Genome-Wide Dissection of the CRF Gene Family in Brassica napus Indicates that BnaCRF8s Specifically Regulate Root Architecture and Phosphate Homeostasis against Phosphate Fluctuation in Plants
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogenetic Relationships of the CRF Family in Brassica napus
2.2. Gene Structure, Protein Features, and Chromosomal Duplication of the CRF Family in Brassica napus
2.3. Expression Profiles of the BnaCRF Genes in Response to Phosphorus Stress
2.4. Subcellular Localization of the BnaCRF8 Genes
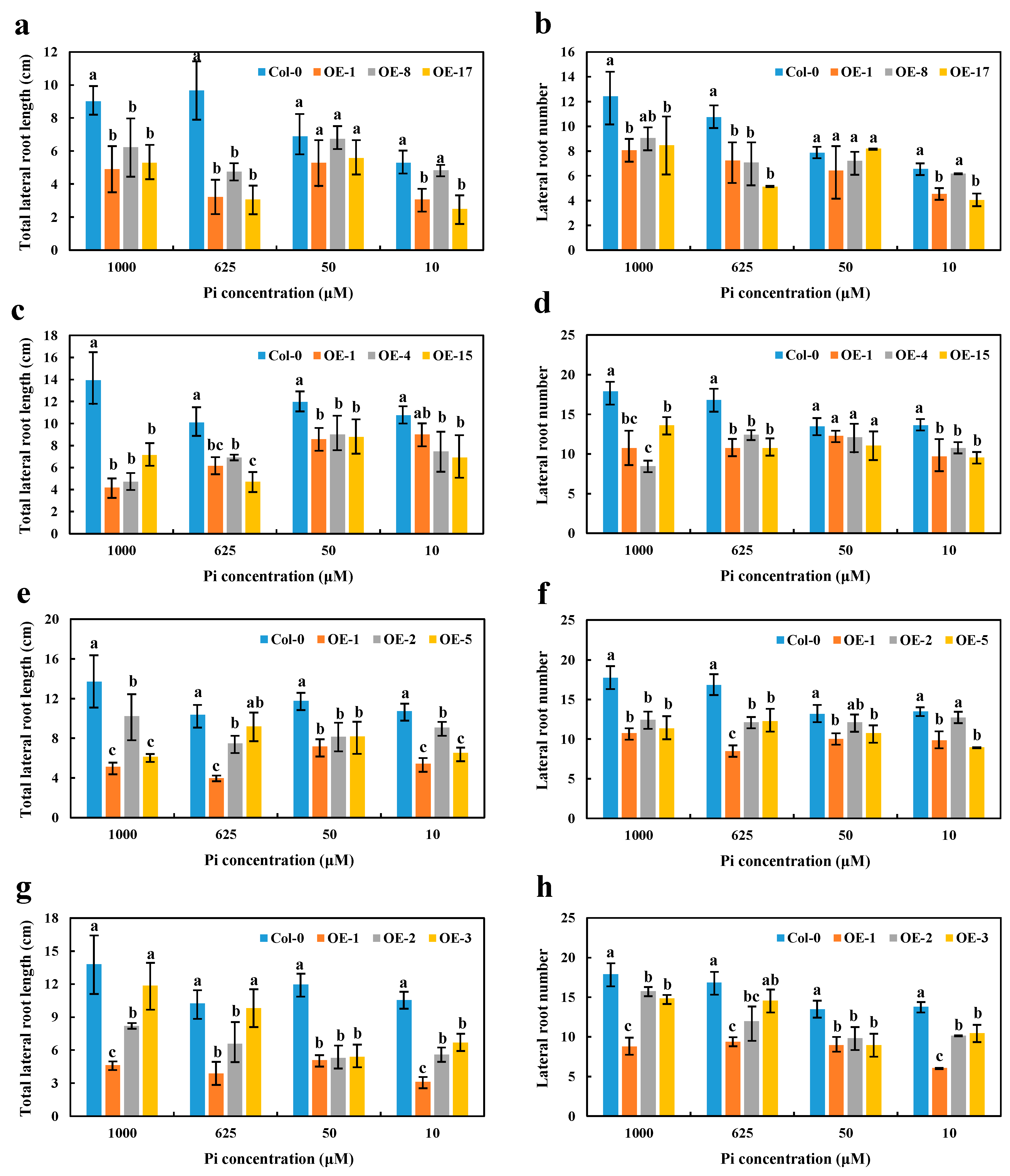
2.5. Phenotypical Characterization of the BnaCRF8 Overexpression Lines in Arabidopsis under Varied Phosphate Conditions
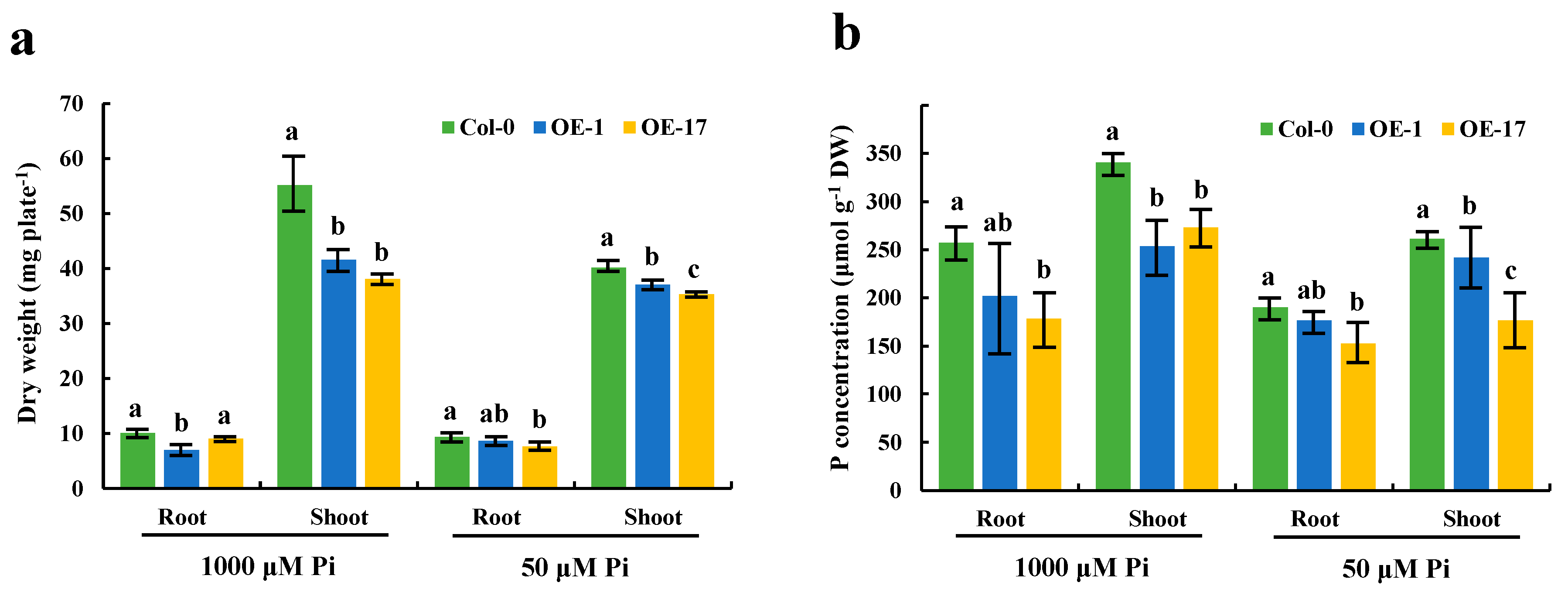
2.6. BnaA7.CRF8 is a Negative Regulator of Phosphate Homeostasis in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Identification of CRF TFs in the Brassica napus Genome
4.2. Phylogenetic and Conserved Motif Analyses of BnaCRF Genes
4.3. Chromosomal Location and Gene Duplication Analyses
4.4. Plant Materials and Treatments
4.5. RNA Isolation and Real-Time Quantitative PCR
4.6. Expression Pattern Analysis of BnaCRF Genes in Response to Pi Starvation using RNA-Seq Data
4.7. Vector Construction and Arabidopsis Transformation
4.8. Transient Expression Assays in Nicotiana benthamiana
4.9. Measurements of Root Traits
4.10. Quantification of Total Phosphorus
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| P | Phosphorus |
| Pi | Phosphate |
| TF | Transcription factor |
| OE | Overexpression |
| PR | Primary root |
| LR | Lateral root |
| CRF | Cytokinin Response Factor |
| AP2 | APETALA2 |
| ERF | ETHYLENE RESPONSIVE FACTOR |
| FPKM | fragments per kilobase of transcript per million fragments mapped |
References
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [Green Version]
- Oldroyd, G.; Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science 2020, 368, eaba0196. [Google Scholar] [CrossRef]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef]
- Bari, R.; Datt Pant, B.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [Green Version]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.Y.; Huang, T.K.; Tseng, C.Y.; Lai, Y.S.; Lin, S.I.; Lin, W.Y.; Chen, J.W.; Chiou, T.J. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 2012, 24, 2168–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essigmann, B.; Güler, S.; Narang, R.A.; Linke, D.; Benning, C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 1950–1955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Härtel, H.; Dormann, P.; Benning, C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 10649–10654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Welti, R.; Wang, X. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 2006, 142, 750–761. [Google Scholar] [CrossRef] [Green Version]
