Recent Advances in the Molecular Biology of Systemic Mastocytosis: Implications for Diagnosis, Prognosis, and Therapy
Abstract
1. Introduction
2. KIT Activating Mutations as the Main Disease Driver
3. Mutant KIT D816V Receptor Induces Abnormal Signaling Pathways
4. In Which Cell Type Does the D816V Mutation Arise?
5. KIT Mutations: Diagnostic and Prognostic Considerations
- -
- RT-PCR combined with restriction fragment length polymorphism (RFLP): genetic analysis by RFLP is one of the most commonly used techniques for the identification of a known sequence variant. The RT–PCR followed by the RFLP assay represents a rapid, cheap, and easy method that allows the detection of KIT D816V mutation with a sensitivity of 0.05% [68,69]. However, it cannot reveal D816 variant mutations, nor can it detect mutations at other codons. Moreover, it does not allow for quantitation of the allele burden.
- -
- Nested RT-PCR followed by D-HPLC of PCR amplicons: this technique allows the identification of all the variants at codon 816. Sensitivity has been reported to be modest at 0.5–1% [70]. The method is relatively time-consuming and requires sample pooling for cost-effectiveness. Moreover, D-HPLC instruments are not widely available.
- -
- Peptide nucleic acid-mediated (PNA)-mediated PCR: this can be used for the detection of all the variants at codon 816 as well as at adjacent codons with a sensitivity of 0.1% [71]. Similar to the previous methods, PNA-mediated PCR is not quantitative. However, this is the recommended method for formalin-fixed, paraffin-embedded tissues [72].
- -
- Allele-specific oligonucleotide (ASO)-quantitative PCR (ASO-qPCR): this is one of the most sensitive methods for the identification and quantitation (allele burden) of the D816V mutation in different substrates. With ASO-qPCR it is possible to detect less than 0.01% KIT D816V mutation-positive cells [73,74,75]. Unfortunately, D816 variant mutations cannot be picked up by this assay.
- -
- Droplet digital PCR (ddPCR): a recent validation study has shown the potential of ddPCR to become the method of choice for D816V detection and quantitation because of its precision, accuracy, and sensitivity (0.01%) [76]. Moreover, ddPCR has proven capable to robustly quantitate the allele burden also in formalin-fixed, paraffin-embedded tissues [77].
| Strategy | Pros | Cons | Lower Detection Limit | Ref |
|---|---|---|---|---|
| PCR + RFLP (Hinf I) | - Simple - Rapid - Reliable - Inexpensive | - Cannot reveal D816 variant mutations - Not quantitative | 0.05% | Fritsche-Polanz, Br J Haematol 2001 [69] |
| Nested RT-PCR + D-HPLC | - Allows to identify all the variants at codon 816 or adjacent positions | - Not quantitative - Time-consuming - Needs dedicated, expensive instrumentation | 0.5–1% | Erben, Ann Hematol 2014 [70] |
| PNA-mediated PCR clamping + melting curve analysis | - Allows to identify all the variants at codon 816 or adjacent positions - Works well also on FFPE tissues | - Not quantitative | 0.1% | Sotlar, Am J Pathol 2003 [71] |
| ASO-qPCR | - Rapid - Relatively inexpensive - Quantitative | - Cannot reveal D816 variant mutations - Standardization and harmonization not yet undertaken | 0.01% | Kristensen, J Mol Diagn 2011 [74] |
| ddPCR | - Simple - Rapid - Relatively inexpensive - Quantitative - Works well also on FFPE tissues | - Cannot reveal D816 variant mutations - Standardization and harmonization not yet undertaken | 0.01% | Greiner, Clin Chem 2018 [76] |
6. Mutations in Genes Other Than KIT
7. Cytogenetic Abnormalities
8. New Clinical and Clinico-Molecular Prognostic Scoring Systems in SM
| Variable | REMA [104] | * MARS [105] | * MAPS [106] | * IPSM [108] | |||
|---|---|---|---|---|---|---|---|
| ISM Only | AdvSM Only | All | Non-AdvSM | AdvSM | |||
| PFS | OS | LFS, OS | OS | PFS, OS | PFS, OS | ||
| WHO class | + | ||||||
| Age | ≥60 years | + | + | + | + | ||
| Anemia | ≤10 g/dL | + | |||||
| ≤11 g/dL | + | ||||||
| Thrombocytopenia | <100 × 109/L | + | + | ||||
| <150 × 109/L | + | ||||||
| Leukocytosis | ≥16 × 109/L | + | |||||
| Serum tryptase | ≥125 ng/mL | + | |||||
| Serum β2m | >2.5 μg/mL | + | |||||
| Serum ALP | >100 U/L | + | |||||
| >ULN | + | ||||||
| Mutations | KIT D816V in BM | + (VAF ≥ 1%) | |||||
| Myeloid mutations (NGS) | + A/R/D (VAF ≥ 30%) | + A/R/D (VAF ≥ 30%) | + (S/A/R) | + (A/R/NRAS) | |||
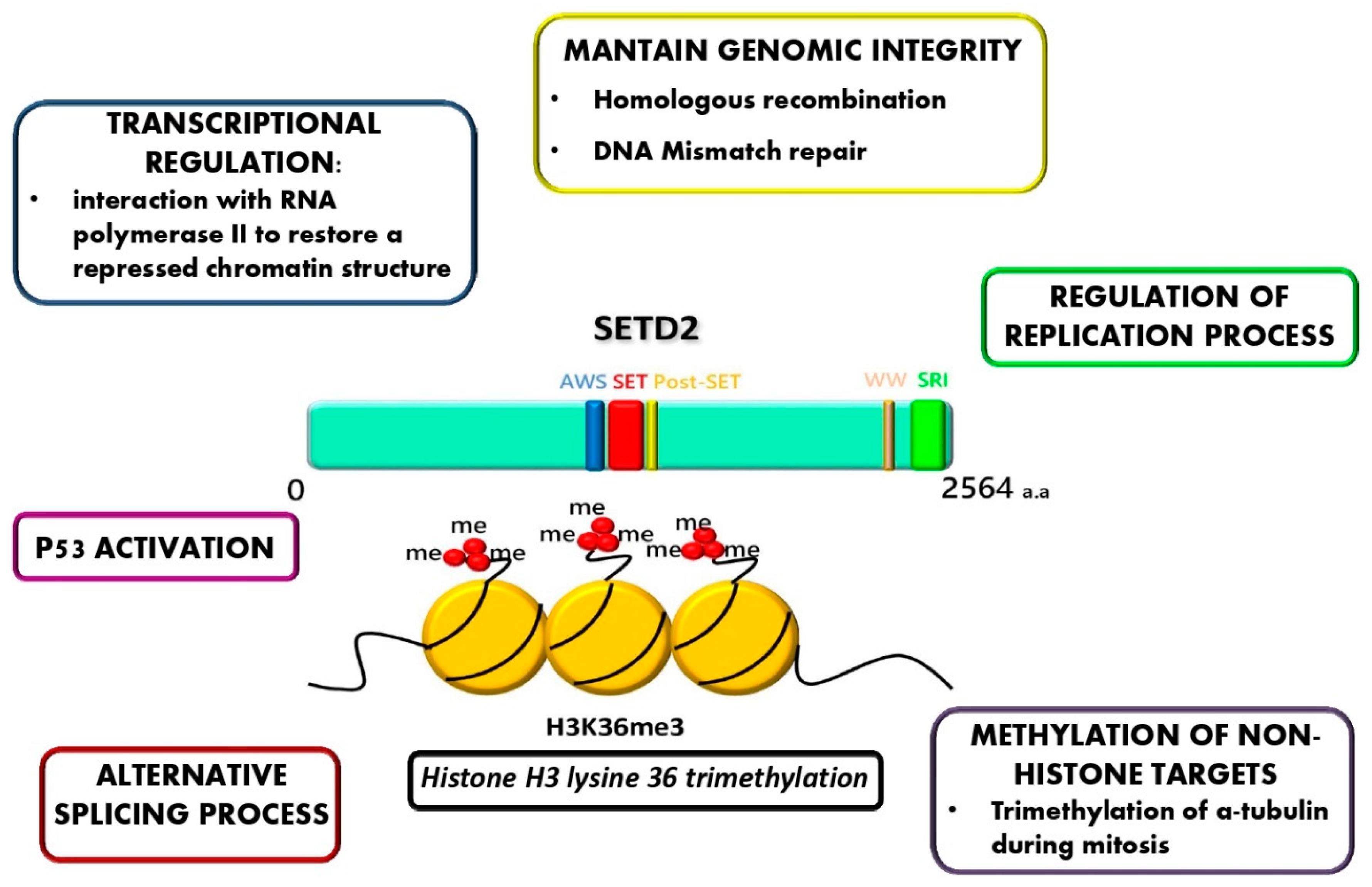
9. Novel Molecular Alterations: SETD2 Non-Genomic Loss of Function in AdvSM
10. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Ehrlich, P. Beitrage zur Kenntnis der granulierten. Bindegewebszellen und der eosinophilen Leukozyten. Arch. Anat. Physiol. 1879, 3, 166–169. [Google Scholar]
- Reber, L.L.; Sibilano, R.; Mukai, K.; Galli, S.J. Potential effector and immunoregulatory functions of mast cells in mucosal immunity. Mucosal Immunol. 2015, 8, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H. WHO classification of tumours of haematopoietic and lymphoid tissues 4th ed (revised).; IARC Press: Lyon, France, 2017; ISBN 928324494X. [Google Scholar]
- Valent, P.; Akin, C.; Metcalfe, D.D. Mastocytosis: 2016 updated WHO classification and novel emerging treatment concepts. Blood J. Am. Soc. Hematol. 2017, 129, 1420–1427. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.-H.; Tefferi, A.; Lasho, T.L.; Finke, C.; Patnaik, M.; Butterfield, J.H.; McClure, R.F.; Li, C.-Y.; Pardanani, A. Systemic mastocytosis in 342 consecutive adults: survival studies and prognostic factors. Blood J. Am. Soc. Hematol. 2009, 113, 5727–5736. [Google Scholar] [CrossRef]
- Pieri, L.; Bonadonna, P.; Elena, C.; Papayannidis, C.; Grifoni, F.I.; Rondoni, M.; Girlanda, S.; Mauro, M.; Magliacane, D.; Elli, E.M. Clinical presentation and management practice of systemic mastocytosis. A survey on 460 Italian patients. Am. J. Hematol. 2016, 91, 692–699. [Google Scholar] [CrossRef]
- Escribano, L.; Álvarez-Twose, I.; Sánchez-Muñoz, L.; Garcia-Montero, A.; Núñez, R.; Almeida, J.; Jara-Acevedo, M.; Teodósio, C.; García-Cosío, M.; Bellas, C. Prognosis in adult indolent systemic mastocytosis: a long-term study of the Spanish Network on Mastocytosis in a series of 145 patients. J. Allergy Clin. Immunol. 2009, 124, 514–521. [Google Scholar] [CrossRef]
- Pardanani, A.; Tefferi, A. Systemic mastocytosis in adults: a review on prognosis and treatment based on 342 Mayo Clinic patients and current literature. Curr. Opin. Hematol. 2010, 17, 125–132. [Google Scholar] [CrossRef]
- Trizuljak, J.; Sperr, W.R.; Nekvindová, L.; Oude Elberink, H.; Gleixner, K.V.; Gorska, A.; Lange, M.; Hartmann, K.; Illerhaus, A.; Bonifacio, M. Clinical Features and Survival of Patients with Indolent Systemic Mastocytosis defined by the Updated WHO Classification. Allergy 2020. [Google Scholar] [CrossRef]
- Tefferi, A.; Shah, S.; Reichard, K.K.; Hanson, C.A.; Pardanani, A. Smoldering mastocytosis: Survival comparisons with indolent and aggressive mastocytosis. Am. J. Hematol. 2019, 94, E1–E2. [Google Scholar] [CrossRef]
- Yarden, Y.; Kuang, W.-J.; Yang-Feng, T.; Coussens, L.; Munemitsu, S.; Dull, T.J.; Chen, E.; Schlessinger, J.; Francke, U.; Ullrich, A. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987, 6, 3341–3351. [Google Scholar] [CrossRef]
- Lev, S.; Yarden, Y.; Givol, D. Receptor functions and ligand-dependent transforming potential of a chimeric kit proto-oncogene. Mol. Cell. Biol. 1990, 10, 6064–6068. [Google Scholar] [CrossRef] [PubMed]
- Besmer, P. The kit ligand encoded at the murine Steel locus: a pleiotropic growth and differentiation factor. Curr. Opin. Cell Biol. 1991, 3, 939–946. [Google Scholar] [CrossRef]
- Broxmeyer, H.E. Interactions of colony stimulating factors, other modulating cytokines and hematopoietic progenitor cells. Laboratory and clinical studies. Leukemia 1992, 6, 38–40. [Google Scholar] [PubMed]
- Giebel, L.B.; Strunk, K.M.; Holmes, S.A.; Spritz, R.A. Organization and nucleotide sequence of the human KIT (mast/stem cell growth factor receptor) proto-oncogene. Oncogene 1992, 7, 2207–2217. [Google Scholar]
- Tsujimura, T.; Morii, E.; Nozaki, M.; Hashimoto, K.; Moriyama, Y.; Takebayashi, K.; Kondo, T.; Kanakura, Y.; Kitamura, Y. Involvement of transcription factor encoded by the mi locus in the expression of c-kit receptor tyrosine kinase in cultured mast cells of mice. Blood 1996, 88, 1225–1233. [Google Scholar] [CrossRef]
- Welham, M.J.; Schrader, J.W. Modulation of c-kit mRNA and protein by hemopoietic growth factors. Mol. Cell. Biol. 1991, 11, 2901–2904. [Google Scholar] [CrossRef][Green Version]
- Blechman, J.M.; Lev, S.; Givol, D.; Yarden, Y. Structure-function analyses of the kit receptor for the steel factor. Stem Cells 1993, 11, 12–21. [Google Scholar] [CrossRef]
- Crosier, P.S.; Ricciardi, S.T.; Hall, L.R.; Vitas, M.R.; Clark, S.C.; Crosier, K.E. Expression of isoforms of the human receptor tyrosine kinase c-kit in leukemic cell lines and acute myeloid leukemia. Blood 1993, 82, 1151–1158. [Google Scholar] [CrossRef]
- Kaufman, E. Cloning and structural analysis of the human c-kit gene. Oncogene 1992, 7, 1259–1266. [Google Scholar]
