Patel, S.; Ganta, T.; Greco, S.J.; Rameshwar, P. The RNA-binding protein Musashi 1 stabilizes the oncotachykinin 1 mRNA in breast cancer cells to promote cell growth. FASEB J. 2015, 30, 149–159. [Google Scholar] [CrossRef]
- Yi, C.; Li, G.; Ivanov, D.N.; Wang, Z.; Velasco, M.X.; Hernandez, G.; Kaundal, S.; Villarreal, J.; Gupta, Y.K.; Qiao, M.; et al. Luteolin inhibits Musashi1 binding to RNA and disrupts cancer phenotypes in glioblastoma cells. RNA Boil. 2018, 15, 1420–1432. [Google Scholar] [CrossRef]
- Kharas, M.G.; Lengner, C.J.; Al-Shahrour, F.; Bullinger, L.; Ball, B.; Zaidi, S.; Morgan, K.; Tam, W.; Paktinat, M.; Okabe, R.; et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010, 16, 903–908. [Google Scholar] [CrossRef]
- Hope, K.J.; Cellot, S.; Ting, S.B.; Macrae, T.; Mayotte, N.; Iscove, N.N.; Sauvageau, G. An RNAi Screen Identifies Msi2 and Prox1 as Having Opposite Roles in the Regulation of Hematopoietic Stem Cell Activity. Cell Stem Cell 2010, 7, 101–113. [Google Scholar] [CrossRef]
- Ito, T.; Kwon, H.Y.; Zimdahl, B.; Congdon, K.L.; Blum, J.; Lento, W.E.; Zhao, C.; Lagoo, A.; Gerrard, G.; Foroni, L.; et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010, 466, 765–768. [Google Scholar] [CrossRef]
- Lan, L.; Xing, M.; Douglas, J.T.; Gao, P.; Hanzlik, R.P.; Xu, L. Human oncoprotein Musashi-2 N-terminal RNA recognition motif backbone assignment and identification of RNA-binding pocket. Oncotarget 2017, 8, 106587–106597. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Winschel, C.; Sonntag, A.-K.; Damm, F.; Wagner, K.; Chaturvedi, A.; Göhring, G.; Schlegelberger, B.; Lübbert, M.; Fiedler, W.; et al. Prognostic significance of expression levels of stem cell regulators MSI2 and NUMB in acute myeloid leukemia. Ann. Hematol. 2012, 92, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Wang, Y.; Chen, B.; Qian, W.; Meng, H.; Tong, H.; Chen, F.; Ma, Q.; Ni, W.; Chen, S.; et al. High expression of Musashi-2 indicates poor prognosis in adult B-cell acute lymphoblastic leukemia. Leuk. Res. 2013, 37, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Aly, R.M.; Ghazy, H.F. Prognostic significance ofMSI2predicts unfavorable outcome in adult B-acute lymphoblastic leukemia. Int. J. Lab. Hematol. 2014, 37, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.Z.; Jia, M.; Luo, Z.B.; Cheng, Y.P.; Xu, X.J.; Zhang, J.Y.; Li, S.S.; Tang, Y.M. Prognostic significance of the Musashi-2 (MSI2) gene in childhood acute lymphoblastic leukemia. Neoplasma 2016, 63, 150–157. [Google Scholar] [CrossRef]
- Byers, R.; Currie, T.; Tholouli, E.; Rodig, S.J.; Kutok, J.L. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood 2011, 118, 2857–2867. [Google Scholar] [CrossRef]
- Pereira, J.K.N.; Traina, F.; Machado-Neto, J.A.; Duarte, A.D.S.S.; Lopes, M.R.; Saad, S.T.O.; Favaro, P. Distinct expression profiles of MSI2 and NUMB genes in myelodysplastic syndromes and acute myeloid leukemia patients. Leuk. Res. 2012, 36, 1300–1303. [Google Scholar] [CrossRef]
- Pece, S.; Confalonieri, S.; Romano, P.R.; Di Fiore, P.P. NUMB-ing down cancer by more than just a NOTCH. Biochim. et Biophys. Acta (BBA) - Rev. Cancer 2011, 1815, 26–43. [Google Scholar] [CrossRef]
- McCubrey, J.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.D.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. et Biophys. Acta (BBA) - Bioenerg. 2006, 1773, 1263–1284. [Google Scholar] [CrossRef]
- Han, Y.; Ye, A.; Zhang, Y.; Cai, Z.; Wang, W.; Sun, L.; Jiang, S.; Wu, J.; Yu, K.; Zhang, S. Musashi-2 Silencing Exerts Potent Activity against Acute Myeloid Leukemia and Enhances Chemosensitivity to Daunorubicin. PLoS ONE 2015, 10, e0136484. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, S.; Wang, J.; Chen, S.; Quan, J.; Xian, J.; Zhang, S.S.; He, J.; Zhang, L. Musashi2 modulates K562 leukemic cell proliferation and apoptosis involving the MAPK pathway. Exp. Cell Res. 2014, 320, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Martelli, A.; Evangelisti, C.; Chiarini, F.; McCubrey, J. The phosphatidylinositol 3-kinase/Akt/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget 2010, 1, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Kharas, M.G.; Okabe, R.; Ganis, J.J.; Gozo, M.; Khandan, T.; Paktinat, M.; Gilliland, D.G.; Gritsman, K. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood 2010, 115, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Lv, H.; Wu, F.; Wang, C.; Li, T.; Lv, G.; Tang, L.; Guo, L.; Tang, S.; Cao, D.; et al. Musashi 2 contributes to the stemness and chemoresistance of liver cancer stem cells via LIN28A activation. Cancer Lett. 2017, 384, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Dong, M.; Chen, C.; Wang, Z.; Li, Y.; Wang, K.; Li, Y.; Zhou, J. Cooperation of Musashi-2, Numb, MDM2, and P53 in drug resistance and malignant biology of pancreatic cancer. FASEB J. 2017, 31, 2429–2438. [Google Scholar] [CrossRef]
- Minuesa, G.; Albanese, S.; Xie, W.; Kazansky, Y.; Worroll, D.; Chow, A.; Schurer, A.; Park, S.-M.; Rotsides, C.Z.; Taggart, J.; et al. Small-molecule targeting of MUSASHI RNA-binding activity in acute myeloid leukemia. Nat. Commun. 2019, 10, 2691. [Google Scholar] [CrossRef]
- Ostareck-Lederer, A.; Ostareck, D.H.; Hentze, M.W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 1998, 23, 409–411. [Google Scholar] [CrossRef]
- Messias, A.; Harnisch, C.; Ostareck-Lederer, A.; Sattler, M.; Ostareck, D.H. The DICE-binding Activity of KH Domain 3 of hnRNP K Is Affected by c-Src-mediated Tyrosine Phosphorylation. J. Mol. Boil. 2006, 361, 470–481. [Google Scholar] [CrossRef]
- Backe, P.H.; Messias, A.; Ravelli, R.B.G.; Sattler, M.; Cusack, S. X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids. Structure 2005, 13, 1055–1067. [Google Scholar] [CrossRef]
- Backe, P.H.; Ravelli, R.B.G.; Garman, E.; Cusack, S. Crystallization, microPIXE and preliminary crystallographic analysis of the complex between the third KH domain of hnRNP K and single-stranded DNA. Acta Crystallogr. Sect. D Boil. Crystallogr. 2004, 60, 784–787. [Google Scholar] [CrossRef]
