Yeast Mitochondrial Translation Initiation Factor 3 Interacts with Pet111p to Promote COX2 mRNA Translation
Abstract
:1. Introduction
2. Results
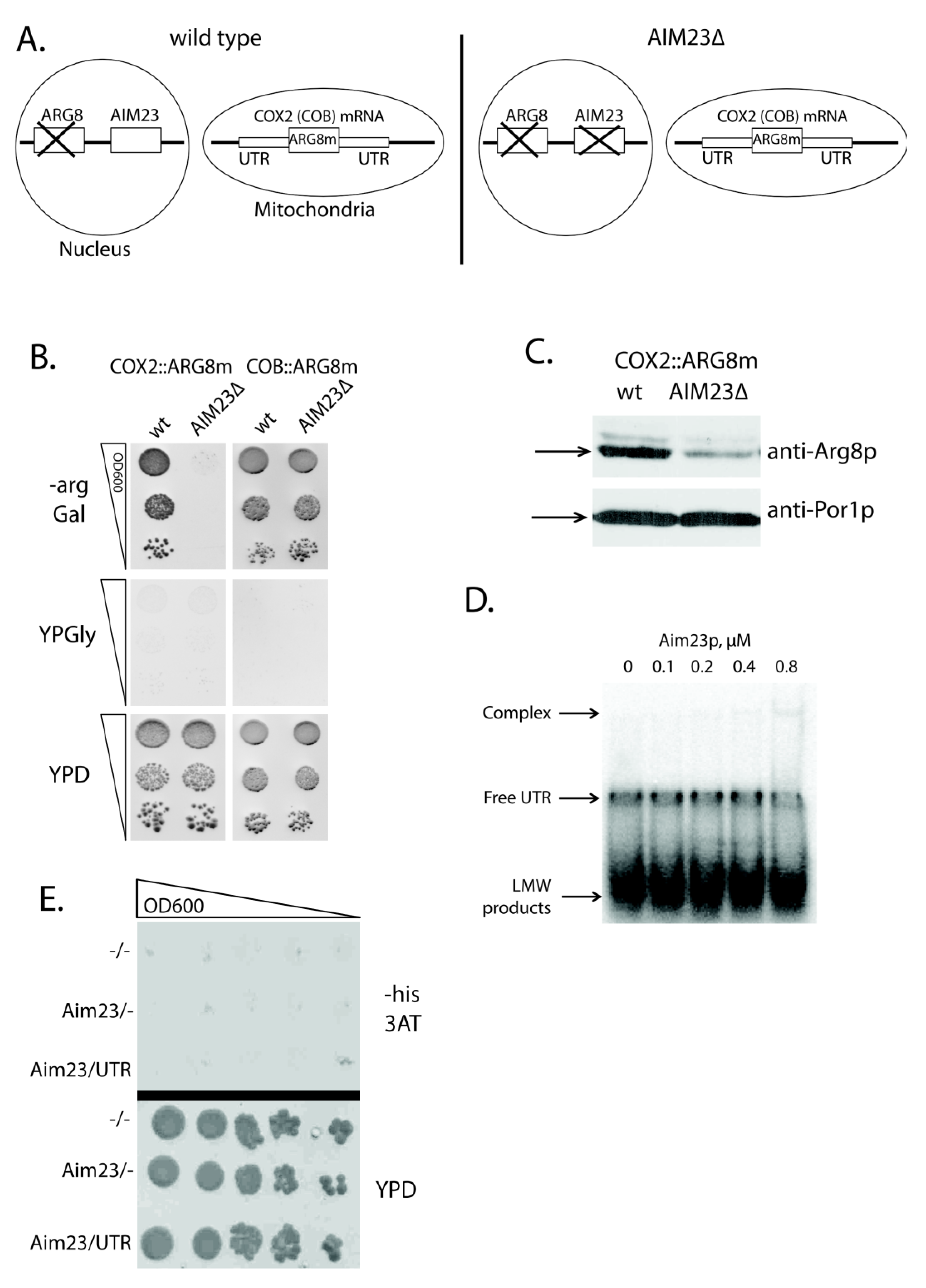
2.1. Aim23p Ensures COX2 mRNA Translation via Its UTRs
2.2. Aim23p Physical Interaction with COX2 mRNA 5′-UTR Is Detectable Neither in Vitro Nor in Vivo
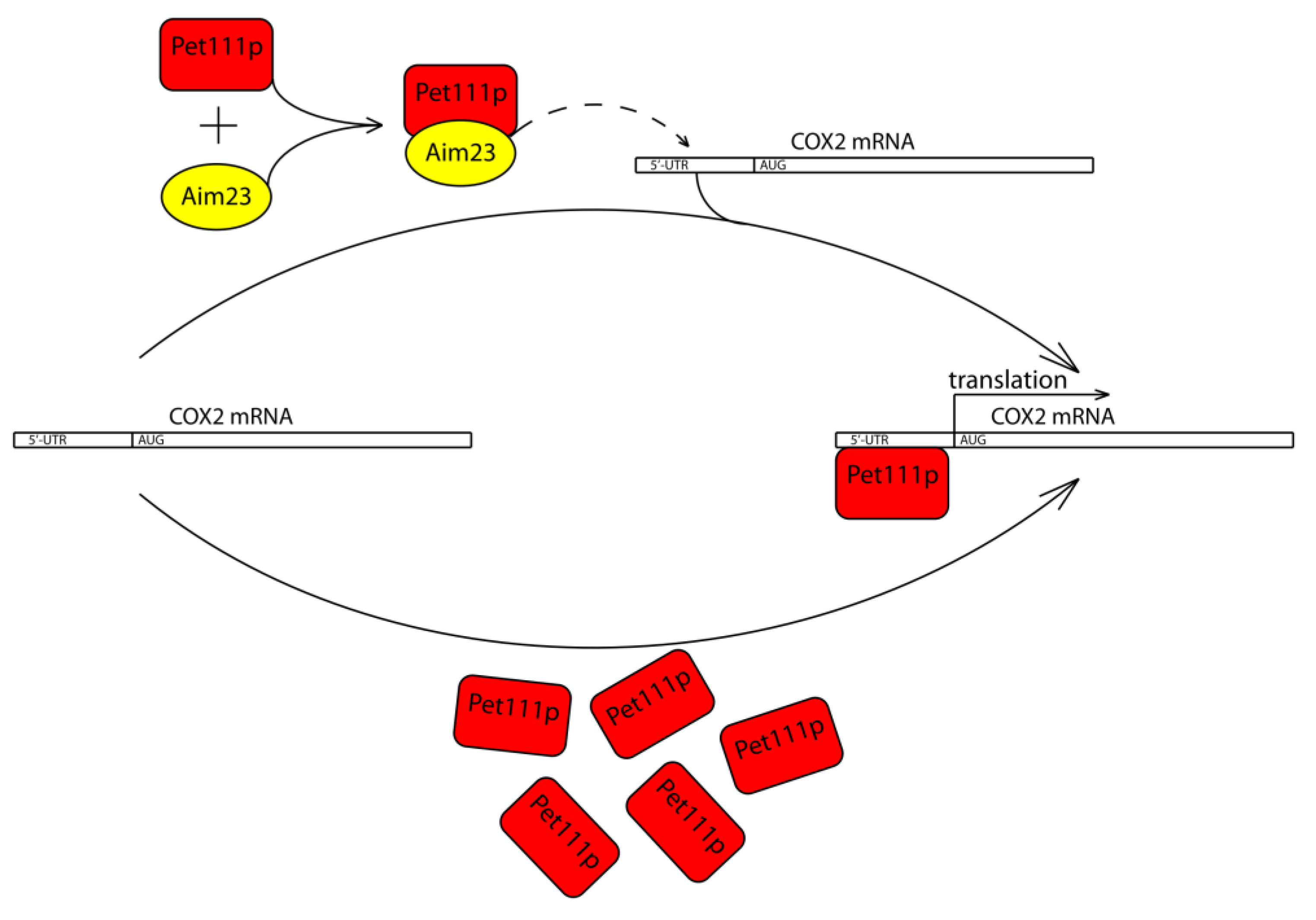
2.3. Aim23p Physically Interacts with Pet111p and Enhances Its Interaction with COX2 mRNA 5′-UTR
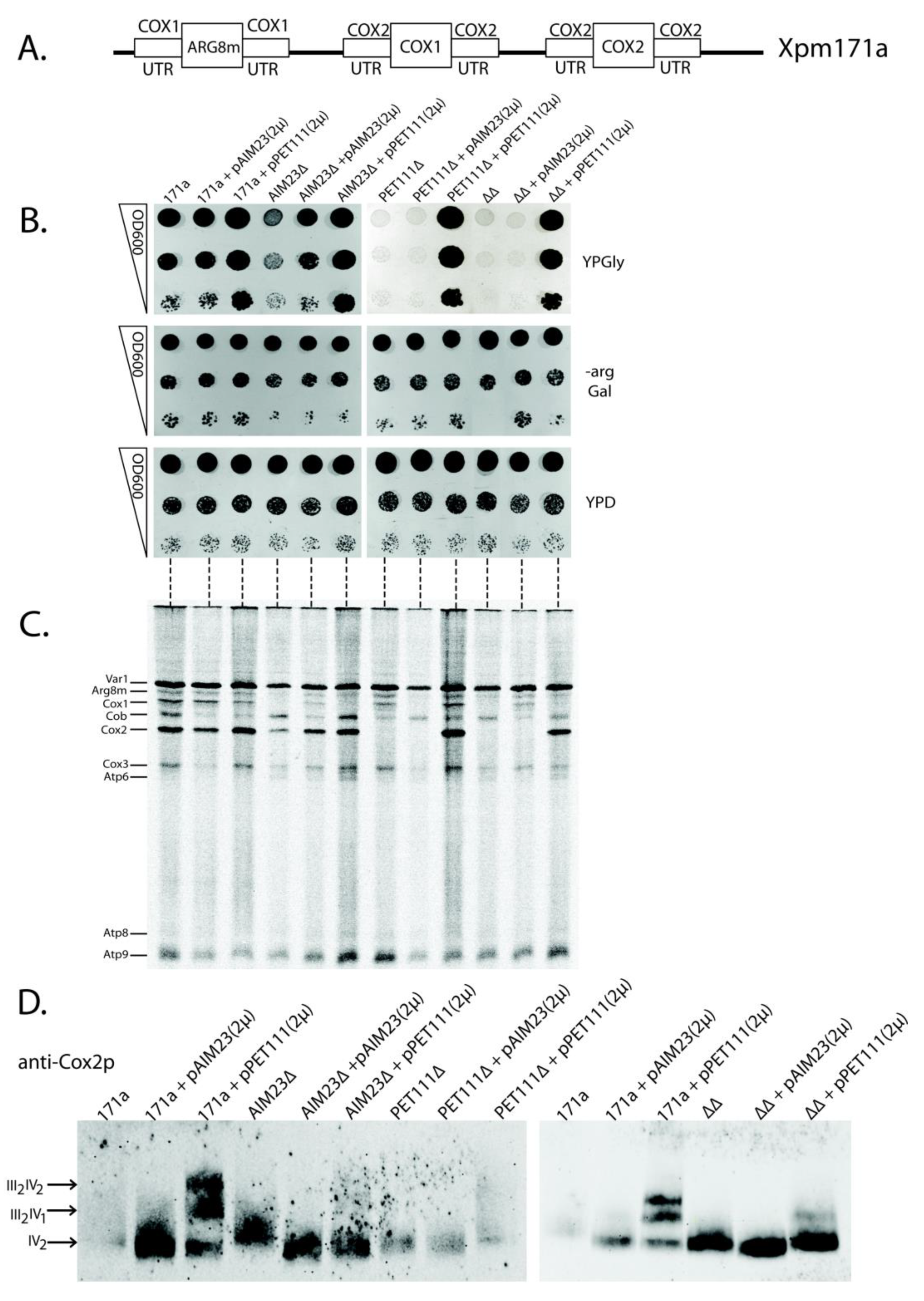
2.4. The Increased Amount of Pet111p May Compensate for Aim23p Absence in the Process of COX2 mRNA Translation
3. Discussion
4. Materials and Methods
4.1. Plasmids, Strains, Oligonucleotides
4.2. Yeast Procedures
- YPD (20 g/L peptone, 10 g/L yeast extract, 20 g/L glucose),
- YPGly (20 g/L peptone, 10 g/L yeast extract, 30 g/L glycerol),
- SC-Arg (2.6 g/L amino acid mix without arginine, 3.4 g yeast nitrogen base, and 10 g/l ammonium sulfate) supplemented with 20 g/L galactose, and
- SC-His 3AT (2.6 g/L amino acid mix without histidine, 3.4 g yeast nitrogen base and 10 g/L ammonium sulfate) supplemented with 20 g/L glucose and 2 mM of 3-Amino-1,2,4-triazole (3-AT).
4.3. Gene Deletions
4.4. Construction of YEplac195-Derivative Plasmids
4.5. Yeast Two- and Three-Hybrid Systems
4.6. Expression and Purification of Recombinant Proteins
4.7. Gel-Shift Assay
4.8. Mitochondrial Translation Analysis
4.9. Mitochondria Isolation
4.10. Blue Native Polyacrylamide Gel Electrophoresis (PAGE)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martin, W.F.; Garg, S.; Zimorski, V. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. L B Biol. Sci. 2015, 370, 20140330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amunts, A.; Brown, A.; Toots, J.; Scheres, S.H.; Ramakrishnan, V. The structure of the human mitochondrial ribosome. Science 2015, 348, 95–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, N.; Brown, A.; Amunts, A.; Ramakrishnan, V. The structure of the yeast mitochondrial ribosome. Science 2017, 355, 528–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kummer, E.; Leibundgut, M.; Rackham, O.; Lee, R.G.; Boehringer, D.; Filipovska, A.; Ban, N. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 2018, 560, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenko, A.; Atkinson, G.C.; Levitskii, S.; Zenkin, N.; Tenson, T.; Hauryliuk, V.; Kamenski, P. Mitochondrial translation initiation machinery: Conservation and diversification. Biochimie 2014, 100, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, G.C.; Kuzmenko, A.; Kamenski, P.; Vysokikh, M.Y.; Lakunina, V.; Tankov, S.; Smirnova, E.; Soosaar, A.; Tenson, T.; Hauryliuk, V. Evolutionary and genetic analyses of mitochondrial translation initiation factors identify the missing mitochondrial IF3 in S. cerevisiae. Nucleic Acids Res. 2012, 40, 6122–6134. [Google Scholar] [CrossRef] [Green Version]
- Koc, E.C.; Spremulli, L.L. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 2002, 277, 35541–35549. [Google Scholar] [CrossRef] [Green Version]
- Derbikova, K.; Kuzmenko, A.; Levitskii, S.; Klimontova, M.; Chicherin, I.; Baleva, M.V.; Krasheninnikov, I.A.; Kamenski, P. Biological and evolutionary significance of terminal extensions of mitochondrial translation initiation factor 3. Int. J. Mol. Sci. 2018, 19, 3861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chicherin, I.V.; Zinina, V.V.; Levitskiy, S.A.; Serebryakova, M.V.; Kamenski, P.A. Aim23p interacts with the yeast mitochondrial ribosomal small subunit. Biochemistry (Moscow) 2019, 84, 40–46. [Google Scholar] [CrossRef]
- Haque, M.E.; Grasso, D.; Spremulli, L.L. The interaction of mammalian mitochondrial translational initiation factor 3 with ribosomes: Evolution of terminal extensions in IF3mt. Nucleic Acids Res. 2008, 36, 589–597. [Google Scholar] [CrossRef]
- Levitskii, S.; Derbikova, K.; Baleva, M.V.; Kuzmenko, A.; Golovin, A.V.; Chicherin, I.; Krasheninnikov, I.A.; Kamenski, P. 60S dynamic state of bacterial ribosome is fixed by yeast mitochondrial initiation factor 3. PeerJ 2018, 6, e5620. [Google Scholar] [CrossRef] [PubMed]