- Lyman, S.D.; Jacobsen, S.E.W. c-kit ligand and Flt3 ligand: stem/progenitor cell factors with overlapping yet distinct activities. Blood J. Am. Soc. Hematol. 1998, 91, 1101–1134. [Google Scholar] [CrossRef]
- Reith, A.D.; Ellis, C.; Lyman, S.D.; Anderson, D.M.; Williams, D.E.; Bernstein, A.; Pawson, T. Signal transduction by normal isoforms and W mutant variants of the Kit receptor tyrosine kinase. EMBO J. 1991, 10, 2451–2459. [Google Scholar] [CrossRef] [PubMed]
- Ashman, L.K.; Griffith, R. Therapeutic targeting of c-KIT in cancer. Expert Opin. Investig. Drugs 2013, 22, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Yavuz, A.S.; Lipsky, P.E.; Yavuz, S.; Metcalfe, D.D.; Akin, C. Evidence for the involvement of a hematopoietic progenitor cell in systemic mastocytosis from single-cell analysis of mutations in the c-kit gene. Blood 2002, 100, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004, 5, 464. [Google Scholar] [CrossRef]
- Lin, Z.-H.; Han, E.M.; Lee, E.S.; Kim, C.W.; Kim, H.K.; Kim, I.; Kim, Y.-S. A distinct expression pattern and point mutation of c-kit in papillary renal cell carcinomas. Mod. Pathol. 2004, 17, 611. [Google Scholar] [CrossRef]
- Hirota, S.; Isozaki, K.; Moriyama, Y.; Hashimoto, K.; Nishida, T.; Ishiguro, S.; Kawano, K.; Hanada, M.; Kurata, A.; Takeda, M. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science (80-) 1998, 279, 577–580. [Google Scholar] [CrossRef]
- Longley, B.J.; Reguera, M.J.; Ma, Y. Classes of c-KIT activating mutations: proposed mechanisms of action and implications for disease classification and therapy. Leuk. Res. 2001, 25, 571–576. [Google Scholar] [CrossRef]
- Ma, Y.; Longley, B.J.; Wang, X.; Blount, J.L.; Langley, K.; Caughey, G.H. Clustering of activating mutations in c-KIT’s juxtamembrane coding region in canine mast cell neoplasms. J. Investig. Dermatol. 1999, 112, 165–170. [Google Scholar] [CrossRef]
- Ma, Y.; Cunningham, M.E.; Wang, X.; Ghosh, I.; Regan, L.; Longley, B.J. Inhibition of spontaneous receptor phosphorylation by residues in a putative α-helix in the KIT intracellular juxtamembrane region. J. Biol. Chem. 1999, 274, 13399–13402. [Google Scholar] [CrossRef]
- Kanakura, Y.; Furitsu, T.; Tsujimura, T.; Butterfield, J.H.; Ashman, L.K.; Ikeda, H.; Kitayama, H.; Kanayama, Y.; Matsuzawa, Y.; Kitamura, Y. Activating mutations of the c-kit proto-oncogene in a human mast cell leukemia cell line. Leukemia 1994, 8, S18–S22. [Google Scholar]
- Kitayama, H.; Kanakura, Y.; Furitsu, T.; Tsujimura, T.; Oritani, K.; Ikeda, H.; Sugahara, H.; Mitsui, H.; Kanayama, Y.; Kitamura, Y. Constitutively activating mutations of c-kit receptor tyrosine kinase confer factor-independent growth and tumorigenicity of factor-dependent hematopoietic cell lines. Blood 1995, 85, 790–798. [Google Scholar] [CrossRef]
- Georgin-Lavialle, S.; Lhermitte, L.; Dubreuil, P.; Chandesris, M.-O.; Hermine, O.; Damaj, G. Mast cell leukemia. Blood J. Am. Soc. Hematol. 2013, 121, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Bodemer, C.; Hermine, O.; Palmérini, F.; Yang, Y.; Grandpeix-Guyodo, C.; Leventhal, P.S.; Hadj-Rabia, S.; Nasca, L.; Georgin-Lavialle, S.; Cohen-Akenine, A. Pediatric mastocytosis is a clonal disease associated with D816V and other activating c-KIT mutations. J. Investig. Dermatol. 2010, 130, 804–815. [Google Scholar] [CrossRef] [PubMed]
- Nagata, H.; Worobec, A.S.; Metcalfe, D.D. Identification of a polymorphism in the transmembrane domain of the protooncogene c-kit in healthy subjects. Exp. Clin. Immunogenet. 1996, 13, 210–214. [Google Scholar] [PubMed]
- Laine, E.; de Beauchêne, I.C.; Perahia, D.; Auclair, C.; Tchertanov, L. Mutation D816V alters the internal structure and dynamics of c-KIT receptor cytoplasmic region: implications for dimerization and activation mechanisms. PLoS Comput. Biol. 2011, 7, e1002068. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Kreisel, F.; Cain, J.; Colson, A.; Tomasson, M.H. Neoplasia driven by mutant c-KIT is mediated by intracellular, not plasma membrane, receptor signaling. Mol. Cell. Biol. 2007, 27, 267–282. [Google Scholar] [CrossRef]
- Chian, R.; Young, S.; Danilkovitch-Miagkova, A.; Rönnstrand, L.; Leonard, E.; Ferrao, P.; Ashman, L.; Linnekin, D. Phosphatidylinositol 3 kinase contributes to the transformation of hematopoietic cells by the D816V c-Kit mutant. Blood J. Am. Soc. Hematol. 2001, 98, 1365–1373. [Google Scholar] [CrossRef]