- Braddock, D.T.; Baber, J.L.; Levens, D.; Clore, G.M. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002, 21, 3476–3485. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, A.V.; Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002, 8, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Paziewska, A.; Wyrwicz, L.S.; Ostrowski, J. The binding activity of yeast RNAs to yeast Hek2p and mammalian hnRNP K proteins, determined using the three-hybrid system. Cell. Mol. Boil. Lett. 2005, 10, 227–235. [Google Scholar]
- Michael, W.M.; Choi, M.; Dreyfuss, G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell 1995, 83, 415–422. [Google Scholar] [CrossRef]
- Michael, W.; Eder, P.S.; Dreyfuss, G. The K nuclear shuttling domain: A novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997, 16, 3587–3598. [Google Scholar] [CrossRef] [PubMed]
- Matunis, M.; Matunis, M.J.; Dreyfuss, G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J. Cell Boil. 1992, 116, 257–269. [Google Scholar] [CrossRef]
- Dejgaard, K.; Leffers, H. Characterisation of the Nucleic-Acid-Binding Activity of KH Domains Different Properties of Different Domains. JBIC J. Boil. Inorg. Chem. 1996, 241, 425–431. [Google Scholar] [CrossRef]
- Hutchins, E.J.; Belrose, J.L.; Szaro, B.G. A novel role for the nuclear localization signal in regulating hnRNP K protein stability in vivo. Biochem. Biophys. Res. Commun. 2016, 478, 772–776. [Google Scholar] [CrossRef]
- Lu, J.; Gao, F. Role and molecular mechanism of heterogeneous nuclear ribonucleoprotein K in tumor development and progression. Biomed. Rep. 2016, 4, 657–663. [Google Scholar] [CrossRef]
- Bomsztyk, K.; Denisenko, O.; Ostrowski, J. hnRNP K: One protein multiple processes. BioEssays 2004, 26, 629–638. [Google Scholar] [CrossRef]
- Carpenter, B.; McKay, M.; Dundas, S.R.; Lawrie, L.C.; Telfer, C.; I Murray, G. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br. J. Cancer 2006, 95, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Shen, A.; Shañas, R.; Bhattacharyya, A.; Lian, F.; Hostetter, G.; Shi, J. Higher expression of the heterogeneous nuclear ribonucleoprotein k in melanoma. Ann. Surg. Oncol. 2010, 17, 2619–2627. [Google Scholar] [CrossRef] [PubMed]
- Daskalaki, W.; Wardelmann, E.; Port, M.; Stock, K.; Steinestel, J.; Huss, S.; Sperveslage, J.; Steinestel, K.; Eder, S. Expression levels of hnRNP K and p21WAF1/CIP1 are associated with resistance to radiochemotherapy independent of p53 pathway activation in rectal adenocarcinoma. Int. J. Mol. Med. 2018, 42, 3269–3277. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, M.; Hornbaker, M.J.; Zhang, X.; Hu, P.; Bueso-Ramos, C.; Post, S.M. Aberrant hnRNP K expression: All roads lead to cancer. Cell Cycle 2016, 15, 1552–1557. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, B.; Mackay, C.; Alnabulsi, A.; Mackay, M.; Telfer, C.; Melvin, W.T.; Murray, G.I. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim. et Biophys. Acta (BBA) - Bioenerg. 2006, 1765, 85–100. [Google Scholar] [CrossRef]
- Shnyreva, M.; Schullery, D.S.; Suzuki, H.; Higaki, Y.; Bomsztyk, K. Interaction of Two Multifunctional Proteins. J. Boil. Chem. 2000, 275, 15498–15503. [Google Scholar] [CrossRef]
- Ritchie, S.A. Identification of the SRC pyrimidine-binding protein (SPy) as hnRNP K: Implications in the regulation of SRC1A transcription. Nucleic Acids Res. 2003, 31, 1502–1513. [Google Scholar] [CrossRef]
- Michelotti, E.F.; Michelotti, G.; I Aronsohn, A.; Levens, D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Boil. 1996, 16, 2350–2360. [Google Scholar] [CrossRef]
- Evans, J.R.; A Mitchell, S.; A Spriggs, K.; Ostrowski, J.; Bomsztyk, K.; Ostarek, D.; E Willis, A. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene 2003, 22, 8012–8020. [Google Scholar] [CrossRef]
- Ostareck, D.H.; Ostareck-Lederer, A.; Wilm, M.; Thiele, B.J.; Mann, M.; Hentze, M.W. mRNA Silencing in Erythroid Differentiation: hnRNP K and hnRNP E1 Regulate 15-Lipoxygenase Translation from the 3′ End. Cell 1997, 89, 597–606. [Google Scholar] [CrossRef]
- Cao, W.; Razanau, A.; Feng, D.; Lobo, V.G.; Xie, J. Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic Acids Res. 2012, 40, 8059–8071. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.-Z.; Liu, J.-X.; Li, C.-F.; Ma, R.; Jie, J.-Z. hnRNPK promotes gastric tumorigenesis through regulating CD44E alternative splicing. Cancer Cell Int. 2019, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Burda, P.; Laslo, P.; Stopka, T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia 2010, 24, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Kastner, P.; Chan, S. PU.1: A crucial and versatile player in hematopoiesis and leukemia. Int. J. Biochem. Cell Boil. 2008, 40, 22–27. [Google Scholar] [CrossRef]
- Nika, E.; Brugnoli, F.; Piazzi, M.; Lambertini, E.; Grassilli, S.; Bavelloni, A.; Piva, R.; Capitani, S.; Bertagnolo, V. hnRNP K in PU.1-containing complexes recruited at the CD11b promoter: A distinct role in modulating granulocytic and monocytic differentiation of AML-derived cells. Biochem. J. 2014, 463, 115–122. [Google Scholar] [CrossRef]
- Gallardo, M.; Lee, H.J.; Zhang, X.; Bueso-Ramos, C.; Pageon, L.R.; McArthur, M.; Multani, A.; Nazha, A.; Manshouri, T.; Parker-Thornburg, J.; et al. hnRNP K Is a Haploinsufficient Tumor Suppressor that Regulates Proliferation and Differentiation Programs in Hematologic Malignancies. Cancer Cell 2015, 28, 486–499. [Google Scholar] [CrossRef]
- Enge, M.; Bao, W.; Hedström, E.; Jackson, S.; Moumen, A.; Selivanova, G. MDM2-Dependent Downregulation of p21 and hnRNP K Provides a Switch between Apoptosis and Growth Arrest Induced by Pharmacologically Activated p53. Cancer Cell 2009, 15, 171–183. [Google Scholar] [CrossRef]
- Moumen, A.; Masterson, P.; O’Connor, M.J.; Jackson, S. hnRNP K: An HDM2 Target and Transcriptional Coactivator of p53 in Response to DNA Damage. Cell 2005, 123, 1065–1078. [Google Scholar] [CrossRef]
