Impact of the Hereditary P301L Mutation on the Correlated Conformational Dynamics of Human Tau Protein Revealed by the Paramagnetic Relaxation Enhancement NMR Experiments
Abstract
1. Introduction
2. Results
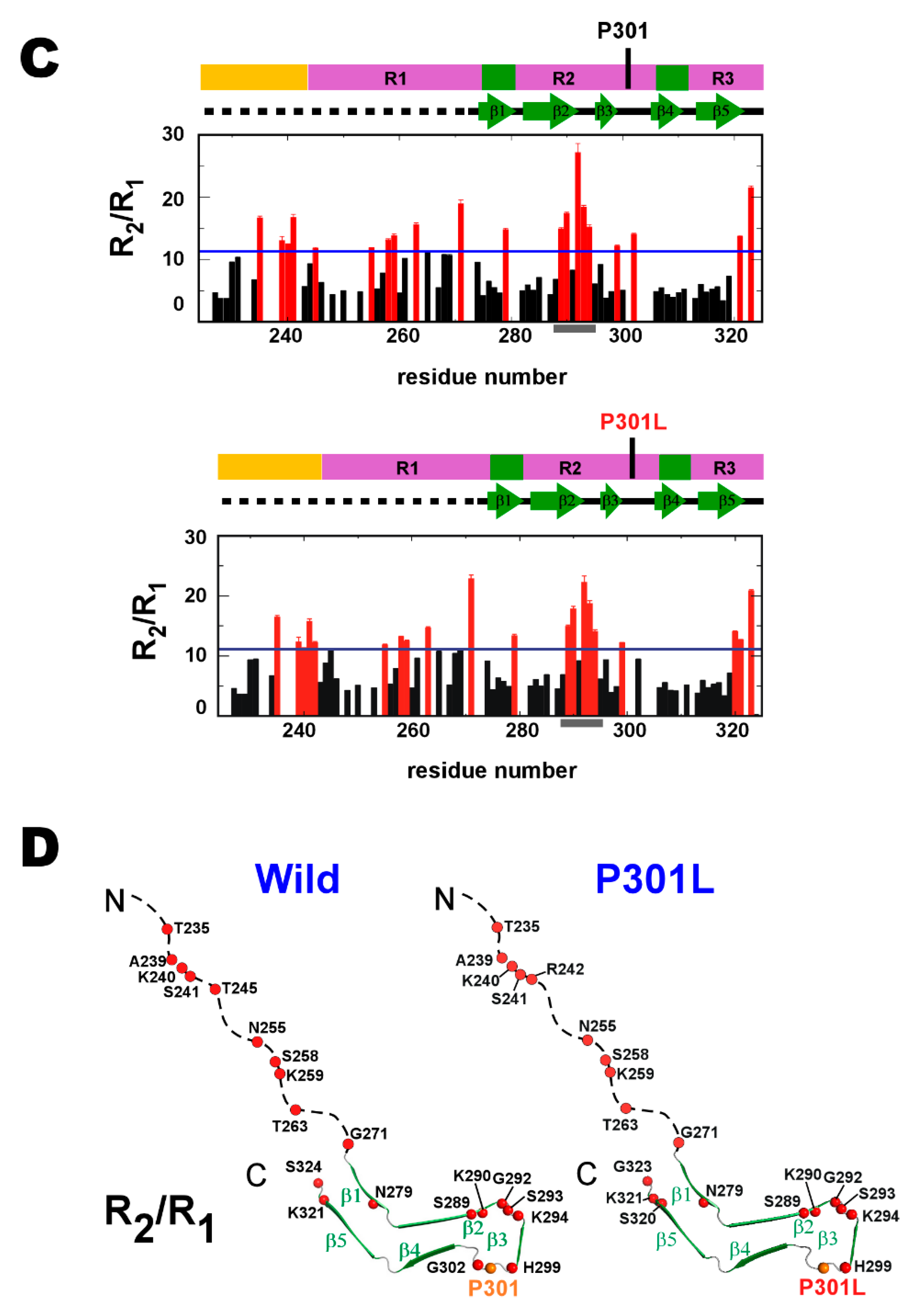
2.1. P301L Mutation Effects on the Backbone Dynamics of TauF4Δ
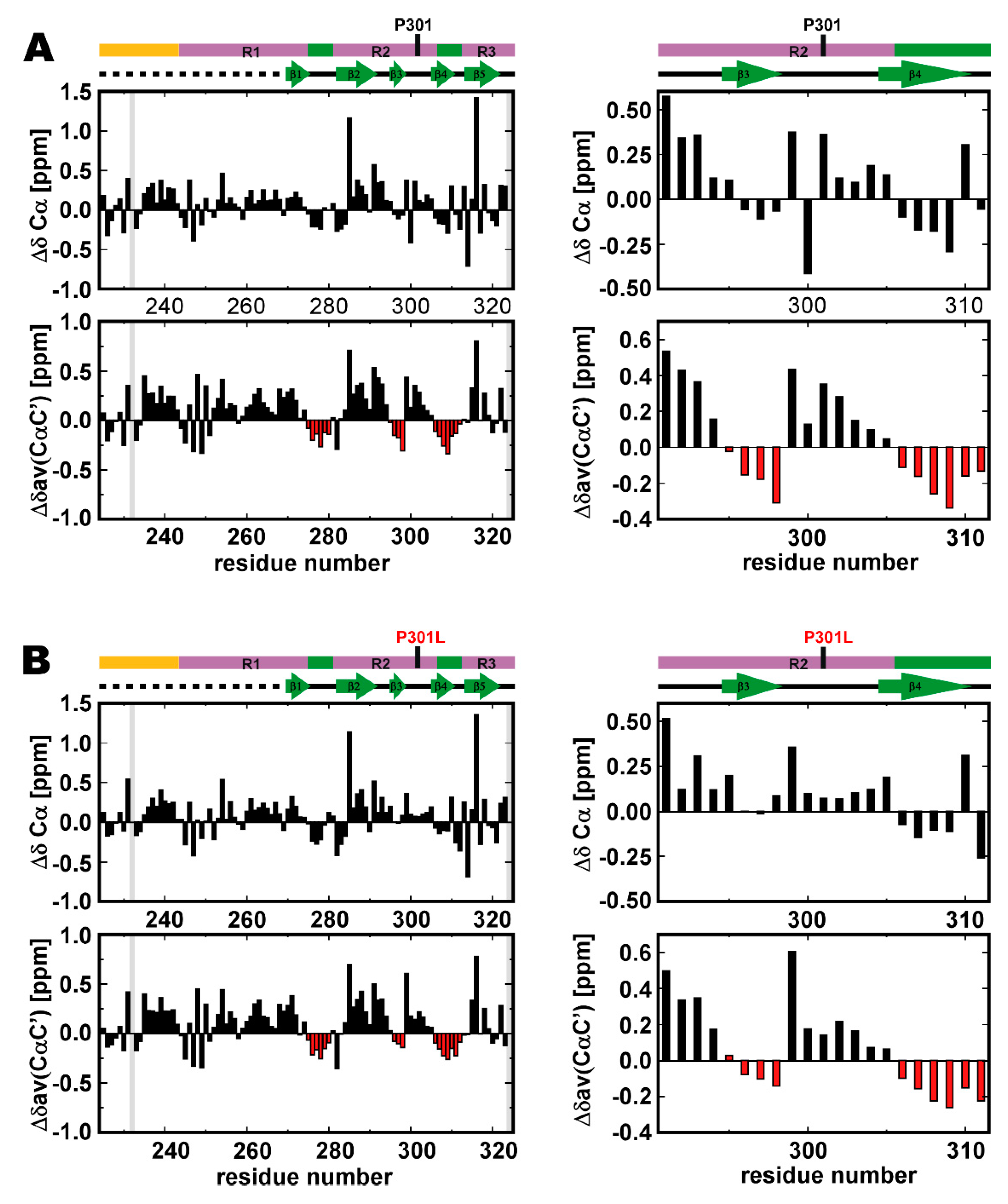
2.2. P301L Reduces the Transiently Populated β-Conformation of R2
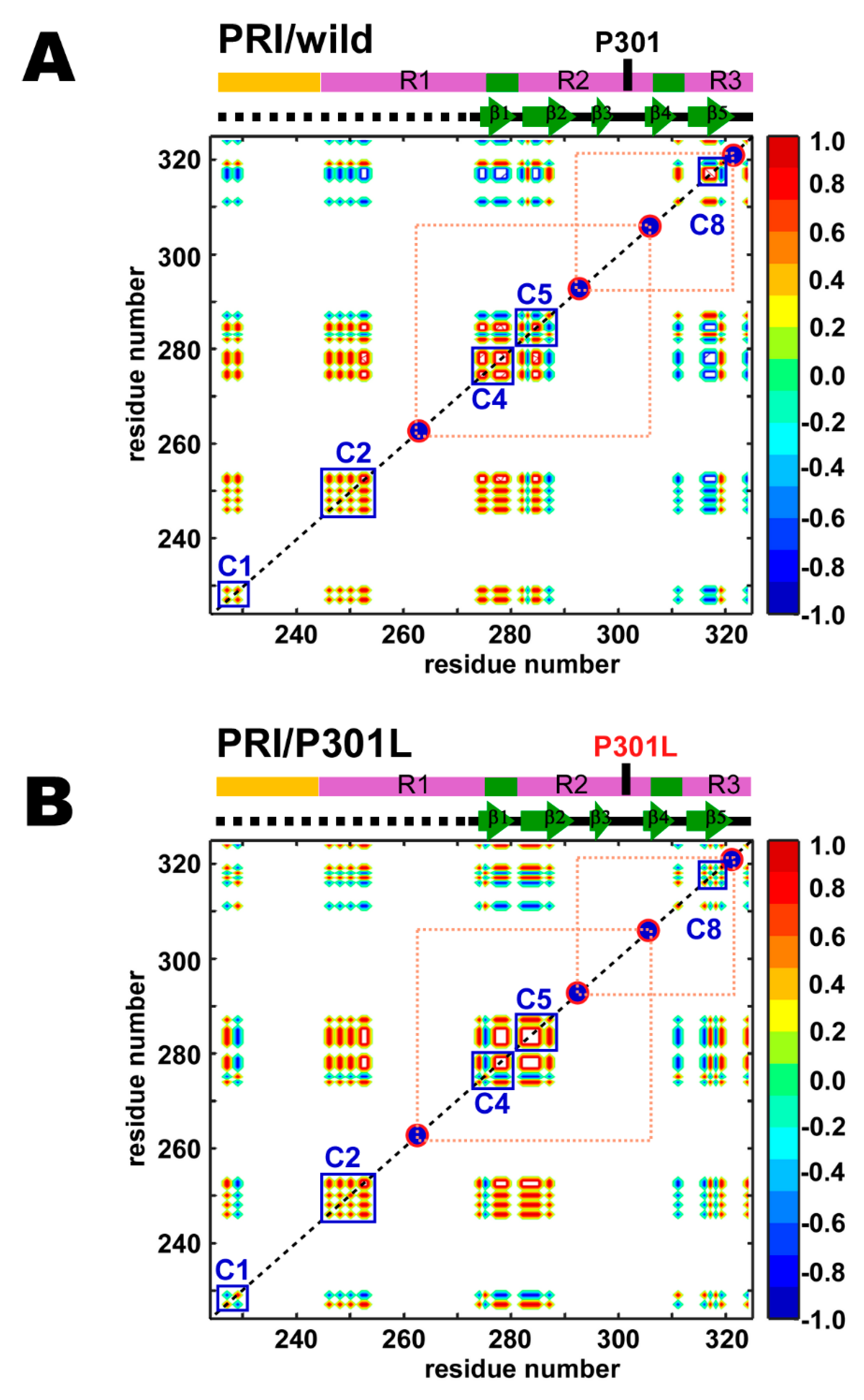
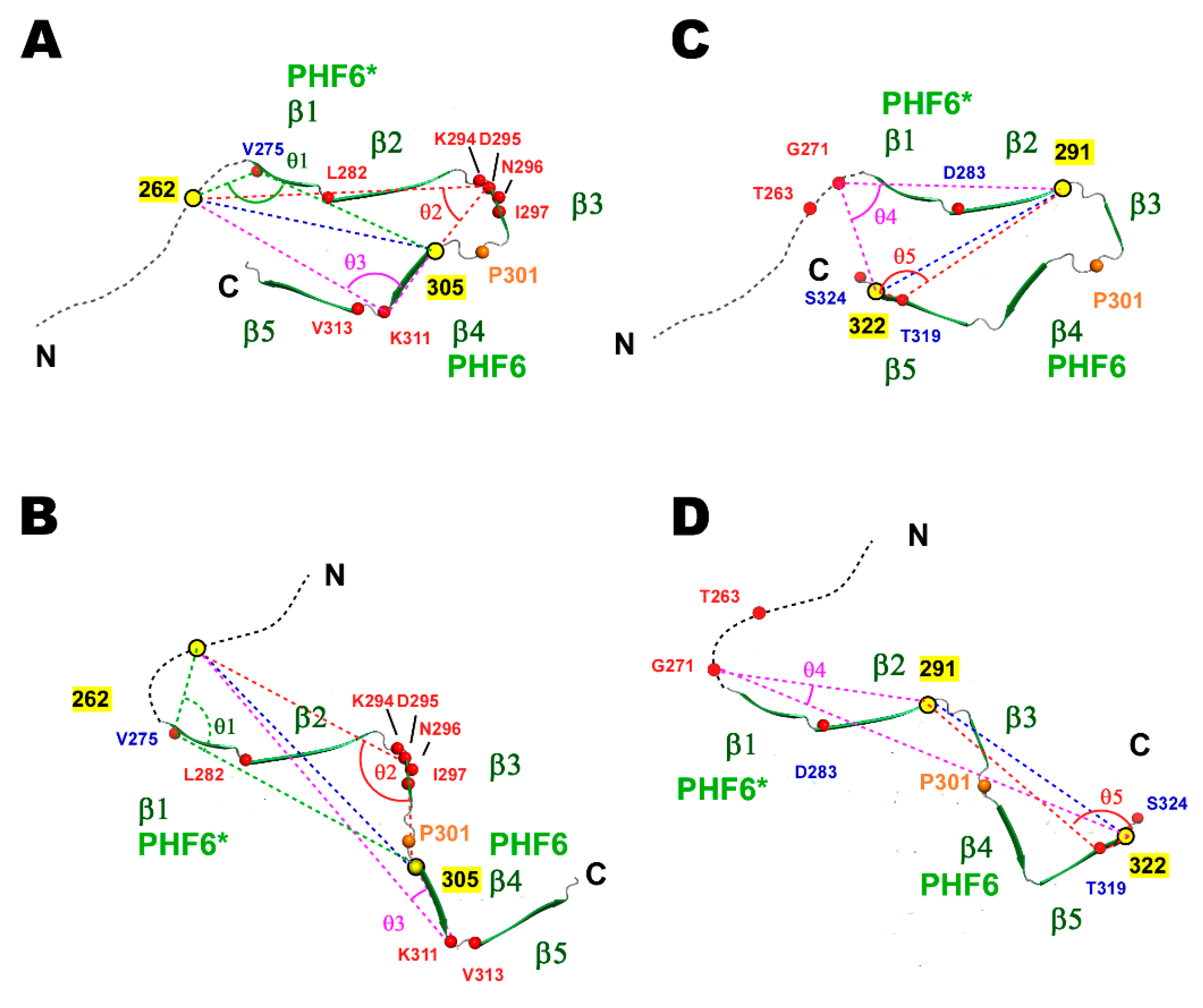
2.3. Correlated Motions Among Residues in TauF4Δ
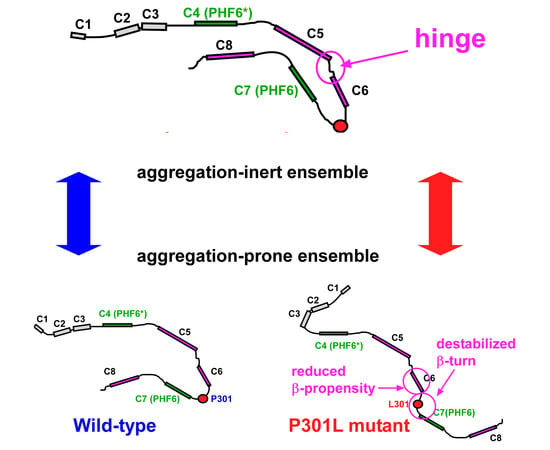
2.4. PRI Data Reveals Conformation Changes Caused by P301L are Exclusively Associated with Transient Structures
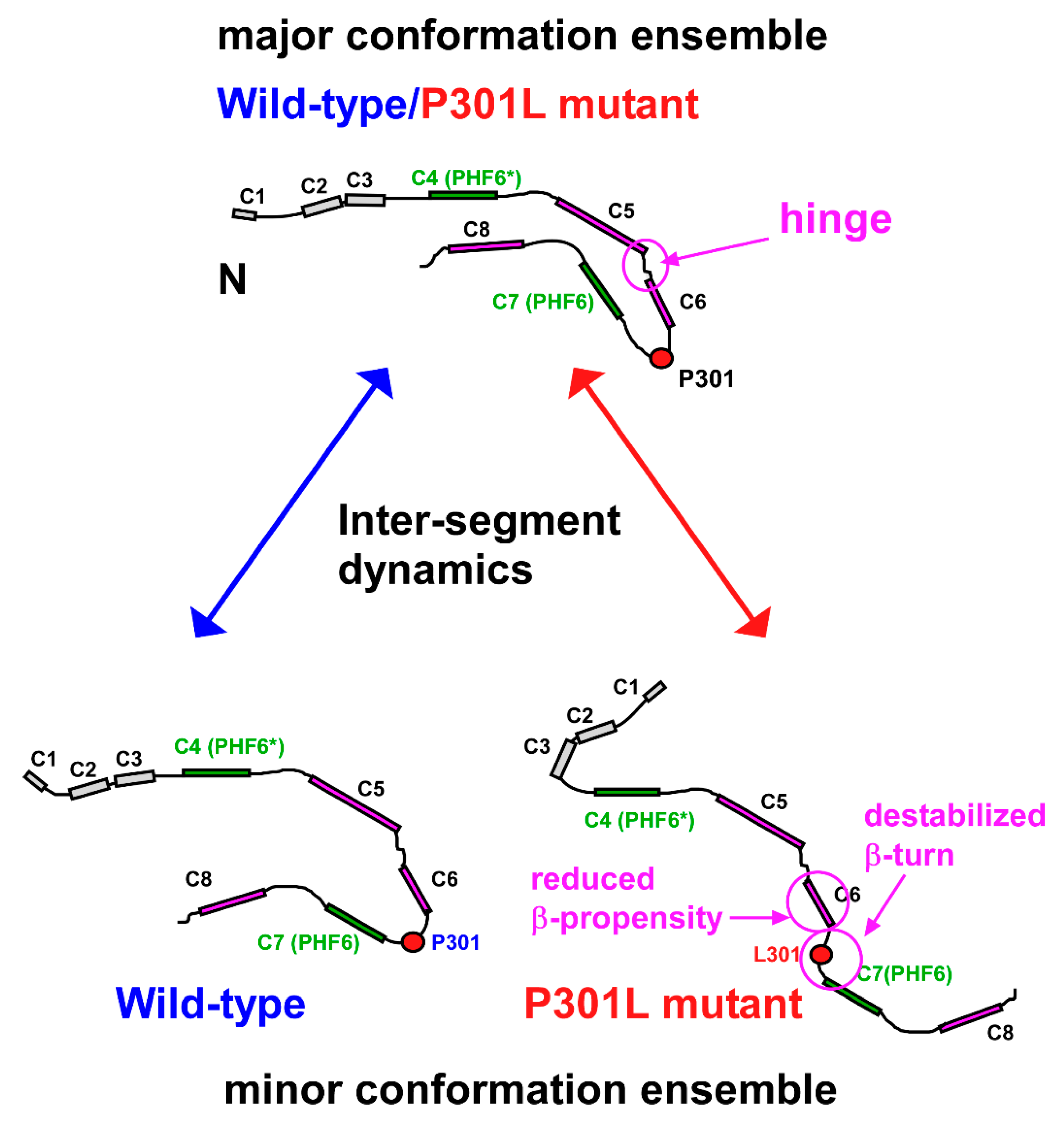
2.5. Structural Changes in the Transient Compact Structures of the P301L Mutant TauF4Δ
3. Discussion
4. Materials and Methods
4.1. TauF4Δ Fragment Preparation
4.2. Backbone NMR Resonance Assignments
4.3. NMR Spin Relaxation Experiments
4.4. Paramagnetic Relaxation Enhancement (PRE) Experiments
4.5. Paramagnetic Relaxation Interference (PRI) Experiments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NMR | Nuclear magnetic resonance |
| AD | Alzheimer’s disease |
| NTF | Neurofibrillary tangle |
| PHF | Paired helical filament |
| PTDP-17 | Frontotemporal dementia and Parkinsonism linked to chromatin 17 |
| MTSL | (1-oxyl-2,2,5,5,-tetramethyl-d-3-pyrroline-3-methyl) methanethiosulfonate |
References
- von Bergen, M.; Barghorn, S.; Müller, S.A.; Pickhardt, M.; Biernat, J.; Mandelkow, E.-M.; Davies, P.; Aebi, U.; Mandelkow, E. The Core of Tau-Paired Helical Filaments Studied by Scanning Transmission Electron Microscopy and Limited Proteolysis. Biochemistry 2006, 45, 6446–6457. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.R. Tauopathies: Classification and clinical update on neurodegenerative diseases associated with microtubule-associated protein tau. Int. Med. J. 2006, 36, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Buée, L.; Bussière, T.; Buée-Scherrer, V.; Delacourte, A.; Hof, P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders11These authors contributed equally to this work. Brain Res. Rev. 2000, 33, 95–130. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Spillantini, M.G.; Hasegawa, M.; Smith, M.J.; Crowther, R.A. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 1996, 383, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Chapter 21 Neurofibrillary pathology of Alzheimer’s disease and other tauopathies. In Progress in Brain Research; Callow, J.A., Ed.; Elsevier: Amsterdam, The Netherlands, 1998; Volume 117, pp. 287–306. [Google Scholar]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Goedert, M. Tau pathology and neurodegeneration. Lancet Neurol. 2013, 12, 609–622. [Google Scholar] [CrossRef]