- Ayyub, S.A.; Aswathy, S.L.; Dobriyal, D.; Aluri, S.; Spremulli, L.L.; Varshney, U. Fidelity of translation in the presence of mammalian mitochondrial initiation factor 3. Mitochondrion 2018, 39, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenko, A.; Derbikova, K.; Salvatori, R.; Tankov, S.; Atkinson, G.C.; Tenson, T.; Ott, M.; Kamenski, P.; Hauryliuk, V. Aim-less translation: Loss of Saccharomyces cerevisiae mitochondrial translation initiation factor mIF3/Aim23 leads to unbalanced protein synthesis. Sci. Rep. 2016, 6, 18749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derbikova, K.S.; Levitsky, S.A.; Chicherin, I.V.; Vinogradova, E.N.; Kamenski, P.A. Activation of yeast mitochondrial translation: Who is in charge? Biochemistry (Moscow) 2018, 83, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Amunts, A.; Brown, A. Organization and regulation of mitochondrial protein synthesis. Annu. Rev. Biochem. 2016, 85, 77–101. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.M.; Woellhaf, M.W.; Bonnefoy, N. Control of protein synthesis in yeast mitochondria: The concept of translational activators. Biochim. Biophys. Acta 2013, 1833, 286–294. [Google Scholar] [CrossRef] [Green Version]
- Bordonne, R.; Dirheimer, G.; Martin, R.P. Expression of the oxi1 and maturase-related RF1 genes in yeast mitochondria. Curr. Genet. 1988, 13, 227–233. [Google Scholar] [CrossRef]
- Simon, M.; Faye, G. Organization and processing of the mitochondrial oxi3/oli2 multigenic transcript in yeast. Mol. Gen. Genet. 1984, 196, 266–274. [Google Scholar] [CrossRef]
- Mulero, J.J.; Fox, T.D. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics 1993, 133, 509–516. [Google Scholar]
- Bonnefoy, N.; Bsat, N.; Fox, T.D. Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol. Cell Biol. 2001, 21, 2359–2372. [Google Scholar] [CrossRef] [Green Version]