- Harir, N.; Boudot, C.; Friedbichler, K.; Sonneck, K.; Kondo, R.; Martin-Lannerée, S.; Kenner, L.; Kerenyi, M.; Yahiaoui, S.; Gouilleux-Gruart, V. Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood J. Am. Soc. Hematol. 2008, 112, 2463–2473. [Google Scholar] [CrossRef]
- Baumgartner, C.; Cerny-Reiterer, S.; Sonneck, K.; Mayerhofer, M.; Gleixner, K.V.; Fritz, R.; Kerenyi, M.; Boudot, C.; Gouilleux, F.; Kornfeld, J.-W. Expression of activated STAT5 in neoplastic mast cells in systemic mastocytosis: subcellular distribution and role of the transforming oncoprotein KIT D816V. Am. J. Pathol. 2009, 175, 2416–2429. [Google Scholar] [CrossRef]
- Tanaka, A.; Konno, M.; Muto, S.; Kambe, N.; Morii, E.; Nakahata, T.; Itai, A.; Matsuda, H. A novel NF-κB inhibitor, IMD-0354, suppresses neoplastic proliferation of human mast cells with constitutively activated c-kit receptors. Blood 2005, 105, 2324–2331. [Google Scholar] [CrossRef]
- Gabillot-Carré, M.; Lepelletier, Y.; Humbert, M.; de Sepuvelda, P.; Hamouda, N.B.; Zappulla, J.P.; Liblau, R.; Ribadeau-Dumas, A.; Machavoine, F.; Letard, S. Rapamycin inhibits growth and survival of D816V-mutated c-kit mast cells. Blood 2006, 108, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Jelacic, T.; Linnekin, D. PKCδ plays opposite roles in growth mediated by wild-type Kit and an oncogenic Kit mutant. Blood 2005, 105, 1923–1929. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodon, J.; Dienstmann, R.; Serra, V.; Tabernero, J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 2013, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Hoermann, G.; Cerny-Reiterer, S.; Perné, A.; Klauser, M.; Hoetzenecker, K.; Klein, K.; Müllauer, L.; Gröger, M.; Nijman, S.M.B.; Klepetko, W. Identification of oncostatin M as a STAT5-dependent mediator of bone marrow remodeling in KIT D816V-positive systemic mastocytosis. Am. J. Pathol. 2011, 178, 2344–2356. [Google Scholar] [CrossRef]
- Morales, J.K.; Falanga, Y.T.; Depcrynski, A.; Fernando, J.; Ryan, J.J. Mast cell homeostasis and the JAK–STAT pathway. Genes Immun. 2010, 11, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Ghosh, J.; Ramdas, B.; Mali, R.S.; Martin, H.; Kobayashi, M.; Vemula, S.; Canela, V.H.; Waskow, E.R.; Visconte, V. Regulation of Stat5 by FAK and PAK1 in oncogenic FLT3-and KIT-driven leukemogenesis. Cell Rep. 2014, 9, 1333–1348. [Google Scholar] [CrossRef]
- Martin, H.; Mali, R.S.; Ma, P.; Chatterjee, A.; Ramdas, B.; Sims, E.; Munugalavadla, V.; Ghosh, J.; Mattingly, R.R.; Visconte, V. Pak and Rac GTPases promote oncogenic KIT–induced neoplasms. J. Clin. Investig. 2013, 123, 4449–4463. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-N.; Brandal, S.; Noel, P.; Wentzel, E.; Mendell, J.T.; McDevitt, M.A.; Kapur, R.; Carter, M.; Metcalfe, D.D.; Takemoto, C.M. KIT signaling regulates MITF expression through miRNAs in normal and malignant mast cell proliferation. Blood J. Am. Soc. Hematol. 2011, 117, 3629–3640. [Google Scholar] [CrossRef]
- Blatt, K.; Herrmann, H.; Mirkina, I.; Hadzijusufovic, E.; Peter, B.; Strommer, S.; Hoermann, G.; Mayerhofer, M.; Hoetzenecker, K.; Klepetko, W. The PI3-kinase/mTOR-targeting drug NVP-BEZ235 inhibits growth and IgE-dependent activation of human mast cells and basophils. PLoS ONE 2012, 7, e29925. [Google Scholar] [CrossRef]
- Parikh, S.A.; Kantarjian, H.M.; Richie, M.A.; Cortes, J.E.; Verstovsek, S. Experience with everolimus (RAD001), an oral mammalian target of rapamycin inhibitor, in patients with systemic mastocytosis. Leuk. Lymphoma 2010, 51, 269–274. [Google Scholar] [CrossRef]
- Smrž, D.; Kim, M.-S.; Zhang, S.; Mock, B.A.; Smržová, Š.; DuBois, W.; Simakova, O.; Maric, I.; Wilson, T.M.; Metcalfe, D.D. mTORC1 and mTORC2 differentially regulate homeostasis of neoplastic and non-neoplastic human mast cells. Blood J. Am. Soc. Hematol. 2011, 118, 6803–6813. [Google Scholar] [CrossRef]
- Mali, R.S.; Ma, P.; Zeng, L.-F.; Martin, H.; Ramdas, B.; He, Y.; Sims, E.; Nabinger, S.; Ghosh, J.; Sharma, N. Role of SHP2 phosphatase in KIT-induced transformation: identification of SHP2 as a druggable target in diseases involving oncogenic KIT. Blood J. Am. Soc. Hematol. 2012, 120, 2669–2678. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Voisset, E.; Lopez, S.; Dubreuil, P.; De Sepulveda, P. The tyrosine kinase FES is an essential effector of KITD816V proliferation signal. Blood J. Am. Soc. Hematol. 2007, 110, 2593–2599. [Google Scholar] [CrossRef] [PubMed]
- Baldus, S.E.; Zirbes, T.K.; Thiele, J.; Eming, S.A.; Henz, B.M.; Hartmann, K. Altered apoptosis and cell cycling of mast cells in bone marrow lesions of patients with systemic mastocytosis. Haematologica 2004, 89, 1525–1527. [Google Scholar] [PubMed]