- Avila, J. Tau aggregation into fibrillar polymers: Taupathies. FEBS Lett. 2000, 476, 89–92. [Google Scholar] [CrossRef]
- Von Bergen, M.; Barghorn, S.; Li, L.; Marx, A.; Biernat, J.; Mandelkow, E.-M.; Mandelkow, E. Mutations of Tau Protein in Frontotemporal Dementia Promote Aggregation of Paired Helical Filaments by Enhancing Local β-Structure. J. Biol. Chem. 2001, 276, 48165–48174. [Google Scholar] [CrossRef]
- Hutton, M.; Lendon, C.L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A.; et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998, 393, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Jakes, R. Mutations causing neurodegenerative tauopathies. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2005, 1739, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Barghorn, S.; Zheng-Fischhöfer, Q.; Ackmann, M.; Biernat, J.; von Bergen, M.; Mandelkow, E.M.; Mandelkow, E. Structure, Microtubule Interactions, and Paired Helical Filament Aggregation by Tau Mutants of Frontotemporal Dementias. Biochemistry 2000, 39, 11714–11721. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G. Tau mutations in frontotemporal dementia FTDP-17 and their relevance for Alzheimer’s disease. Biochim. Biophys. Acta 2000, 1502, 110–121. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Mukrasch, M.D.; Bibow, S.; Korukottu, J.; Jeganathan, S.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Structural Polymorphism of 441-Residue Tau at Single Residue Resolution. PLoS Biol. 2009, 7, e1000034. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, D.; Barré, P.; Kobaslija, M.; Chan, D.; Li, X.; Heend, L. Residual Structure in the Repeat Domain of Tau: Echoes of Microtubule Binding and Paired Helical Filament Formation. Biochemistry 2005, 44, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Mukrasch, M.D.; von Bergen, M.; Klos-Witkowska, A.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Structural and Microtubule Binding Properties of Tau Mutants of Frontotemporal Dementias. Biochemistry 2007, 46, 2574–2582. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef]