- Bonnefoy, N.; Fox, T.D. Directed alteration of Saccharomyces cerevisiae mitochondrial DNA by biolistic transformation and homologous recombination. Methods Mol. Biol. 2007, 372, 153–166. [Google Scholar]
- Bonnefoy, N.; Fox, T.D. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol. Gen. Genet 2000, 262, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Gruschke, S.; Kehrein, K.; Rompler, K.; Grone, K.; Israel, L.; Imhof, A.; Herrmann, J.M.; Ott, M. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol 2011, 193, 1101–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SenGupta, D.J.; Zhang, B.; Kraemer, B.; Pochart, P.; Fields, S.; Wickens, M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 1996, 93, 8496–8501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.L.; Hofmann, K.B.; Cowan, A.T.; Temiakov, D.; Cramer, P.; Anikin, M. Yeast mitochondrial protein Pet111p binds directly to two distinct targets in COX2 mRNA, suggesting a mechanism of translational activation. J. Biol. Chem. 2019, 294, 7528–7536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLellan, T. Electrophoresis buffers for polyacrylamide gels at various pH. Anal. Biochem. 1982, 126, 94–99. [Google Scholar] [CrossRef]
- Fields, S.; Song, O. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Payne, M.J.; Finnegan, P.M.; Smooker, P.M.; Lukins, H.B. Characterization of a second nuclear gene, AEP1, required for expression of the mitochondrial OLI1 gene in Saccharomyces cerevisiae. Curr. Genet. 1993, 24, 126–135. [Google Scholar] [CrossRef]
- Ziaja, K.; Michaelis, G.; Lisowsky, T. Nuclear control of the messenger RNA expression for mitochondrial ATPase subunit 9 in a new yeast mutant. J. Mol. Biol. 1993, 229, 909–916. [Google Scholar] [CrossRef]
- Fiori, A.; Perez-Martinez, X.; Fox, T.D. Overexpression of the COX2 translational activator, Pet111p, prevents translation of COX1 mRNA and cytochrome c oxidase assembly in mitochondria of Saccharomyces cerevisiae. Mol. Microbiol. 2005, 56, 1689–1704. [Google Scholar] [CrossRef]