- Cerveró, C.; Escribano, L.; San Miguel, J.F.; Díaz-Agustín, B.; Bravo, P.; Villarrubia, J.; García-Sanz, R.; Velasco, J.L.; Herrera, P.; Vargas, M. Expression of bcl-2 by human bone marrow mast cells and its overexpression in mast cell leukemia. Am. J. Hematol. 1999, 60, 191–195. [Google Scholar] [CrossRef]
- Hartmann, K.; Artuc, M.; Baldus, S.E.; Zirbes, T.K.; Hermes, B.; Thiele, J.; Mekori, Y.A.; Henz, B.M. Expression of Bcl-2 and Bcl-xL in cutaneous and bone marrow lesions of mastocytosis. Am. J. Pathol. 2003, 163, 819–826. [Google Scholar] [CrossRef]
- Aichberger, K.J.; Mayerhofer, M.; Gleixner, K.V.; Krauth, M.-T.; Gruze, A.; Pickl, W.F.; Wacheck, V.; Selzer, E.; Müllauer, L.; Agis, H. Identification of MCL1 as a novel target in neoplastic mast cells in systemic mastocytosis: inhibition of mast cell survival by MCL1 antisense oligonucleotides and synergism with PKC412. Blood 2007, 109, 3031–3041. [Google Scholar] [CrossRef]
- Aichberger, K.J.; Gleixner, K.V.; Mirkina, I.; Cerny-Reiterer, S.; Peter, B.; Ferenc, V.; Kneidinger, M.; Baumgartner, C.; Mayerhofer, M.; Gruze, A. Identification of proapoptotic Bim as a tumor suppressor in neoplastic mast cells: role of KIT D816V and effects of various targeted drugs. Blood J. Am. Soc. Hematol. 2009, 114, 5342–5351. [Google Scholar] [CrossRef]
- Peter, B.; Cerny-Reiterer, S.; Hadzijusufovic, E.; Schuch, K.; Stefanzl, G.; Eisenwort, G.; Gleixner, K.V.; Hoermann, G.; Mayerhofer, M.; Kundi, M. The pan-Bcl-2 blocker obatoclax promotes the expression of Puma, Noxa, and Bim mRNA and induces apoptosis in neoplastic mast cells. J. Leukoc. Biol. 2014, 95, 95–104. [Google Scholar] [CrossRef]
- Garcia-Montero, A.C.; Jara-Acevedo, M.; Teodosio, C.; Sanchez, M.L.; Nunez, R.; Prados, A.; Aldanondo, I.; Sanchez, L.; Dominguez, M.; Botana, L.M. KIT mutation in mast cells and other bone marrow hematopoietic cell lineages in systemic mast cell disorders: a prospective study of the Spanish Network on Mastocytosis (REMA) in a series of 113 patients. Blood 2006, 108, 2366–2372. [Google Scholar] [CrossRef]
- Sotlar, K.; Colak, S.; Bache, A.; Berezowska, S.; Krokowski, M.; Bültmann, B.; Valent, P.; Horny, H. Variable presence of KITD816V in clonal haematological non-mast cell lineage diseases associated with systemic mastocytosis (SM–AHNMD). J. Pathol. A J. Pathol. Soc. Gt. Britain Irel. 2010, 220, 586–595. [Google Scholar]
- Zhao, W.; Bueso-Ramos, C.E.; Verstovsek, S.; Barkoh, B.A.; Khitamy, A.A.; Jones, D. Quantitative profiling of codon 816 KIT mutations can aid in the classification of systemic mast cell disease. Leukemia 2007, 21, 1574–1576. [Google Scholar] [CrossRef]
- Taylor, M.L.; Sehgal, D.; Raffeld, M.; Obiakor, H.; Akin, C.; Mage, R.G.; Metcalfe, D.D. Demonstration that mast cells, T cells, and B cells bearing the activating kit mutation D816V occur in clusters within the marrow of patients with mastocytosis. J. Mol. Diagn. 2004, 6, 335–342. [Google Scholar] [CrossRef]
- Valent, P.; Horny, H.-P.; Escribano, L.; Longley, B.J.; Li, C.Y.; Schwartz, L.B.; Marone, G.; Nuñez, R.; Akin, C.; Sotlar, K. Diagnostic criteria and classification of mastocytosis: a consensus proposal. Leuk. Res. 2001, 25, 603–625. [Google Scholar] [CrossRef]
- Jawhar, M.; Schwaab, J.; Schnittger, S.; Sotlar, K.; Horny, H.P.; Metzgeroth, G.; Müller, N.; Schneider, S.; Naumann, N.; Walz, C. Molecular profiling of myeloid progenitor cells in multi-mutated advanced systemic mastocytosis identifies KIT D816V as a distinct and late event. Leukemia 2015, 29, 1115. [Google Scholar] [CrossRef]
- Nemeth, K.; Wilson, T.M.; Ren, J.J.; Sabatino, M.; Stroncek, D.M.; Krepuska, M.; Bai, Y.; Robey, P.G.; Metcalfe, D.D.; Mezey, E. Impaired function of bone marrow stromal cells in systemic mastocytosis. Stem Cell Res. 2015, 15, 42–53. [Google Scholar] [CrossRef]
- Valent, P.; Akin, C.; Escribano, L.; Födinger, M.; Hartmann, K.; Brockow, K.; Castells, M.; Sperr, W.R.; Kluin-Nelemans, H.C.; Hamdy, N.A.T. Standards and standardization in mastocytosis: consensus statements on diagnostics, treatment recommendations and response criteria. Eur. J. Clin. Investig. 2007, 37, 435–453. [Google Scholar] [CrossRef]
- Fritsche-Polanz, R.; Jordan, J.; Feix, A.; Sperr, W.R.; Sunder-Plassmann, G.; Valent, P.; Födinger, M. Mutation analysis of C-KIT in patients with myelodysplastic syndromes without mastocytosis and cases of systemic mastocytosis. Br. J. Haematol. 2001, 113, 357–364. [Google Scholar] [CrossRef]