- Zhang, W.; Falcon, B.; Murzin, A.G.; Fan, J.; Crowther, R.A.; Goedert, M.; Scheres, S.H. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. Elife 2019, 8. [Google Scholar] [CrossRef]
- Von Bergen, M.; Barghorn, S.; Biernat, J.; Mandelkow, E.-M.; Mandelkow, E. Tau aggregation is driven by a transition from random coil to beta sheet structure. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2005, 1739, 158–166. [Google Scholar] [CrossRef]
- Von Bergen, M.; Friedhoff, P.; Biernat, J.; Heberle, J.; Mandelkow, E.M.; Mandelkow, E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming beta structure. Proc. Natl. Acad. Sci. USA 2000, 97, 5129–5134. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.W.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Drombosky, K.W.; Hou, Z.; Sari, L.; Kashmer, O.M.; Ryder, B.D.; Perez, V.A.; Woodard, D.R.; Lin, M.M.; Diamond, M.I.; et al. Tau local structure shields an amyloid-forming motif and controls aggregation propensity. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Goux, W.J.; Kopplin, L.; Nguyen, A.D.; Leak, K.; Rutkofsky, M.; Shanmuganandam, V.D.; Sharma, D.; Inouye, H.; Kirschner, D.A. The Formation of Straight and Twisted Filaments from Short Tau Peptides. J. Biol. Chem. 2004, 279, 26868–26875. [Google Scholar] [CrossRef] [PubMed]
- Mukrasch, M.D.; Biernat, J.; von Bergen, M.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Sites of Tau Important for Aggregation Populate β-Structure and Bind to Microtubules and Polyanions. J. Biol. Chem. 2005, 280, 24978–24986. [Google Scholar] [CrossRef]
- Kuret, J.; Congdon, E.E.; Li, G.; Yin, H.; Yu, X.; Zhong, Q. Evaluating triggers and enhancers of tau fibrillization. Microsc. Res. Tech. 2005, 67, 141–155. [Google Scholar] [CrossRef]
- Chirita, C.N.; Congdon, E.E.; Yin, H.; Kuret, J. Triggers of Full-Length Tau Aggregation: A Role for Partially Folded Intermediates. Biochemistry 2005, 44, 5862–5872. [Google Scholar] [CrossRef]
- Pérez, M.; Valpuesta, J.M.; Medina, M.; Montejo de Garcini, E.; Avila, J. Polymerization of τ into Filaments in the Presence of Heparin: The Minimal Sequence Required for τ—τ Interaction. J. Neurochem. 1996, 67, 1183–1190. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Crowther, R.A. Effects of frontotemporal dementia FTDP-17 mutations on heparin-induced assembly of tau filaments. FEBS Lett. 1999, 450, 306–311. [Google Scholar] [CrossRef]