- Perez-Martinez, X.; Broadley, S.A.; Fox, T.D. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003, 22, 5951–5961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schagger, H.; Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000, 19, 1777–1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostojic, J.; Panozzo, C.; Bourand-Plantefol, A.; Herbert, C.J.; Dujardin, G.; Bonnefoy, N. Ribosome recycling defects modify the balance between the synthesis and assembly of specific subunits of the oxidative phosphorylation complexes in yeast mitochondria. Nucleic Acids Res. 2016, 44, 5785–5797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudler, D.L.; Hughes, L.A.; Perks, K.L.; Richman, T.R.; Kuznetsova, I.; Ermer, J.A.; Abudulai, L.N.; Shearwood, A.J.; Viola, H.M.; Hool, L.C.; et al. Fidelity of translation initiation is required for coordinated respiratory complex assembly. Sci. Adv. 2019, 5, eaay2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chicherin, I.V.; Baleva, M.V.; Levitskii, S.A.; Dashinimaev, E.B.; Krasheninnikov, I.A.; Kamenski, P. Initiation factor 3 is dispensable for mitochondrial translation in cultured human cells. Sci. Rep. 2020, 10, 7110. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Kehrein, K.; Schilling, R.; Moller-Hergt, B.V.; Wurm, C.A.; Jakobs, S.; Lamkemeyer, T.; Langer, T.; Ott, M. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 2015, 10, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Krause, K.; Lopes de Souza, R.; Roberts, D.G.; Dieckmann, C.L. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell 2004, 15, 2674–2683. [Google Scholar] [CrossRef]
- Juergens, H.; Varela, J.A.; de Vries, A.R.G.; Perli, T.; Gast, V.J.M.; Gyurchev, N.Y.; Rajkumar, A.S.; Mans, R.; Pronk, J.T.; Morrissey, J.P.; et al. Genome editing in Kluyveromyces and Ogataea yeasts using a broad-host-range Cas9/gRNA co-expression plasmid. FEMS Yeast Res. 2018, 18, foy012. [Google Scholar] [CrossRef]
- Gietz, R.D.; Sugino, A. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 1988, 74, 527–534. [Google Scholar] [CrossRef]