- Fukuda, T.; Naiki, T.; Saito, M.; Irie, K. hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions. Genes Cells 2009, 14, 113–128. [Google Scholar] [CrossRef]
- Notari, M.; Neviani, P.; Santhanam, R.; Blaser, B.W.; Chang, J.-S.; Galietta, A.; Willis, A.E.; Roy, D.C.; Caligiuri, M.A.; Marcucci, G.; et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood 2006, 107, 2507–2516. [Google Scholar] [CrossRef]
- Ceballos, E.; Muñoz-Alonso, M.J.; Berwanger, B.; Acosta, J.C.; Hernàndez, R.; Krause, M.; Hartmann, O.; Eilers, M.; León, J. Inhibitory effect of c-Myc on p53-induced apoptosis in leukemia cells. Microarray analysis reveals defective induction of p53 target genes and upregulation of chaperone genes. Oncogene 2005, 24, 4559–4571. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Lee, M.H.; Park, J.H.; Kang, S.H.; Yoo, H.M.; Ka, S.H.; Oh, Y.M.; Jeon, Y.J.; Chung, C.H. SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 2012, 31, 4441–4452. [Google Scholar] [CrossRef] [PubMed]
- Créancier, L.; Prats, H.; Zanibellato, C.; Amalric, F.; Bugler, B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol. Boil. Cell 1993, 4, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Köhn, M.; Misiak, D.; Bäumer, N.; Cui, C.; Göllner, S.; et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nature 2017, 19, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Scherl, A.; Couté, Y.; Déon, C.; Callé, A.; Kindbeiter, K.; Sanchez, J.-C.; Greco, A.; Hochstrasser, D.; Diaz, J.-J. Functional Proteomic Analysis of Human Nucleolus. Mol. Boil. Cell 2002, 13, 4100–4109. [Google Scholar] [CrossRef] [PubMed]
- Cong, R.; Das, S.; Ugrinova, I.; Kumar, S.; Mongelard, F.; Wong, J.; Bouvet, P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012, 40, 9441–9454. [Google Scholar] [CrossRef]
- Qiu, W.; Zhou, F.; Zhang, Q.; Sun, X.; Shi, X.; Liang, Y.; Wang, X.; Yue, L. Overexpression of nucleolin and different expression sites both related to the prognosis of gastric cancer. APMIS 2013, 121, 919–925. [Google Scholar] [CrossRef]
- Wolfson, E.; Solomon, S.; Schmukler, E.; Goldshmit, Y.; Pinkas-Kramarski, R. Nucleolin and ErbB2 inhibition reduces tumorigenicity of ErbB2-positive breast cancer. Cell Death Dis. 2018, 9, 47. [Google Scholar] [CrossRef]
- Otake, Y.; Soundararajan, S.; Sengupta, T.K.; Kio, E.A.; Smith, J.C.; Pineda-Roman, M.; Stuart, R.; Spicer, E.K.; Fernandes, D.J. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood 2006, 109, 3069–3075. [Google Scholar] [CrossRef]
- Shen, N.; Yan, F.; Pang, J.; Wu, L.-C.; Al-Kali, A.; Litzow, M.R.; Liu, S. A nucleolin-DNMT1 regulatory axis in acute myeloid leukemogenesis. Oncotarget 2014, 5, 5494–5509. [Google Scholar] [CrossRef]
- Marcel, V.; Catez, F.; Berger, C.M.; Perrial, E.; Pleşa, A.; Thomas, X.; Mattei, E.; Hayette, S.; Saintigny, P.; Bouvet, P.; et al. Expression Profiling of Ribosome Biogenesis Factors Reveals Nucleolin as a Novel Potential Marker to Predict Outcome in AML Patients. PLoS ONE 2017, 12, e0170160. [Google Scholar] [CrossRef]
- Takagi, M.; Absalon, M.J.; McLure, K.G.; Kastan, M.B. Regulation of p53 Translation and Induction after DNA Damage by Ribosomal Protein L26 and Nucleolin. Cell 2005, 123, 49–63. [Google Scholar] [CrossRef]
- Ishimaru, D.; Ramalingam, S.; Sengupta, T.K.; Bandyopadhyay, S.; Dellis, S.; Tholanikunnel, B.G.; Fernandes, D.J.; Spicer, E.K. Regulation of Bcl-2 Expression by HuR in HL60 Leukemia Cells and A431 Carcinoma Cells. Mol. Cancer Res. 2009, 7, 1354–1366. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, D.; Zuraw, L.; Ramalingam, S.; Sengupta, T.K.; Bandyopadhyay, S.; Reuben, A.; Fernandes, D.J.; Spicer, E.K. Mechanism of Regulation of bcl-2 mRNA by Nucleolin and A+U-rich Element-binding Factor 1 (AUF1). J. Boil. Chem. 2010, 285, 27182–27191. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, T.K.; Bocharova, O.V.; Makarava, N.; Breydo, L.; Anderson, M.; Salnikov, V.V.; Baskakov, I.V. Identification of Nucleolin as an AU-rich Element Binding Protein Involved inbcl-2mRNA Stabilization. J. Boil. Chem. 2003, 279, 10855–10863. [Google Scholar] [CrossRef] [PubMed]
- Willimott, S.; Wagner, S.D. Post-transcriptional and post-translational regulation of Bcl2. Biochem. Soc. Trans. 2010, 38, 1571–1575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, H.; Jiang, B.; Liang, P.; Liu, M.; Deng, G.; Xiao, X. Nucleolin/C23 is a negative regulator of hydrogen peroxide-induced apoptosis in HUVECs. Cell Stress Chaperon- 2009, 15, 249–257. [Google Scholar] [CrossRef]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic Nucleophosmin in Acute Myelogenous Leukemia with a Normal Karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef]
- Šašinková, M.; Holoubek, A.; Otevřelová, P.; Kuželová, K.; Brodská, B. AML-associated mutation of nucleophosmin compromises its interaction with nucleolin. Int. J. Biochem. Cell Boil. 2018, 103, 65–73. [Google Scholar] [CrossRef]
- Martelli, M.P.; Gionfriddo, I.; Mezzasoma, F.; Milano, F.; Pierangeli, S.; Mulas, F.; Pacini, R.; Tabarrini, A.; Pettirossi, V.; Rossi, R.; et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood 2015, 125, 3455–3465. [Google Scholar] [CrossRef]
- Grant, S. ATRA and ATO team up against NPM1. Blood 2015, 125, 3369–3371. [Google Scholar] [CrossRef] [PubMed]
- Vasaturo, M.; Cotugno, R.; Fiengo, L.; Vinegoni, C.; Piaz, F.D.; De Tommasi, N. The anti-tumor diterpene oridonin is a direct inhibitor of Nucleolin in cancer cells. Sci. Rep. 2018, 8, 16735. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Buckler, A.J.; Glaser, T.; Call, K.M.; Pelletier, J.; Sohn, R.L.; Douglass, E.C.; Housman, D.E. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms’ tumor. Cell 1990, 61, 1257–1269. [Google Scholar] [CrossRef]