- Yotsumoto, K.; Saito, T.; Asada, A.; Oikawa, T.; Kimura, T.; Uchida, C.; Ishiguro, K.; Uchida, T.; Hasegawa, M.; Hisanaga, S.-I. Effect of Pin1 or Microtubule Binding on Dephosphorylation of FTDP-17 Mutant Tau. J. Biol. Chem. 2009, 284, 16840–16847. [Google Scholar] [CrossRef]
- Holmes, B.B.; Furman, J.L.; Mahan, T.E.; Yamasaki, T.R.; Mirbaha, H.; Eades, W.C.; Belaygorod, L.; Cairns, N.J.; Holtzman, D.M.; Diamond, M.I. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, E4376–E4385. [Google Scholar] [CrossRef]
- Aoyagi, H.; Hasegawa, M.; Tamaoka, A. Fibrillogenic Nuclei Composed of P301L Mutant Tau Induce Elongation of P301L Tau but Not Wild-type Tau. J. Biol. Chem. 2007, 282, 20309–20318. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, T.K.; Kalverda, A.P.; Thompson, G.S.; Radford, S.E. Mechanisms of amyloid formation revealed by solution NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2015, 88–89, 86–104. [Google Scholar] [CrossRef] [PubMed]
- Mirbaha, H.; Chen, D.; Morazova, O.A.; Ruff, K.M.; Sharma, A.M.; Liu, X.; Goodarzi, M.; Pappu, R.V.; Colby, D.W.; Mirzaei, H.; et al. Inert and seed-competent tau monomers suggest structural origins of aggregation. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Eschmann, N.A.; Georgieva, E.R.; Ganguly, P.; Borbat, P.P.; Rappaport, M.D.; Akdogan, Y.; Freed, J.H.; Shea, J.-E.; Han, S. Signature of an aggregation-prone conformation of tau. Sci. Rep. 2017, 7, 44739. [Google Scholar] [CrossRef]
- Zhu, S.; Shala, A.; Bezginov, A.; Sljoka, A.; Audette, G.; Wilson, D.J. Hyperphosphorylation of Intrinsically Disordered Tau Protein Induces an Amyloidogenic Shift in Its Conformational Ensemble. PLoS ONE 2015, 10, e0120416. [Google Scholar] [CrossRef]
- Berendsen, H.; Hayward, S. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 2000, 10, 165–169. [Google Scholar] [CrossRef]
- García, A.E. Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 1992, 68, 2696–2699. [Google Scholar] [CrossRef]
- de Groot, B.L.; Hayward, S.; van Aalten, D.M.; Amadei, A.; Berendsen, H.J. Domain motions in bacteriophage T4 lysozyme: A comparison between molecular dynamics and crystallographic data. Proteins 1998, 31, 116–127. [Google Scholar] [CrossRef]
- Kurzbach, D.; Beier, A.; Vanas, A.; Flamm, A.G.; Platzer, G.; Schwarz, T.C.; Konrat, R. NMR probing and visualization of correlated structural fluctuations in intrinsically disordered proteins. Phys. Chem. Chem. Phys. 2017, 19, 10651–10656. [Google Scholar] [CrossRef]
- Clore, G.M.; Tang, C.; Iwahara, J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 2007, 17, 603–616. [Google Scholar] [CrossRef]