- Svistoonoff, S.; Creff, A.; Reymond, M.; Sigoillot-Claude, C.; Ricaud, L.; Blanchet, A.; Nussaume, L.; Desnos, T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 2007, 39, 792–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho-Cristóbal, J.J.; Rexach, J.; Conéjéro, G.; Al-Ghazi, Y.; Nacry, P.; Doumas, P. PRD, an Arabidopsis AINTEGUMENTA-like gene, is involved in root architectural changes in response to phosphate starvation. Planta 2008, 228, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J. CYTOKININ RESPONSE FACTORs gating environmental signals and hormones. Trends Plant Sci. 2016, 21, 993–996. [Google Scholar] [CrossRef]
- Rashotte, A.M.; Goertzen, L.R. The CRF domain defines cytokinin response factor proteins in plants. BMC Plant Biol. 2010, 10, 74. [Google Scholar] [CrossRef] [Green Version]
- Ramaiah, M.; Jain, A.; Raghothama, K.G. ETHYLENE RESPONSE FACTOR070 regulates root development and phosphate starvation-mediated responses. Plant Physiol. 2014, 164, 1484–1498. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.; Cho, C.; Lee, M.R.; Van Binh, N.; Kim, J. CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant Cell 2016, 28, 1828–1843. [Google Scholar] [CrossRef] [Green Version]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Zhao, K.; Gao, Y.; Miao, L.; Chen, C.; Deng, H.; Liu, Z.; Yu, X. Comparative analyses of cytokinin response factors in Brassica diploids and amphidiploids and insights into the evolution of Brassica species. BMC Genomics 2018, 19, 728. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Brooks, M.D.; Cirrone, J.; Pasquino, A.V.; Alvarez, J.M.; Swift, J.; Mittal, S.; Juang, C.L.; Varala, K.; Gutiérrez, R.A.; Krouk, G.; et al. Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat. Commun. 2019, 10, 1569. [Google Scholar] [CrossRef] [Green Version]
- Hammond, J.P.; Bennett, M.J.; Bowen, H.C.; Broadley, M.R.; Eastwood, D.C.; May, S.T.; Rahn, C.; Swarup, R.; Woolaway, K.E.; White, P.J. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003, 132, 578–596. [Google Scholar] [CrossRef] [Green Version]