- Kim, M.K.-H.; McGarry, T.J.; Broin, P.Ó.; Flatow, J.M.; Golden, A.A.-J.; Licht, J.D. An integrated genome screen identifies the Wnt signaling pathway as a major target of WT1. Proc. Natl. Acad. Sci. USA 2009, 106, 11154–11159. [Google Scholar] [CrossRef] [PubMed]
- A Davies, J.; Ladomery, M.; Hohenstein, P.; Michael, L.; Shafe, A.; Spraggon, L.; Hastie, N.D. Development of an siRNA-based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumour suppressor is required for nephron differentiation. Hum. Mol. Genet. 2003, 13, 235–246. [Google Scholar] [CrossRef]
- Ladomery, M.; Dellaire, G. Multifunctional zinc finger proteins in development and disease. Ann. Hum. Genet. 2002, 66, 331–342. [Google Scholar] [CrossRef]
- Wilm, B.; Muñoz-Chápuli, R. The Role of WT1 in Embryonic Development and Normal Organ Homeostasis. Adv. Struct. Saf. Stud. 2016, 1467, 23–39. [Google Scholar] [CrossRef]
- Huff, V. Wilms’ tumours: About tumour suppressor genes, an oncogene and a chameleon gene. Nat. Rev. Cancer 2011, 11, 111–121. [Google Scholar] [CrossRef]
- Fraizer, G.; Leahy, R.; Priyadarshini, S.; Graham, K.; DeLacerda, J.; Diaz, M. Suppression of prostate tumor cell growth in vivo by WT1, the Wilms’ tumor suppressor gene. Int. J. Oncol. 2004, 24, 461–471. [Google Scholar] [CrossRef]
- Haber, D.; Park, S.; Maheswaran, S.; Englert, C.; Re, G.; Hazen-Martin, D.; Sens, D.; Garvín, A. WT1-mediated growth suppression of Wilms tumor cells expressing a WT1 splicing variant. Science 1993, 262, 2057–2059. [Google Scholar] [CrossRef]
- McMaster, M.L.; Gessler, M.; Stanbridge, E.J.; E Weissman, B. WT1 expression alters tumorigenicity of the G401 kidney-derived cell line. Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1995, 6, 1609–1617. [Google Scholar]
- Luo, X.N.; Reddy, J.C.; Yeyati, P.L.; Idris, A.H.; Hosono, S.; A Haber, D.; Licht, J.D.; Atweh, G.F. The tumor suppressor gene WT1 inhibits ras-mediated transformation. Oncogene 1995, 11, 743–750. [Google Scholar] [PubMed]
- Yi-Ning, Y.; Xiao-Rui, W.; Chu-Xian, Z.; Chun, W.; You-Wen, Q. Prognostic significance of diagnosed WT1 level in acute myeloid leukemia: A meta-analysis. Ann. Hematol. 2015, 94, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Malagola, M.; Skert, C.; Ruggeri, G.; Turra, A.; Ribolla, R.; Cancelli, V.; Cattina, F.; Alghisi, E.; Bernardi, S.; Perucca, S.; et al. Peripheral Blood WT1 Expression Predicts Relapse in AML Patients Undergoing Allogeneic Stem Cell Transplantation. BioMed Res. Int. 2014, 2014, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ido, K.; Nakamae, M.; Koh, H.; Okamura, H.; Nanno, S.; Nishimoto, M.; Takeoka, Y.; Hirose, A.; Nakashima, Y.; Hashimoto, Y.; et al. The Proportional Relationship Between Pretransplant WT1 mRNA Levels and Risk of Mortality After Allogeneic Hematopoietic Cell Transplantation in Acute Myeloid Leukemia Not in Remission. Transplantation 2019, 103, 2201–2210. [Google Scholar] [CrossRef]
- Chapuis, A.G.; Egan, D.N.; Bar, M.; Schmitt, T.M.; McAfee, M.S.; Paulson, K.G.; Voillet, V.; Gottardo, R.; Ragnarsson, G.B.; Bleakley, M.; et al. T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat. Med. 2019, 25, 1064–1072. [Google Scholar] [CrossRef]
- Hastie, N.D. Life, Sex, and WT1 Isoforms— Three Amino Acids Can Make All the Difference. Cell 2001, 106, 391–394. [Google Scholar] [CrossRef]
- Ullmark, T.; Montano, G.; Gullberg, U. DNA and RNA binding by the Wilms’ tumour gene 1 (WT1) protein +KTS and ?KTS isoforms-From initial observations to recent global genomic analyses. Eur. J. Haematol. 2018, 100, 229–240. [Google Scholar] [CrossRef]
- Cesaro, E.; Montano, G.; Rosati, A.; Crescitelli, R.; Izzo, P.; Turco, M.C.; Costanzo, P. WT1 protein is a transcriptional activator of the antiapoptotic bag3 gene. Leukemia 2010, 24, 1204–1206. [Google Scholar] [CrossRef][Green Version]
- Rosati, A.; Graziano, V.; De Laurenzi, V.; Pascale, M.; Turco, M.C. BAG3: A multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011, 2, e141. [Google Scholar] [CrossRef]
- Rosati, A.; Ammirante, M.; Gentilella, A.; Basile, A.; Festa, M.; Pascale, M.; Marzullo, L.; Belisario, M.A.; Alessandra, T.; Franceschelli, S.; et al. Apoptosis inhibition in cancer cells: A novel molecular pathway that involves BAG3 protein. Int. J. Biochem. Cell Boil. 2007, 39, 1337–1342. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Thomas, D.; Yu, L.; Gentles, A.J.; Jung, N.; Corces, M.R.; Chan, S.M.; Reinisch, A.; Feinberg, A.P.; Dill, D.L.; et al. Mutant WT1 is associated with DNA hypermethylation of PRC2 targets in AML and responds to EZH2 inhibition. Blood 2015, 125, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-P.; Xiao, M.; Chen, X.; Chen, L.; Xu, Y.; Lv, L.; Wang, P.; Yang, H.; Ma, S.; Lin, H.; et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell 2015, 57, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.E.; Abdel-Wahab, O.; Lu, C.; Ward, P.; Patel, J.; Shih, A.; Li, Y.; Bhagwat, N.; VasanthaKumar, A.; Fernandez, H.F.; et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010, 18, 553–567. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Lugthart, S.; Li, Y.; Erpelinck-Verschueren, C.; Deng, X.; Christos, P.J.; Schifano, E.; Booth, J.; Van Putten, W.; Skrabanek, L.; et al. DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia. Cancer Cell 2010, 17, 13–27. [Google Scholar] [CrossRef]
- Lu, K.-H.; Li, W.; Liu, X.-H.; Sun, M.; Zhang, M.-L.; Wu, W.-Q.; Xie, W.-P.; Hou, Y. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013, 13, 461. [Google Scholar] [CrossRef]
- Lyu, Y.; Lou, J.; Yang, Y.; Feng, J.; Hao, Y.; Huang, S.; Yin, L.; Xu, J.; Huang, D.; Ma, B.; et al. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia 2017, 31, 2543–2551. [Google Scholar] [CrossRef]
- Pronier, E.; Bowman, R.L.; Ahn, J.; Glass, J.; Kandoth, C.; Merlinsky, T.R.; Whitfield, J.T.; Durham, B.H.; Gruet, A.; Somasundara, A.V.H.; et al. Genetic and epigenetic evolution as a contributor to WT1-mutant leukemogenesis. Blood 2018, 132, 1265–1278. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A.; Chávez-Valencia, V. FLT3–ITD and its current role in acute myeloid leukaemia. Med Oncol. 2017, 34, 179. [Google Scholar] [CrossRef]