- Clore, G.M.; Iwahara, J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009, 109, 4108–4139. [Google Scholar] [CrossRef] [PubMed]
- Kurzbach, D.; Vanas, A.; Flamm, A.G.; Tarnoczi, N.; Kontaxis, G.; Maltar-Strmecki, N.; Widder, K.; Hinderberger, D.; Konrat, R. Detection of correlated conformational fluctuations in intrinsically disordered proteins through paramagnetic relaxation interference. Phys. Chem. Chem. Phys. PCCP 2016, 18, 5753–5758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beier, A.; Schwarz, T.C.; Kurzbach, D.; Platzer, G.; Tribuzio, F.; Konrat, R. Modulation of Correlated Segment Fluctuations in IDPs upon Complex Formation as an Allosteric Regulatory Mechanism. J. Mol. Biol. 2018, 430, 2439–2452. [Google Scholar] [CrossRef] [PubMed]
- Kurzbach, D.; Kontaxis, G.; Coudevylle, N.; Konrat, R. NMR Spectroscopic Studies of the Conformational Ensembles of Intrinsically Disordered Proteins. In Intrinsically Disordered Proteins Studied by NMR Spectroscopy; Felli, I.C., Pierattelli, R., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 149–185. [Google Scholar] [CrossRef]
- Konrat, R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 2014, 241, 74–85. [Google Scholar] [CrossRef]
- Palmer Iii, A.G. Probing molecular motion by NMR. Curr. Opin. Struct. Biol. 1997, 7, 732–737. [Google Scholar] [CrossRef]
- Palmer, A.G., 3rd. Nmr probes of molecular dynamics: Overview and comparison with other techniques. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 129–155. [Google Scholar] [CrossRef]
- Křížová, H.; Žídek, L.; Stone, M.; Novotny, M.; Sklenář, V. Temperature-dependent spectral density analysis applied to monitoring backbone dynamics of major urinary protein-I complexed with the pheromone 2-sec-butyl-4,5-dihydrothiazole*. J. Biomol. NMR 2004, 28, 369–384. [Google Scholar] [CrossRef]
- Andrec, M.; Montelione, G.T.; Levy, R.M. Lipari–Szabo mapping: A graphical approach to Lipari–Szabo analysis of NMR relaxation data using reduced spectral density mapping. J. Biomol. NMR 2000, 18, 83–100. [Google Scholar] [CrossRef]
- Lefèvre, J.F.; Dayie, K.T.; Peng, J.W.; Wagner, G. Internal Mobility in the Partially Folded DNA Binding and Dimerization Domains of GAL4: NMR Analysis of the N−H Spectral Density Functions†. Biochemistry 1996, 35, 2674–2686. [Google Scholar] [CrossRef]
- d’Auvergne, E.; Gooley, P. Optimisation of NMR dynamic models II. A new methodology for the dual optimisation of the model-free parameters and the Brownian rotational diffusion tensor. J. Biomol. NMR 2008, 40, 121–133. [Google Scholar] [CrossRef]