- Misson, J.; Raghothama, K.G.; Jain, A.; Jouhet, J.; Block, M.A.; Bligny, R.; Ortet, P.; Creff, A.; Somerville, S.; Rolland, N.; et al. A genome-wide transcriptional analyses using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 2005, 102, 11934–11939. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.D.; Liao, Y.Y.; Yang, T.J.; Pan, C.Y.; Buckhout, T.J.; Schmidt, W. Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 2011, 155, 1383–1402. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, J.A.; Yang, S.S.; Miller, S.S.; Bucciarelli, B.; Liu, J.; Rydeen, A.; Bozsoki, Z.; Uhde-Stone, C.; Tu, Z.J.; Allan, D.; et al. An RNA-Seq transcriptome analyses of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 2013, 161, 705–724. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Zhuang, Q.; Zhu, S.; Xiao, B.; Liang, C.; Liao, H.; Tian, J. Genome wide transcriptome analyses reveals complex regulatory mechanisms underlying phosphate homeostasis in soybean nodules. Int. J. Mol. Sci. 2018, 19, 2924. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Gupta, S.; Rashotte, A.M. Solanum lycopersicum cytokinin response factor (SlCRF) genes: Characterization of CRF domain-containing ERF genes in tomato. J. Exp. Bot. 2012, 63, 973–982. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Rashotte, A.M. Expression patterns and regulation of SlCRF3 and SlCRF5 in response to cytokinin and abiotic stresses in tomato (Solanum lycopersicum). J. Plant Physiol. 2014, 171, 349–358. [Google Scholar] [CrossRef]
- Zwack, P.J.; De Clercq, I.; Howton, T.C.; Hallmark, H.T.; Hurny, A.; Keshishian, E.A.; Parish, A.M.; Benkova, E.; Mukhtar, M.S.; Van Breusegem, F.; et al. Cytokinin response factor 6 represses cytokinin-associated genes during oxidative stress. Plant Physiol. 2016, 172, 1249–1258. [Google Scholar] [CrossRef] [Green Version]
- Cucinotta, M.; Manrique, S.; Guazzotti, A.; Quadrelli, N.E.; Mendes, M.A.; Benkova, E.; Colombo, L. Cytokinin response factors integrate auxin and cytokinin pathways for female reproductive organ development. Development 2016, 143, 4419–4424. [Google Scholar] [CrossRef] [Green Version]
- Raines, T.; Shanks, C.; Cheng, C.Y.; McPherson, D.; Argueso, C.T.; Kim, H.J.; Franco-Zorrilla, J.M.; López-Vidriero, I.; Solano, R.; Vaňková, R.; et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016, 85, 134–147. [Google Scholar] [CrossRef]
- Qin, L.; Wang, L.; Guo, Y.; Li, Y.; Ümüt, H.; Wang, Y. An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance. Plant Sci. 2017, 265, 154–166. [Google Scholar] [CrossRef]
- Varala, K.; Marshall-Colón, A.; Cirrone, J.; Brooks, M.D.; Pasquino, A.V.; Léran, S.; Mittal, S.; Rock, T.M.; Edwards, M.B.; Kim, G.J.; et al. Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. USA 2018, 115, 6494–6499. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Wang, Z.; Wang, X.; Liu, D. Blue light-triggered chemical reactions underlie phosphate deficiency-induced inhibition of root elongation of Arabidopsis seedlings grown in Petri dishes. Mol. Plant. 