- Annesley, C.E.; Rabik, C.; Duffield, A.S.; Rau, R.E.; Magoon, D.; Li, L.; Huff, V.; Small, D.; Loeb, D.M.; Brown, P. Knock-in of the Wt1 R394W mutation causes MDS and cooperates with Flt3/ITD to drive aggressive myeloid neoplasms in mice. Oncotarget 2018, 9, 35313–35326. [Google Scholar] [CrossRef]
- Bourkoula, K.; Englert, C.; Giaisi, M.; Köhler, R.; Krammer, P.H.; Li-Weber, M. The Wilms’ tumor suppressor WT1 enhances CD95L expression and promotes activation-induced cell death in leukemic T cells. Int. J. Cancer 2013, 134, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Golks, A.; Becker, M.; Muller, W.; Frey, C.R.; Novak, R.; Melamed, R.; Kiefer, F.; Krammer, P.H.; Arnold, R. Caspase-cleaved HPK1 induces CD95L-independent activation-induced cell death in T and B lymphocytes. Blood 2007, 110, 3968–3977. [Google Scholar] [CrossRef] [PubMed]
- Baou, M.; Jewell, A.; Murphy, J.J. TIS11 Family Proteins and Their Roles in Posttranscriptional Gene Regulation. J. Biomed. Biotechnol. 2009, 2009, 634520. [Google Scholar] [CrossRef] [PubMed]
- Blackshear, P.J.; Phillips, R.S.; Ghosh, S.; Ramos, S.V.; Richfield, E.K.; Lai, W.S. Zfp36l3, a Rodent X Chromosome Gene Encoding a Placenta-Specific Member of the Tristetraprolin Family of CCCH Tandem Zinc Finger Proteins. Boil. Reprod. 2005, 73, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Blackshear, P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002, 30, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Blackshear, P.J.; Lai, W.S.; Kennington, E.A.; Brewer, G.; Wilson, G.; Guan, X.; Zhou, P.; Barrowman, J.; Wang, W.; Zhang, Y.; et al. Characteristics of the Interaction of a Synthetic Human Tristetraprolin Tandem Zinc Finger Peptide with AU-rich Element-containing RNA Substrates. J. Boil. Chem. 2003, 278, 19947–19955. [Google Scholar] [CrossRef]
- Stumpo, D.J.; Broxmeyer, H.E.; Ward, T.; Cooper, S.; Hangoc, G.; Chung, Y.J.; Shelley, W.C.; Richfield, E.K.; Ray, M.K.; Yoder, M.C.; et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 2009, 114, 2401–2410. [Google Scholar] [CrossRef]
- Hodson, D.J.; Janas, M.L.; Galloway, A.; Bell, S.E.; Andrews, S.; Li, C.M.; Pannell, R.; Siebel, C.W.; Macdonald, H.R.; De Keersmaecker, K.; et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat. Immunol. 2010, 11, 717–724. [Google Scholar] [CrossRef]
- A Johnson, B.; Geha, M.; Blackwell, T.K. Similar but distinct effects of the tristetraprolin/TIS11 immediate-early proteins on cell survival. Oncogene 2000, 19, 1657–1664. [Google Scholar] [CrossRef]
- Iwanaga, E.; Nanri, T.; Mitsuya, H.; Asou, N. Mutation in the RNA binding protein TIS11D/ZFP36L2 is associated with the pathogenesis of acute leukemia. Int. J. Oncol. 2011, 38, 25–31. [Google Scholar] [CrossRef]
- Ciais, D.; Cherradi, N.; Bailly, S.; Grenier, E.; Berra, E.; Pouysségur, J.; Lamarre, J.; Feige, J.-J. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene 2004, 23, 8673–8680. [Google Scholar] [CrossRef] [PubMed]
- Zekavati, A.; Nasir, A.; Alcaraz, A.; Aldrovandi, M.; Marsh, P.; Norton, J.D.; Murphy, J.J. Post-Transcriptional Regulation of BCL2 mRNA by the RNA-Binding Protein ZFP36L1 in Malignant B Cells. PLoS ONE 2014, 9, e102625. [Google Scholar] [CrossRef] [PubMed]
- Kampen, K.R.; Ter Elst, A.; De Bont, E.S.J.M. Vascular endothelial growth factor signaling in acute myeloid leukemia. Cell. Mol. Life Sci. 2012, 70, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Kale, J.; Osterlund, E.J.; Andrews, D.W. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2017, 25, 65–80. [Google Scholar] [CrossRef]
- Schmidlin, M.; Lu, M.; A Leuenberger, S.; Stoecklin, G.; Mallaun, M.; Gross, B.; Gherzi, R.; Hess, D.; A Hemmings, B.; Moroni, C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004, 23, 4760–4769. [Google Scholar] [CrossRef]
- Benjamin, D.; Schmidlin, M.; Min, L.; Gross, B.; Moroni, C. BRF1 Protein Turnover and mRNA Decay Activity Are Regulated by Protein Kinase B at the Same Phosphorylation Sites. Mol. Cell. Boil. 2006, 26, 9497–9507. [Google Scholar] [CrossRef]
- Maitra, S.; Chou, C.-F.; Luber, C.A.; Lee, K.-Y.; Mann, M.; Chen, C.-Y. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA 2008, 14, 950–959. [Google Scholar] [CrossRef]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef]
- Meyerson, M.; Harlow, E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Boil. 1994, 14, 2077–2086. [Google Scholar] [CrossRef]
- Brennan, S.E.; Kuwano, Y.; Alkharouf, N.; Blackshear, P.J.; Gorospe, M.; Wilson, G. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009, 69, 5168–5176. [Google Scholar] [CrossRef]
- Ma, Z.; Morris, S.W.; Valentine, V.; Martin, L.; Herbrick, J.-A.; Cui, X.; Bouman, D.; Li, Y.; Mehta, P.K.; Nizetic, D.; et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 2001, 28, 220–221. [Google Scholar] [CrossRef] [PubMed]
- Mercher, T.; Coniat, M.B.-L.; Monni, R.; Mauchauffé, M.; Nguyen-Khac, F.; Gressin, L.; Mugneret, F.; Leblanc, T.; Dastugue, N.; Berger, R.; et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 2001, 98, 5776–5779. [Google Scholar] [CrossRef] [PubMed]
- Dastugue, N.; Lafage-Pochitaloff, M.; Pagès, M.-P.; Radford, I.; Bastard, C.; Talmant, P.; Mozziconacci, M.-J.; Leonard, C.; Bilhou-Nabera, C.; Cabrol, C.; et al. Cytogenetic profile of childhood and adult megakaryoblastic leukemia (M7): A study of the Groupe Francais de Cytogenetique Hematologique (GFCH). Blood 2002, 100, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, J.; Dastugue, N.; A Haas, O.; Harbott, J.; A Heere, N.; Huret, J.L.; Landman-Parker, J.; Lebeau, M.M.; Leonard, C.; Mann, G.; et al. Nineteen cases of the t(1;22)(p13;q13) acute megakaryblastic leukaemia of infants/children and a review of 39 cases: Report from a t(1;22) study group. Leukemia 2000, 14, 216–218. [Google Scholar] [CrossRef]