- d’Auvergne, E.; Gooley, P. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR 2008, 40, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 58, 62–87. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Sykes, B.D. Chemical shifts as a tool for structure determination. In Methods in Enzymology; Thomas, L., James, N.J.O., Eds.; Academic Press: London, UK, 1994; Volume 239, pp. 363–392. [Google Scholar]
- Wishart, D.S.; Case, D.A. Use of chemical shifts in macromolecular structure determination. In Methods in Enzymology; Thomas, L., James, V.D.U.S., Eds.; Academic Press: Amsterdam, The Netherlands, 2002; Volume 338, pp. 3–34. [Google Scholar]
- Camilloni, C.; De Simone, A.; Vranken, W.F.; Vendruscolo, M. Determination of Secondary Structure Populations in Disordered States of Proteins Using Nuclear Magnetic Resonance Chemical Shifts. Biochemistry 2012, 51, 2224–2231. [Google Scholar] [CrossRef] [PubMed]
- Tamiola, K.; Acar, B.; Mulder, F.A.A. Sequence-Specific Random Coil Chemical Shifts of Intrinsically Disordered Proteins. J. Am. Chem. Soc. 2010, 132, 18000–18003. [Google Scholar] [CrossRef] [PubMed]
- Tamiola, K.; Frans. Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem. Soc. Trans. 2012, 40, 1014–1020. [Google Scholar] [CrossRef]
- Kurzbach, D.; Canet, E.; Flamm, A.G.; Jhajharia, A.; Weber, E.M.M.; Konrat, R.; Bodenhausen, G. Investigation of Intrinsically Disordered Proteins through Exchange with Hyperpolarized Water. Angew. Chem. Int. Ed. 2017, 56, 389–392. [Google Scholar] [CrossRef]
- Jensen, M.R.; Ruigrok, R.W.H.; Blackledge, M. Describing intrinsically disordered proteins at atomic resolution by NMR. Curr. Opin. Struct. Biol. 2013, 23, 426–435. [Google Scholar] [CrossRef]
- Sengupta, A.; Novak, M.; Grundke-Iqbal, I.; Iqbal, K. Regulation of phosphorylation of tau by cyclin-dependent kinase 5 and glycogen synthase kinase-3 at substrate level. FEBS Lett. 2006, 580, 5925–5933. [Google Scholar] [CrossRef]
- Goode, B.L.; Denis, P.E.; Panda, D.; Radeke, M.J.; Miller, H.P.; Wilson, L.; Feinstein, S.C. Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol. Biol. Cell 1997, 8, 353–365. [Google Scholar] [CrossRef]
- Sibille, N.; Sillen, A.; Leroy, A.; Wieruszeski, J.-M.; Mulloy, B.; Landrieu, I.; Lippens, G. Structural Impact of Heparin Binding to Full-Length Tau As Studied by NMR Spectroscopy. Biochemistry 2006, 45, 12560–12572. [Google Scholar] [CrossRef]