- Harper, J.W.; Adami, G.R.; Wei, N.; Keyomarsi, K.; Elledge, S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993, 75, 805–816. [Google Scholar] [CrossRef]
- Fromont-Racine, M.; Rain, J.C.; Legrain, P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997, 16, 277–282. [Google Scholar] [CrossRef]
- Bernstein, D.S.; Buter, N.; Stumpf, C.; Wickens, M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 2002, 26, 123–141. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Wolberger, C. A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal. Biochem. 2011, 418, 102–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, W.B. Host specificity of DNA produced by Escherichia coli: Bacterial mutations affecting the restriction and modification of DNA. J. Mol. Biol. 1966, 16, 118–133. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006, 313, 107–120. [Google Scholar]
- Anderson, S.L.; Lin, A.P.; McAlister-Henn, L. Analysis of interactions with mitochondrial mRNA using mutant forms of yeast NAD(+)-specific isocitrate dehydrogenase. Biochemistry 2005, 44, 16776–16784. [Google Scholar] [CrossRef] [Green Version]
- Gregg, C.; Kyryakov, P.; Titorenko, V.I. Purification of mitochondria from yeast cells. J. Vis. Exp. 2009, 30, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Wittig, I.; Braun, H.P.; Schagger, H. Blue native PAGE. Nat. Protoc. 2006, 1, 418–428. [Google Scholar] [CrossRef]

| Name, Reference | Description |
|---|---|
| pAIM23-KanMX4 [13] | pRS416 plasmid containing KanMX4 cassette with AIM23 5’- and 3’- flanks. Constructed in our lab previously. |
| pUG-natNT2 [39] | Plasmid containing NatN2 cassette. A gift from D.Knorre. |
| YEplac195 [40] | Shuttle episomal vector with 2-micron replication origin. A gift from D.Knorre. |
| pAIM23(2µ) | YEplac195 vector with AIM23 gene and its genomic flanks cloned. |
| pPET111(2µ) | YEplac195 vector with PET111 gene and its genomic flanks cloned. |
| pACT2 [41] | Vector for 2- and 3-hybrid systems. A gift from N.Entelis. |
| pACT2-AIM23 | pACT2 with AIM23 gene cloned. |