- De Rooij, J.D.; Branstetter, C.; Ma, J.; Li, Y.; Walsh, M.P.; Cheng, J.; Obulkasim, A.; Dang, J.; Easton, J.; Verboon, L.J.; et al. Pediatric non–Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat. Genet. 2017, 49, 451–456. [Google Scholar] [CrossRef]
- Schweitzer, J.; Zimmermann, M.; Rasche, M.; Von Neuhoff, C.; Creutzig, U.; Dworzak, M.; Reinhardt, D.; Klusmann, J.-H. Improved outcome of pediatric patients with acute megakaryoblastic leukemia in the AML-BFM 04 trial. Ann. Hematol. 2015, 94, 1327–1336. [Google Scholar] [CrossRef]
- Hsiao, H.-H.; Yang, M.-Y.; Liu, Y.-C.; Tseng, S.-B.; Chao, M.-C.; Liu, T.-C.; Lin, S.-F.; Hsiao, H.-P. RBM15-MKL1 (OTT-MAL) fusion transcript in an adult acute myeloid leukemia patient. Am. J. Hematol. 2005, 79, 43–45. [Google Scholar] [CrossRef]
- Wiellette, E.L.; Harding, K.W.; A Mace, K.; Ronshaugen, M.R.; Wang, F.Y.; McGinnis, W. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development 1999, 126, 5373–5385. [Google Scholar]
- Zhao, X.; Su, H.; Liu, Y. Split End Family RNA Binding Proteins: Novel Tumor Suppressors Coupling Transcriptional Regulation with RNA Processing. Cancer Transl. Med. 2015, 1, 21. [Google Scholar] [CrossRef]
- Niu, C.; Zhang, J.; Breslin, P.; Onciu, M.; Ma, Z.; Morris, S.W. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood 2009, 114, 2087–2096. [Google Scholar] [CrossRef]
- Raffel, G.D.; Chu, G.C.; Jesneck, J.L.; Cullen, D.E.; Bronson, R.T.; Bernard, O.A.; Gilliland, D. Ott1 (Rbm15) Is Essential for Placental Vascular Branching Morphogenesis and Embryonic Development of the Heart and Spleen. Mol. Cell. Boil. 2008, 29, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tran, N.-T.; Su, H.; Wang, R.; Lu, Y.; Tang, H.; Aoyagi, S.; Guo, A.; Khodadadi-Jamayran, A.; Zhou, D.; et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. eLife 2015, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Rainis, L.; Bercovich, D.; Strehl, S.; Teigler-Schlegel, A.; Stark, B.; Trka, J.; Amariglio, N.; Biondi, A.; Muler, I.; Rechavi, G.; et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood 2003, 102, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sabath, D.F.; Kuter, D.J. cloning and functional characterization of a novel c-mpl variant expressed in human cd34 cells and platelets. Cytokine 2000, 12, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Hitzler, J.; Cheung, J.; Li, Y.; Scherer, S.W.; Zipursky, A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood 2003, 101, 4301–4304. [Google Scholar] [CrossRef] [PubMed]
- Klusmann, J.-H.; Godinho, F.J.; Heitmann, K.; Maroz, A.; Koch, M.L.; Reinhardt, D.; Orkin, S.H.; Li, Z. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010, 24, 1659–1672. [Google Scholar] [CrossRef]
- Gialesaki, S.; Mahnken, A.K.; Schmid, L.; Labuhn, M.; Bhayadia, R.; Heckl, D.; Klusmann, J.-H. GATA1s exerts developmental stage-specific effects in human hematopoiesis. Haematologica 2018, 103, e336–e340. [Google Scholar] [CrossRef]
- Maroz, A.; Stachorski, L.; Emmrich, S.; Reinhardt, K.; Xu, J.; Shao, Z.; Käbler, S.; Dertmann, T.; Hitzler, J.; Roberts, I.; et al. GATA1s induces hyperproliferation of eosinophil precursors in Down syndrome transient leukemia. Leukemia 2013, 28, 1259–1270. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Q.; Zhang, D.-E.; Zhou, C.; Xing, H.; Tian, Z.; Rao, Q.; Wang, M.; Wang, J. Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis. Leukemia 2009, 23, 739–745. [Google Scholar] [CrossRef][Green Version]
- Xiao, N.; Laha, S.; Das, S.P.; Morlock, K.; Jesneck, J.L.; Raffel, G.D. Ott1 (Rbm15) regulates thrombopoietin response in hematopoietic stem cells through alternative splicing of c-Mpl. Blood 2015, 125, 941–948. [Google Scholar] [CrossRef]
- Kuwahara, K.; Barrientos, T.; Pipes, G.C.T.; Li, S.; Olson, E. Muscle-Specific Signaling Mechanism That Links Actin Dynamics to Serum Response Factor. Mol. Cell. Boil. 2005, 25, 3173–3181. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Morita, T. Importance of dimer formation of myocardin family members in the regulation of their nuclear export. Cell Struct. Funct. 2013, 38, 123–134. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Du, K.L.; Chen, M.; Li, J.; Lepore, J.J.; Mericko, P.; Parmacek, M.S. Megakaryoblastic Leukemia Factor-1 Transduces Cytoskeletal Signals and Induces Smooth Muscle Cell Differentiation from Undifferentiated Embryonic Stem Cells. J. Boil. Chem. 2004, 279, 17578–17586. [Google Scholar] [CrossRef] [PubMed]
- I Dickstein, J.; Davis, E.M.; Roulston, D. Localization of the chromosome 22 breakpoints in two cases of acute megakaryoblastic leukemia with t(1;22)(p13;q13). Cancer Genet. Cytogenet. 2001, 129, 150–154. [Google Scholar] [CrossRef]
- Lee, J.-H.; Skalnik, D.G. Rbm15-Mkl1 Interacts with the Setd1b Histone H3-Lys4 Methyltransferase via a SPOC Domain That Is Required for Cytokine-Independent Proliferation. PLoS ONE 2012, 7, e42965. [Google Scholar] [CrossRef]
- Sawada, T.; Nishiyama, C.; Kishi, T.; Sasazuki, T.; Komazawa-Sakon, S.; Xue, X.; Piao, J.-H.; Ogata, H.; Nakayama, J.-I.; Taki, T.; et al. Fusion of OTT to BSAC Results in Aberrant Up-regulation of Transcriptional Activity. J. Boil. Chem. 2008, 283, 26820–26828. [Google Scholar] [CrossRef]
- Macdonald, D.; Reiter, A.; Cross, N.C.P. The 8p11 Myeloproliferative Syndrome: A Distinct Clinical Entity Caused by Constitutive Activation of FGFR1. Acta Haematol. 2002, 107, 101–107. [Google Scholar] [CrossRef]
- Cross, N.C.P.; Reiter, A. Fibroblast Growth Factor Receptor and Platelet-Derived Growth Factor Receptor Abnormalities in Eosinophilic Myeloproliferative Disorders. Acta Haematol. 2008, 119, 199–206. [Google Scholar] [CrossRef]
- Jackson, C.C.; Medeiros, L.J.; Miranda, R.N. 8p11 myeloproliferative syndrome: A review. Hum. Pathol. 2010, 41, 461–476. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-Scale Proteomic Analysis of the Human Spliceosome. Genome Res. 2002, 12, 1231–1245. [Google Scholar] [CrossRef]