- Lippens, G.; Sillen, A.; Landrieu, I.; Amniai, L.; Sibille, N.; Barbier, P.; Leroy, A.; Hanoulle, X.; Wieruszeski, J.-M. Tau Aggregation in Alzheimer’s Disease. Prion 2007, 1, 21–25. [Google Scholar] [CrossRef]
- Amniai, L.; Barbier, P.; Sillen, A.; Wieruszeski, J.-M.; Peyrot, V.; Lippens, G.; Landrieu, I. Alzheimer disease specific phosphoepitopes of Tau interfere with assembly of tubulin but not binding to microtubules. FASEB J. 2009, 23, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Mylonas, E.; Hascher, A.; Bernadó, P.; Blackledge, M.; Mandelkow, E.; Svergun, D.I. Domain Conformation of Tau Protein Studied by Solution Small-Angle X-ray Scattering†. Biochemistry 2008, 47, 10345–10353. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, S.; von Bergen, M.; Brutlach, H.; Steinhoff, H.-J.; Mandelkow, E. Global Hairpin Folding of Tau in Solution. Biochemistry 2006, 45, 2283–2293. [Google Scholar] [CrossRef]
- Nath, A.; Sammalkorpi, M.; DeWitt, D.C.; Trexler, A.J.; Elbaum-Garfinkle, S.; O’Hern, C.S.; Rhoades, E. The Conformational Ensembles of α-Synuclein and Tau: Combining Single-Molecule FRET and Simulations. Biophys. J. 2012, 103, 1940–1949. [Google Scholar] [CrossRef] [PubMed]
- Wischik, C.M.; Novak, M.; Thogersen, H.C.; Edwards, P.C.; Runswick, M.J.; Jakes, R.; Walker, J.E.; Milstein, C.; Roth, M.; Klug, A. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. USA 1988, 85, 4506–4510. [Google Scholar] [CrossRef] [PubMed]
- Friedhoff, P.; Von Bergen, M.; Mandelkow, E.-M.; Mandelkow, E. Structure of tau protein and assembly into paired helical filaments. Biochim. Biophys. Acta (BBA) Mole. Basis Dis. 2000, 1502, 122–132. [Google Scholar] [CrossRef]
- Kikhney, A.G.; Svergun, D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015, 589, 2570–2577. [Google Scholar] [CrossRef]
- Han, D.; Qureshi, H.Y.; Lu, Y.; Paudel, H.K. Familial FTDP-17 Missense Mutations Inhibit Microtubule Assembly-promoting Activity of Tau by Increasing Phosphorylation at Ser202in Vitro. J. Biol. Chem. 2009, 284, 13422–13433. [Google Scholar] [CrossRef]
- Morris, M.; Knudsen, G.M.; Maeda, S.; Trinidad, J.C.; Ioanoviciu, A.; Burlingame, A.L.; Mucke, L. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat. Neurosci. 2015, 18, 1183–1189. [Google Scholar] [CrossRef]