- Erben, P.; Schwaab, J.; Metzgeroth, G.; Horny, H.-P.; Jawhar, M.; Sotlar, K.; Fabarius, A.; Teichmann, M.; Schneider, S.; Ernst, T. The KIT D816V expressed allele burden for diagnosis and disease monitoring of systemic mastocytosis. Ann. Hematol. 2014, 93, 81–88. [Google Scholar] [CrossRef]
- Sotlar, K.; Escribano, L.; Landt, O.; Möhrle, S.; Herrero, S.; Torrelo, A.; Lass, U.; Horny, H.P.; Bültmann, B. One-step detection of c-kit point mutations using PNA-mediated PCR-clamping and hybridization probes. Am. J. Pathol 2003, 162, 737–746. [Google Scholar] [CrossRef]
- Sotlar, K. C-kit mutational analysis in paraffin material. In Hematological Malignancies; Springer: Berlin, Germany, 2013; pp. 59–78. [Google Scholar]
- Kristensen, T.; Vestergaard, H.; Bindslev-Jensen, C.; Møller, M.B.; Broesby-Olsen, S.; (MastOUH), M.C.O.U.H. Sensitive KIT D816V mutation analysis of blood as a diagnostic test in mastocytosis. Am. J. Hematol. 2014, 89, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, T.; Vestergaard, H.; Møller, M.B. Improved detection of the KIT D816V mutation in patients with systemic mastocytosis using a quantitative and highly sensitive real-time qPCR assay. J. Mol. Diagn. 2011, 13, 180–188. [Google Scholar] [CrossRef]
- Kristensen, T.; Broesby-Olsen, S.; Vestergaard, H.; Bindslev-Jensen, C.; Møller, M.B.; (MastOUH), M.C.O.U.H. Circulating KIT D 816 V mutation-positive non-mast cells in peripheral blood are characteristic of indolent systemic mastocytosis. Eur. J. Haematol. 2012, 89, 42–46. [Google Scholar] [CrossRef]
- Greiner, G.; Gurbisz, M.; Ratzinger, F.; Witzeneder, N.; Simonitsch-Klupp, I.; Mitterbauer-Hohendanner, G.; Mayerhofer, M.; Müllauer, L.; Sperr, W.R.; Valent, P. Digital PCR: a sensitive and precise method for KIT D816V quantification in mastocytosis. Clin. Chem. 2018, 64, 547–555. [Google Scholar] [CrossRef]
- Greiner, G.; Gurbisz, M.; Ratzinger, F.; Witzeneder, N.; Class, S.V.; Eisenwort, G.; Simonitsch-Klupp, I.; Esterbauer, H.; Mayerhofer, M.; Müllauer, L. Molecular quantification of tissue disease burden is a new biomarker and independent predictor of survival in mastocytosis. Haematologica 2020, 105, 366–374. [Google Scholar] [CrossRef]
- Hoermann, G.; Gleixner, K.V.; Dinu, G.E.; Kundi, M.; Greiner, G.; Wimazal, F.; Hadzijusufovic, E.; Mitterbauer, G.; Mannhalter, C.; Valent, P. The KIT D 816 V allele burden predicts survival in patients with mastocytosis and correlates with the WHO type of the disease. Allergy 2014, 69, 810–813. [Google Scholar] [CrossRef]
- Jara-Acevedo, M.; Teodosio, C.; Sanchez-Muñoz, L.; Álvarez-Twose, I.; Mayado, A.; Caldas, C.; Matito, A.; Morgado, J.M.; Muñoz-González, J.I.; Escribano, L. Detection of the KIT D816V mutation in peripheral blood of systemic mastocytosis: diagnostic implications. Mod. Pathol. 2015, 28, 1138–1149. [Google Scholar] [CrossRef]
- Broesby-Olsen, S.; Oropeza, A.R.; Bindslev-Jensen, C.; Vestergaard, H.; Møller, M.B.; Siebenhaar, F.; Kristensen, T.; Mortz, C.G. Recognizing mastocytosis in patients with anaphylaxis: value of KIT D816V mutation analysis of peripheral blood. J. Allergy Clin. Immunol. 2015, 135, 262–264. [Google Scholar] [CrossRef]
- Valent, P.; Escribano, L.; Broesby-Olsen, S.; Hartmann, K.; Grattan, C.; Brockow, K.; Niedoszytko, M.; Nedoszytko, B.; Oude Elberink, J.N.G.; Kristensen, T. Proposed diagnostic algorithm for patients with suspected mastocytosis: a proposal of the European Competence Network on Mastocytosis. Allergy 2014, 69, 1267–1274. [Google Scholar] [CrossRef]
- Arock, M.; Sotlar, K.; Akin, C.; Broesby-Olsen, S.; Hoermann, G.; Escribano, L.; Kristensen, T.K.; Kluin-Nelemans, H.C.; Hermine, O.; Dubreuil, P. KIT mutation analysis in mast cell neoplasms: recommendations of the European Competence Network on Mastocytosis. Leukemia 2015, 29, 1223. [Google Scholar] [CrossRef]
- Kristensen, T.; Broesby-Olsen, S.; Vestergaard, H.; Bindslev-Jensen, C.; Møller, M.B.; (MastOUH), M.C.O.U.H. Targeted ultradeep next-generation sequencing as a method for KIT D 816 V mutation analysis in mastocytosis. Eur. J. Haematol. 2016, 96, 381–388. [Google Scholar] [CrossRef]
- Teodosio, C.; Garcia-Montero, A.C.; Jara-Acevedo, M.; Álvarez-Twose, I.; Sánchez-Muñoz, L.; Almeida, J.; Morgado, J.M.; Matito, A.; Escribano, L.; Orfao, A. An immature immunophenotype of bone marrow mast cells predicts for multilineage D816V KIT mutation in systemic mastocytosis. Leukemia 2012, 26, 951–958. [Google Scholar] [CrossRef]