- Zhou, Z.; Sim, J.; Griffith, J.; Reed, R. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl. Acad. Sci. USA 2002, 99, 12203–12207. [Google Scholar] [CrossRef] [PubMed]
- Dettwiler, S.; Aringhieri, C.; Cardinale, S.; Keller, W.; Barabino, S.M.L. Distinct Sequence Motifs within the 68-kDa Subunit of Cleavage Factor ImMediate RNA Binding, Protein-Protein Interactions, and Subcellular Localization. J. Boil. Chem. 2004, 279, 35788–35797. [Google Scholar] [CrossRef] [PubMed]
- Bastien, J.; Rochette-Egly, C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 2004, 328, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Such, E.; Cervera, J.; Valencia, A.; Barragán, E.; Ibáñez, M.; Luna, I.; Fuster, Ó.; Perez-Sirvent, M.L.; Senent, L.; Sempere, A.; et al. A novel NUP98/RARG gene fusion in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood 2011, 117, 242–245. [Google Scholar] [CrossRef]
- Ha, J.-S.; Do, R.Y.; Ki, C.-S.; Lee, C.; Kim, D.-H.; Lee, W.; Ryoo, N.-H.; Jeon, D.-S. Identification of a novel PML-RARG fusion in acute promyelocytic leukemia. Leukemia 2017, 31, 1992–1995. [Google Scholar] [CrossRef]
- Zhao, J.; Liang, J.-W.; Xue, H.-L.; Shen, S.-H.; Chen, J.; Tang, Y.-J.; Yu, L.-S.; Liang, H.-H.; Gu, L.-J.; Tang, J.-Y.; et al. The genetics and clinical characteristics of children morphologically diagnosed as acute promyelocytic leukemia. Leukemia 2018, 33, 1387–1399. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, M.; Borthakur, G.; Luan, S.; Huang, X.; Tang, G.; Xu, Q.; Ji, D.; Boyer, A.D.; Li, F.; et al. Acute myeloid leukemia with a novel CPSF6-RARG variant is sensitive to homoharringtonine and cytarabine chemotherapy. Am. J. Hematol. 2019, 95, E48–E51. [Google Scholar] [CrossRef]
- De Braekeleer, M. Variant Philadelphia translocations in chronic myeloid leukemia. Cytogenet. Genome Res. 1987, 44, 215–222. [Google Scholar] [CrossRef]
- Morel, F.; Herry, A.; Le Bris, M.-J.; Morice, P.; Bouquard, P.; Abgrall, J.-F.; Berthou, C.; De Braekeleer, M. Contribution of fluorescence in situ hybridization analyses to the characterization of masked and complex Philadelphia chromosome translocations in chronic myelocytic leukemia. Cancer Genet. Cytogenet. 2003, 147, 115–120. [Google Scholar] [CrossRef]
- Bernt, K.M.; Hunger, S.P. Current Concepts in Pediatric Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia. Front. Oncol. 2014, 4, 54. [Google Scholar] [CrossRef]
- Suryanarayan, K.; Hunger, S.P.; Kohler, S.; Carroll, A.J.; Crist, W.; Link, M.P.; Cleary, M.L. Consistent involvement of the bcr gene by 9;22 breakpoints in pediatric acute leukemias. Blood 1991, 77, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Heckl, D.; Kowalczyk, M.S.; Yudovich, D.; Belizaire, R.; Puram, R.V.; McConkey, M.E.; Thielke, A.; Aster, J.C.; Regev, A.; Ebert, B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014, 32, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Heckl, D.; Charpentier, E. Toward Whole-Transcriptome Editing with CRISPR-Cas9. Mol. Cell 2015, 58, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Lu, S.X.; Pastore, A.; Chen, X.; Imig, J.; Lee, S.C.-W.; Hockemeyer, K.; Ghebrechristos, Y.E.; Yoshimi, A.; Inoue, D.; et al. Targeting an RNA-Binding Protein Network in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 369–384.e7. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Masuda, T.; Canver, M.; Seiler, M.; Semba, Y.; Shboul, M.; Al-Raqad, M.; Maeda, M.; Schoonenberg, V.A.C.; Cole, M.; et al. Genome-wide CRISPR-Cas9 Screen Identifies Leukemia-Specific Dependence on a Pre-mRNA Metabolic Pathway Regulated by DCPS. Cancer Cell 2018, 33, 386–400.e5. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef]
- Labuhn, M.; Adams, F.F.; Ng, M.; Knoess, S.; Schambach, A.; Charpentier, E.; Schwarzer, A.; Mateo, J.L.; Klusmann, J.-H.; Heckl, D. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 2018, 46, 1375–1385. [Google Scholar] [CrossRef]
- Reimer, J.; Knöß, S.; Labuhn, M.; Charpentier, E.M.; Göhring, G.; Schlegelberger, B.; Klusmann, J.-H.; Heckl, D. CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica 2017, 102, 1558–1566. [Google Scholar] [CrossRef]
- Hoell, J.I.; Larsson, E.; Runge, S.; Nusbaum, J.; Duggimpudi, S.; Farazi, T.A.; Hafner, M.; Borkhardt, A.; Sander, C.; Tuschl, T. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Boil. 2011, 18, 1428–1431. [Google Scholar] [CrossRef]
- Duggimpudi, S.; Kloetgen, A.; Maney, S.K.; Münch, P.C.; Hezaveh, K.; Shaykhalishahi, H.; Hoyer, W.; McHardy, A.C.; A Lang, P.; Borkhardt, A.; et al. Transcriptome-wide analysis uncovers the targets of the RNA-binding protein MSI2 and effects of MSI2′s RNA-binding activity on IL-6 signaling. J. Boil. Chem. 2018, 293, 15359–15369. [Google Scholar] [CrossRef]
- Hsu, K.-F.; Shen, M.-R.; Huang, Y.-F.; Cheng, Y.-M.; Lin, S.-H.; Chow, N.-H.; Cheng, S.-W.; Chou, C.-Y.; Ho, C. Overexpression of the RNA-binding proteins Lin28B and IGF2BP3 (IMP3) is associated with chemoresistance and poor disease outcome in ovarian cancer. Br. J. Cancer 2015, 113, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Tong, R.; Zhang, J.; Wang, C.; Li, Q.; Wang, L.; Ju, M. Inhibition of miR-574-5p suppresses cell growth and metastasis and enhances chemosensitivity by targeting RNA binding protein QKI in cervical cancer cells. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, W.; Liu, G.; Tang, W. MicroRNA-24 regulates the growth and chemosensitivity of the human colorectal cancer cells by targeting RNA-binding protein DND1. J Buon 2019, 24, 1476–1481. [Google Scholar] [PubMed]
- Hong, S. RNA Binding Protein as an Emerging Therapeutic Target for Cancer Prevention and Treatment. J. Cancer Prev. 2017, 22, 203–210. [Google Scholar] [CrossRef]
- Guo, Y.; Shan, Q.-Q.; Gong, Y.; Lin, J.; Yang, X. Anti-leukemia effect of oridonin on T-cell acute lymphoblastic leukemia. Sichuan da xue xue bao. Yi xue ban = J. Sichuan Univ. Med Sci. Ed. 2014, 45, 903–907. [Google Scholar]


 ) or activating (→).