- Mukrasch, M.D.; Markwick, P.; Biernat, J.; von Bergen, M.; Bernadó, P.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M.; Blackledge, M. Highly Populated Turn Conformations in Natively Unfolded Tau Protein Identified from Residual Dipolar Couplings and Molecular Simulation. J. Am. Chem. Soc. 2007, 129, 5235–5243. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, J.; Fairbrother, W.J.; Palmer, A.G., III; Skelton, N.J. Protein NMR Spectroscopy; Academic Press, Inc.: New York, NY, USA, 1996. [Google Scholar]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Iwahara, J.; Koshiba, S.; Tomizawa, T.; Tochio, N.; Güntert, P.; Kigawa, T.; Yokoyama, S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR 2007, 39, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B. Using NMRView to Visualize and Analyze the NMR Spectra of Macromolecules. In Protein NMR Techniques; Downing, A.K., Ed.; Humana Press: Totowa, NJ, USA, 2004; Volume 278, pp. 313–352. [Google Scholar]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2007, 36, D402–D408. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- Iwahara, J.; Tang, C.; Marius Clore, G. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 2007, 184, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Flory, P.J. Statistical Mechanics of Chain Molecules; Oxford University Press: New York, NY, USA, 1989. [Google Scholar]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawasaki, R.; Tate, S.-i. Impact of the Hereditary P301L Mutation on the Correlated Conformational Dynamics of Human Tau Protein Revealed by the Paramagnetic Relaxation Enhancement NMR Experiments. Int. J. Mol. Sci. 2020, 21, 3920. https://doi.org/10.3390/ijms21113920
Kawasaki R, Tate S-i. Impact of the Hereditary P301L Mutation on the Correlated Conformational Dynamics of Human Tau Protein Revealed by the Paramagnetic Relaxation Enhancement NMR Experiments. International Journal of Molecular Sciences. 2020; 21(11):3920. https://doi.org/10.3390/ijms21113920
Chicago/Turabian StyleKawasaki, Ryosuke, and Shin-ichi Tate. 2020. "Impact of the Hereditary P301L Mutation on the Correlated Conformational Dynamics of Human Tau Protein Revealed by the Paramagnetic Relaxation Enhancement NMR Experiments" International Journal of Molecular Sciences 21, no. 11: 3920. https://doi.org/10.3390/ijms21113920
APA StyleKawasaki, R., & Tate, S.-i. (2020). Impact of the Hereditary P301L Mutation on the Correlated Conformational Dynamics of Human Tau Protein Revealed by the Paramagnetic Relaxation Enhancement NMR Experiments. International Journal of Molecular Sciences, 21(11), 3920. https://doi.org/10.3390/ijms21113920