| pAS2ΔΔ [42] | Vector for 2-hybrid system. A gift from N.Entelis. |
| pAS2ΔΔ-PET111 | pAS2ΔΔ with PET111 gene cloned. |
| pAS2ΔΔ-AEP1 | pAS2ΔΔ with AEP1 gene cloned. |
| pIIIA/MS2-1 [43] | Vector for 3-hybrid system. A gift from N.Entelis. |
| pIIIA-UTR | pIIIA/MS2 with 5′-UTR of the COX2 mRNA cloned. |
| pET21d [44] | Vector for recombinant protein expression in E. coli. A gift from V.Hauryliuk. |
| pET21d-PET111 | pET21d with PET111 gene cloned. |
| pET28a-AIM23 [6] | pET28a vector with AIM23 gene cloned. Constructed in our lab previously. |
| Strain | Genotype/Description |
|---|---|
| Saccharomyces cerevisiae | |
| XPM171a [31] | Matα, lys2, leu2-3,112, arg8::hisG, ura3-52 [ρ+, cox1∆::ARG8m, cox2D::COX1c, COX2]. A gift from M. Ott. |
| ∆AIM23 | XPM171a, AIM23 gene disrupted with KanMX4 cassette. |
| ∆PET111 | XPM171a, PET111 gene disrupted with NatNT2 cassette. |
| ∆∆ | XPM171a with AIM23 and PET111 genes disrupted with the above-mentioned cassettes. |
| Y190 [41] | MATa gal4 gal80 his3 trp1–901 ade2–101 ura3–52 leu2–3, 112 URA3∷GAL1∷lacZ LYS2∷GAL4(UAS)∷HIS3 cyhR Vector for 2-hybrid system. A gift from N. Entelis. |
| L40 coat [24] | MATa, ura3-52, leu2-3, 112, his3-200, trp1-1, ade2, LYS2::(lexA op)-HIS3, URA3::(lexA op)-lacZ, LexA MS2 coat (TRP1) Vector for 3-hybrid system. A gift from N. Entelis. |
| Escherichia coli | |
| B834 [45] | hsdS metE gal ompT For recombinant proteins expression. A gift from D.Knorre. |
| Name | Sequence |
|---|---|
| AEP1_pAS2_Fw | 5′-ATGCGGATCCATGATTACTACAGTG |
| AEP1_pAS2_Rv | 5′-ATGCCTGCAGTTATGGGCGTAAAGCTTC |
| AIM23_A | 5’-TGGGTGTTGATA |
| AIM23_D | 5’-TAGTATGGATGA |
| AIM23_PACT2_Fw | 5′- ATGCCCATGGATGTTAAAAGTTCC |
| AIM23_PACT2_Rv | 5′-ATGCCTCGAGTTACATTTCATTCATTT |
| Aim23_YE_Fw | 5′-ATGCAAGCTTCCTCGTGTAAATGAAATCAAAGAGG |
| AIM23_YE_Rv | 5′-ATGCGTCGACCATGCTCATAAATCCTGAC |
| ICO141 | 5’-CACTACCTATTAAATTTAAACAATTGCTTACGAGAACTTAGACATGGAGGCCCAGAATACC |
| ICO142 | 5’-ATTTACACGTGAGAGAAAGGAAGGTAAATAACTGAAAAGACCGGTAGAGGTGTGGTCAATAAGAGC |