- Jawhar, M.; Schwaab, J.; Naumann, N.; Horny, H.-P.; Sotlar, K.; Haferlach, T.; Metzgeroth, G.; Fabarius, A.; Valent, P.; Hofmann, W.-K. Response and progression on midostaurin in advanced systemic mastocytosis: KIT D816V and other molecular markers. Blood J. Am. Soc. Hematol. 2017, 130, 137–145. [Google Scholar] [CrossRef]
- Broesby-Olsen, S.; Kristensen, T.; Vestergaard, H.; Brixen, K.; Møller, M.B.; Bindslev-Jensen, C. MastOUH, M.C.O.U.H. KIT D816V mutation burden does not correlate to clinical manifestations of indolent systemic mastocytosis. J. Allergy Clin. Immunol. 2013, 132, 723–728. [Google Scholar] [CrossRef]
- Zappulla, J.P.; Dubreuil, P.; Desbois, S.; Létard, S.; Hamouda, N.B.; Daëron, M.; Delsol, G.; Arock, M.; Liblau, R.S. Mastocytosis in mice expressing human Kit receptor with the activating Asp816Val mutation. J. Exp. Med. 2005, 202, 1635–1641. [Google Scholar] [CrossRef]
- Muñoz-González, J.I.; Jara-Acevedo, M.; Alvarez-Twose, I.; Merker, J.D.; Teodosio, C.; Hou, Y.; Henriques, A.; Roskin, K.M.; Sanchez-Muñoz, L.; Tsai, A.G. Impact of somatic and germline mutations on the outcome of systemic mastocytosis. Blood Adv. 2018, 2, 2814–2828. [Google Scholar] [CrossRef]
- Pardanani, A.; Lasho, T.; Elala, Y.; Wassie, E.; Finke, C.; Reichard, K.K.; Chen, D.; Hanson, C.A.; Ketterling, R.P.; Tefferi, A. Next-generation sequencing in systemic mastocytosis: derivation of a mutation-augmented clinical prognostic model for survival. Am. J. Hematol. 2016, 91, 888–893. [Google Scholar] [CrossRef]
- Schwaab, J.; Schnittger, S.; Sotlar, K.; Walz, C.; Fabarius, A.; Pfirrmann, M.; Kohlmann, A.; Grossmann, V.; Meggendorfer, M.; Horny, H.-P. Comprehensive mutational profiling in advanced systemic mastocytosis. Blood 2013, 122, 2460–2466. [Google Scholar] [CrossRef]
- Kohlmann, A.; Grossmann, V.; Klein, H.-U.; Schindela, S.; Weiss, T.; Kazak, B.; Dicker, F.; Schnittger, S.; Dugas, M.; Kern, W. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J. Clin. Oncol. 2010, 28, 3858–3865. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef]
- Meggendorfer, M.; Roller, A.; Haferlach, T.; Eder, C.; Dicker, F.; Grossmann, V.; Kohlmann, A.; Alpermann, T.; Yoshida, K.; Ogawa, S. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood J. Am. Soc. Hematol. 2012, 120, 3080–3088. [Google Scholar] [CrossRef]
- Lundberg, P.; Karow, A.; Nienhold, R.; Looser, R.; Hao-Shen, H.; Nissen, I.; Girsberger, S.; Lehmann, T.; Passweg, J.; Stern, M. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood J. Am. Soc. Hematol. 2014, 123, 2220–2228. [Google Scholar] [CrossRef]
- Jawhar, M.; Schwaab, J.; Schnittger, S.; Meggendorfer, M.; Pfirrmann, M.; Sotlar, K.; Horny, H.P.; Metzgeroth, G.; Kluger, S.; Naumann, N. Additional mutations in SRSF2, ASXL1 and/or RUNX1 identify a high-risk group of patients with KIT D816V+ advanced systemic mastocytosis. Leukemia 2016, 30, 136. [Google Scholar] [CrossRef]
- Jawhar, M.; Schwaab, J.; Hausmann, D.; Clemens, J.; Naumann, N.; Henzler, T.; Horny, H.P.; Sotlar, K.; Schoenberg, S.O.; Cross, N.C.P. Splenomegaly, elevated alkaline phosphatase and mutations in the SRSF2/ASXL1/RUNX1 gene panel are strong adverse prognostic markers in patients with systemic mastocytosis. Leukemia 2016, 30, 2342–2350. [Google Scholar] [CrossRef]
- Gandhi Damaj, M.J.; Chandesris, O.; Hanssens, K.; Soucie, E.; Canioni, D.; Kolb, B.; Durieu, I.; Gyan, E.; Livideanu, C.; Chèze, S. ASXL1 but not TET2 mutations adversely impact overall survival of patients suffering systemic mastocytosis with associated clonal hematologic non-mast-cell diseases. PLoS ONE 2014, 9. [Google Scholar]
- Pardanani, A.; Lasho, T.L.; Finke, C.; Zblewski, D.; Abdelrahman, R.; Wassie, E.; Gangat, N.; Hanson, C.A.; Ketterling, R.P.; Tefferi, A. ASXL1 and CBL mutations are independently predictive of inferior survival in advanced systemic mastocytosis. Blood 2015, 126, 828. [Google Scholar] [CrossRef]
- Naumann, N.; Jawhar, M.; Schwaab, J.; Kluger, S.; Lübke, J.; Metzgeroth, G.; Popp, H.D.; Khaled, N.; Horny, H.; Sotlar, K. Incidence and prognostic impact of cytogenetic aberrations in patients with systemic mastocytosis. Genes Chromosom. Cancer 2018, 57, 252–259. [Google Scholar] [CrossRef]
- Shah, S.; Pardanani, A.; Elala, Y.C.; Lasho, T.L.; Patnaik, M.M.; Reichard, K.K.; Hanson, C.A.; Ketterling, R.P.; Tefferi, A. Cytogenetic abnormalities in systemic mastocytosis: WHO subcategory-specific incidence and prognostic impact among 348 informative cases. Am. J. Hematol. 2018, 93, 1461–1466. [Google Scholar] [CrossRef]