) or activating (→).
 ) or activating (→).
) or activating (→).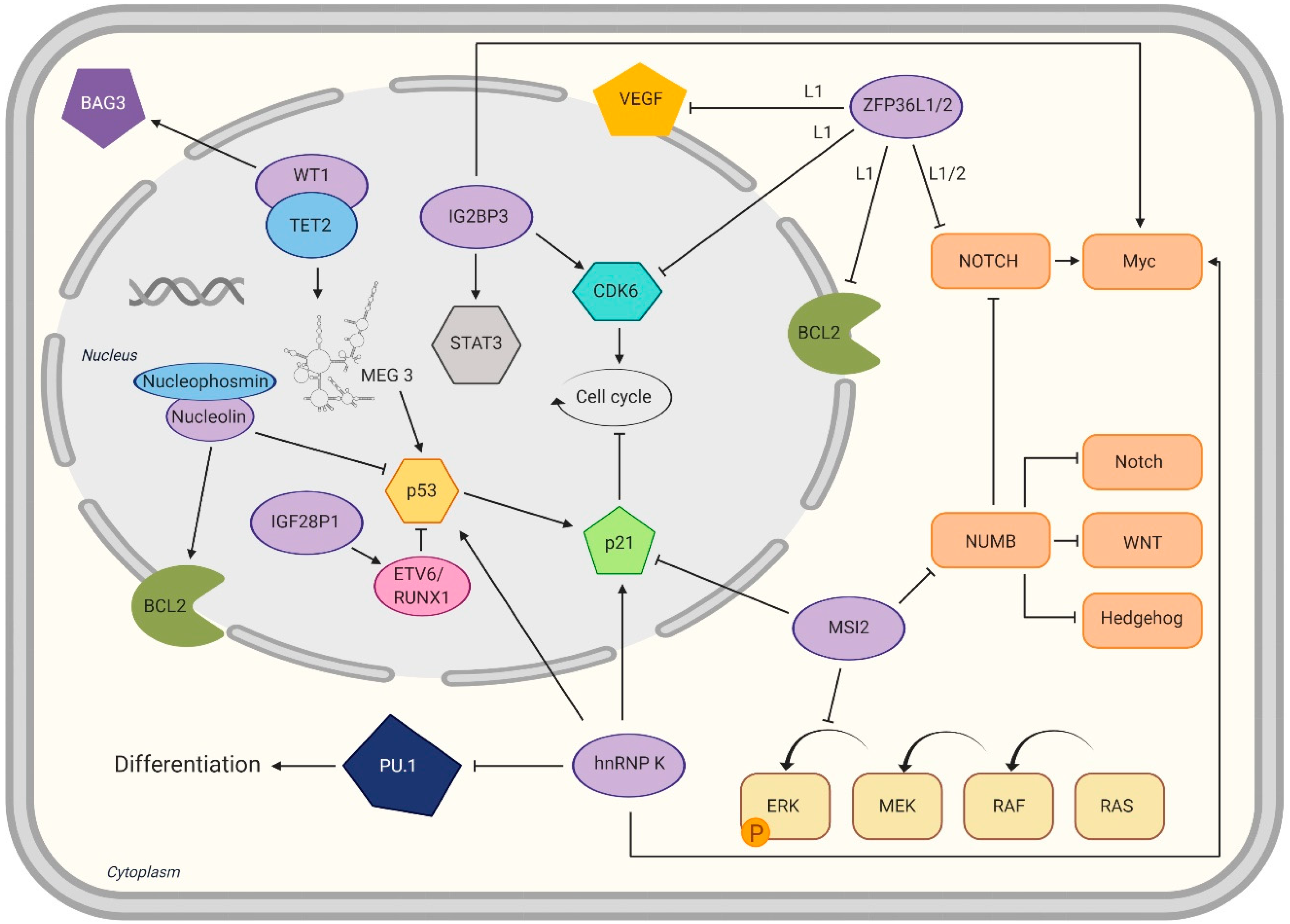

| RBP | Affected Leukemia Type | Localization | Proliferation Apoptosis Differentiation | Clinical Prognosis (When Altered) | ||
|---|---|---|---|---|---|---|
| P | A | D | ||||
| IGF2BP | AML/ALL | Nucleus | ↑ | Poor | ||
| MSI2 | AML/ALL | Cytosol | ↑ | ↓ | Poor | |
| RBM15 | - | Nucleoplasm | ↓ | ↑ | n/a- | |
| RBM15-MKL1 | AML/ALL | Nucleus | ↑ | Poor | ||
| hnRNP K | AML | Nucleoplasm/Cytosol | ↓ | ↑ | Unknown | |
| Nucleolin | AML | Nucleoplasm, Nucleoli | ↑ | Poor | ||
| ZFP36L1/2 | AML/ALL | Nucleus, Cytoplasm | ↓ | ↑ | Unknown | |
| WT1 | ALL | Nucleoplasm | ↑ | Poor | ||
| RBP | Targeting Drug | Mechanism of Action |
|---|---|---|
| IGF2BP1 | BTYNB | Allosteric inhibitor of Igf2BP1 in melanoma and ovarial cancer |
| IGF2BP3 | I-BET151 | dose-dependent reduction of IGF2BP3 and stagnation of the cells in G1-S phase |
| MSI2 | Ro 08-2750 | Competitive inhibition of RNA binding at the RRM1 |
| NCL | Oridonin | Direct inhibitor of NCL |
| AS1411 | Inactivation of NCL and NFκB leads to hypomethylation and activation of caspase signaling |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schuschel, K.; Helwig, M.; Hüttelmaier, S.; Heckl, D.; Klusmann, J.-H.; Hoell, J.I. RNA-Binding Proteins in Acute Leukemias. Int. J. Mol. Sci. 2020, 21, 3409. https://doi.org/10.3390/ijms21103409
Schuschel K, Helwig M, Hüttelmaier S, Heckl D, Klusmann J-H, Hoell JI. RNA-Binding Proteins in Acute Leukemias. International Journal of Molecular Sciences. 2020; 21(10):3409. https://doi.org/10.3390/ijms21103409
Chicago/Turabian StyleSchuschel, Konstantin, Matthias Helwig, Stefan Hüttelmaier, Dirk Heckl, Jan-Henning Klusmann, and Jessica I Hoell. 2020. "RNA-Binding Proteins in Acute Leukemias" International Journal of Molecular Sciences 21, no. 10: 3409. https://doi.org/10.3390/ijms21103409
APA StyleSchuschel, K., Helwig, M., Hüttelmaier, S., Heckl, D., Klusmann, J.-H., & Hoell, J. I. (2020). RNA-Binding Proteins in Acute Leukemias. International Journal of Molecular Sciences, 21(10), 3409. https://doi.org/10.3390/ijms21103409