| ICO61 | 5′-TTGCGCTAGCACTGAGTTGATCAAAAAAAAGC |
| ICO62 | 5′-AAGCCTCGAGCTCCTCCTCCTTTTTATTCTC |
| kanB | 5’-CTGCAGCGAGGAGCCGTAAT |
| kanC | 5’-TGATTTTGATGACGAGCGTAAT |
| KanMX_aim23_mod_fw | 5’-CCCGCGACGGTAAGAACTTTA |
| KanMX_aim23_mod_rev | 5’-GAATCCTGGTACTTTAATGATAAG |
| NAT_B | 5’-ATGCCCCTGAGCTGCGCACG |
| NAT_C | 5’-GAGTAACTCTTTCCTGTAGG |
| PET111_A | 5’-GTACATTTGTTGAAGGAG |
| PET111_D | 5’-GCCAATCAAGTACTGCC |
| PET111_pAS2_Fw | 5′-ATGCGGATCCATGTTACAACGGAG |
| PET111_pAS2_Rv | 5′-ATGCCTGCAGTTACTCCTCCTCCTTTTT |
| Pet111_YE_Fw | 5′-ATGCGGATCCCCGGACCTTACGAGTTCTTCG |
| Pet111_YE_Rv | 5′-ATGCGGTACCGCCCTCCTTCAAACTATTCG |
| pIIIA_UTR_Fw | 5′-ATGCGGGCCCAGTATTAACATATTATAAATAG |
| pIIIA_UTR_Rv | 5′-ATGCGGGCCCTTTAATAAATCTTAACC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chicherin, I.; Levitskii, S.; Baleva, M.V.; Krasheninnikov, I.A.; Patrushev, M.V.; Kamenski, P. Yeast Mitochondrial Translation Initiation Factor 3 Interacts with Pet111p to Promote COX2 mRNA Translation. Int. J. Mol. Sci. 2020, 21, 3414. https://doi.org/10.3390/ijms21103414
Chicherin I, Levitskii S, Baleva MV, Krasheninnikov IA, Patrushev MV, Kamenski P. Yeast Mitochondrial Translation Initiation Factor 3 Interacts with Pet111p to Promote COX2 mRNA Translation. International Journal of Molecular Sciences. 2020; 21(10):3414. https://doi.org/10.3390/ijms21103414
Chicago/Turabian StyleChicherin, Ivan, Sergey Levitskii, Maria V. Baleva, Igor A. Krasheninnikov, Maxim V. Patrushev, and Piotr Kamenski. 2020. "Yeast Mitochondrial Translation Initiation Factor 3 Interacts with Pet111p to Promote COX2 mRNA Translation" International Journal of Molecular Sciences 21, no. 10: 3414. https://doi.org/10.3390/ijms21103414
APA StyleChicherin, I., Levitskii, S., Baleva, M. V., Krasheninnikov, I. A., Patrushev, M. V., & Kamenski, P. (2020). Yeast Mitochondrial Translation Initiation Factor 3 Interacts with Pet111p to Promote COX2 mRNA Translation. International Journal of Molecular Sciences, 21(10), 3414. https://doi.org/10.3390/ijms21103414