2019, 12, 1515–1523. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analyses. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analyses version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analyses of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.Y.; Shi, L.; Ye, X.S.; Wang, Y.H.; Xu, F.S. Identification of phosphorous efficient germplasm in oilseed rape. J. Plant Nutr. 2009, 32, 1148–1163. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Zhang, H.; Wang, S.; Ye, X.; Shi, L.; Xu, F.; Ding, G. Molecular identification of the phosphate transporter family 1 (PHT1) genes and their expression profiles in response to phosphorus deprivation and other abiotic stresses in Brassica napus. PLoS ONE 2019, 14, 0220374. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Pidkowich, M.S.; Nguyen, H.T.; Heilmann, I.; Ischebeck, T.; Shanklin, J. Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc. Natl. Acad. Sci. USA 2007, 104, 4742–4747. [Google Scholar] [CrossRef] [Green Version]







| Gene Name | Molecular Function | References |
|---|---|---|
| PHT1;1 | Pi uptake | [5] |
| PHT1;4 | Pi uptake | [5] |
| PHO1 | Pi loading into xylem | [8] |
| SQD1 | Sulfolipid biosynthesis under Pi limitation | [9] |
| PLDζ2 | Phosphatidylcholine hydrolysis and DGDG accumulation under Pi stress | [11] |
| LPR1/LPR2 | Root growth regulation under Pi stress | [12] |
| PRD | Root growth regulation under Pi stress | [13] |
| PHR1 | A central transcription factor involved in Pi starvation signaling | [14] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Zhang, H.; Shi, L.; Xu, F.; Ding, G. Genome-Wide Dissection of the CRF Gene Family in Brassica napus Indicates that BnaCRF8s Specifically Regulate Root Architecture and Phosphate Homeostasis against Phosphate Fluctuation in Plants. Int. J. Mol. Sci. 2020, 21, 3660. https://doi.org/10.3390/ijms21103660
Wang S, Zhang H, Shi L, Xu F, Ding G. Genome-Wide Dissection of the CRF Gene Family in Brassica napus Indicates that BnaCRF8s Specifically Regulate Root Architecture and Phosphate Homeostasis against Phosphate Fluctuation in Plants. International Journal of Molecular Sciences. 2020; 21(10):3660. https://doi.org/10.3390/ijms21103660
Chicago/Turabian StyleWang, Sheliang, Hao Zhang, Lei Shi, Fangsen Xu, and Guangda Ding. 2020. "Genome-Wide Dissection of the CRF Gene Family in Brassica napus Indicates that BnaCRF8s Specifically Regulate Root Architecture and Phosphate Homeostasis against Phosphate Fluctuation in Plants" International Journal of Molecular Sciences 21, no. 10: 3660. https://doi.org/10.3390/ijms21103660