- Kluin-Nelemans, H.C.; Reiter, A.; Illerhaus, A.; van Anrooij, B.; Hartmann, K.; Span, L.F.R.; Gorska, A.; Niedoszytko, M.; Lange, M.; Scaffidi, L. Prognostic impact of eosinophils in mastocytosis: analysis of 2350 patients collected in the ECNM Registry. Leukemia 2020, 34, 1090–1101. [Google Scholar] [CrossRef]
- Mannelli, F.; Gesullo, F.; Rotunno, G.; Pacilli, A.; Bencini, S.; Annunziato, F.; Zanotti, R.; Scaffidi, L.; Giona, F.; Santopietro, M. Myelodysplasia as assessed by multiparameter flow cytometry refines prognostic stratification provided by genotypic risk in systemic mastocytosis. Am. J. Hematol. 2019, 94, 845–852. [Google Scholar] [CrossRef]
- Pardanani, A.; Reichard, K.K.; Zblewski, D.; Abdelrahman, R.A.; Wassie, E.A.; Morice Ii, W.G.; Brooks, C.; Grogg, K.L.; Hanson, C.A.; Tefferi, A. CD123 immunostaining patterns in systemic mastocytosis: differential expression in disease subgroups and potential prognostic value. Leukemia 2016, 30, 914–918. [Google Scholar] [CrossRef]
- Muñoz-González, J.I.; Álvarez-Twose, I.; Jara-Acevedo, M.; Henriques, A.; Viñas, E.; Prieto, C.; Sánchez-Muñoz, L.; Caldas, C.; Mayado, A.; Matito, A. Frequency and prognostic impact of KIT and other genetic variants in indolent systemic mastocytosis. Blood J. Am. Soc. Hematol. 2019, 134, 456–468. [Google Scholar] [CrossRef]
- Jawhar, M.; Schwaab, J.; Álvarez-Twose, I.; Shoumariyeh, K.; Naumann, N.; Lübke, J.; Perkins, C.; Muñoz-González, J.I.; Meggendorfer, M.; Kennedy, V. MARS: Mutation-Adjusted Risk Score for advanced systemic mastocytosis. J. Clin. Oncol. 2019. [Google Scholar] [CrossRef]
- Pardanani, A.; Shah, S.; Mannelli, F.; Elala, Y.C.; Guglielmelli, P.; Lasho, T.L.; Patnaik, M.M.; Gangat, N.; Ketterling, R.P.; Reichard, K.K. Mayo alliance prognostic system for mastocytosis: clinical and hybrid clinical-molecular models. Blood Adv. 2018, 2, 2964–2972. [Google Scholar] [CrossRef]
- Pardanani, A.; Lasho, T.L.; Reichard, K.K.; Hanson, C.A.; Tefferi, A. World Health Organization class-independent risk categorization in mastocytosis. Blood Cancer J. 2019, 9, 1–3. [Google Scholar] [CrossRef]
- Sperr, W.R.; Kundi, M.; Alvarez-Twose, I.; van Anrooij, B.; Elberink, J.N.G.O.; Gorska, A.; Niedoszytko, M.; Gleixner, K.V.; Hadzijusufovic, E.; Zanotti, R. International prognostic scoring system for mastocytosis (IPSM): a retrospective cohort study. Lancet Haematol. 2019, 6, e638–e649. [Google Scholar] [CrossRef]
- Martinelli, G.; Mancini, M.; De Benedittis, C.; Rondoni, M.; Papayannidis, C.; Manfrini, M.; Meggendorfer, M.; Calogero, R.; Guadagnuolo, V.; Fontana, M.C. SETD2 and histone H3 lysine 36 methylation deficiency in advanced systemic mastocytosis. Leukemia 2018, 32, 139. [Google Scholar] [CrossRef]
- Edmunds, J.W.; Mahadevan, L.C.; Clayton, A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008, 27, 406–420. [Google Scholar] [CrossRef]
- Carvalho, S.; Raposo, A.C.; Martins, F.B.; Grosso, A.R.; Sridhara, S.C.; Rino, J.; Carmo-Fonseca, M.; de Almeida, S.F. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013, 41, 2881–2893. [Google Scholar] [CrossRef]
- De Almeida, S.F.; Grosso, A.R.; Koch, F.; Fenouil, R.; Carvalho, S.; Andrade, J.; Levezinho, H.; Gut, M.; Eick, D.; Gut, I. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 2011, 18, 977. [Google Scholar] [CrossRef]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.-M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef]
- Carvalho, S.; Vítor, A.C.; Sridhara, S.C.; Martins, F.B.; Raposo, A.C.; Desterro, J.M.P.; Ferreira, J.; de Almeida, S.F. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 2014, 3, e02482. [Google Scholar] [CrossRef]
- Park, I.Y.; Powell, R.T.; Tripathi, D.N.; Dere, R.; Ho, T.H.; Blasius, T.L.; Chiang, Y.-C.; Davis, I.J.; Fahey, C.C.; Hacker, K.E. Dual chromatin and cytoskeletal remodeling by SETD2. Cell 2016, 166, 950–962. [Google Scholar] [CrossRef]
- Kanu, N.; Grönroos, E.; Martinez, P.; Burrell, R.A.; Goh, X.Y.; Bartkova, J.; Maya-Mendoza, A.; Mistrík, M.; Rowan, A.J.; Patel, H. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 2015, 34, 5699–5708. [Google Scholar] [CrossRef]
- Li, J.; Duns, G.; Westers, H.; Sijmons, R.; van den Berg, A.; Kok, K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget 2016, 7, 50719. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; He, F.; Zeng, H.; Ling, S.; Chen, A.; Wang, Y.; Yan, X.; Wei, W.; Pang, Y.; Cheng, H. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 2014, 46, 287. [Google Scholar] [CrossRef]
- Liu, W.; Fu, Q.; An, H.; Chang, Y.; Zhang, W.; Zhu, Y.; Xu, L.; Xu, J. Decreased expression of SETD2 predicts unfavorable prognosis in patients with nonmetastatic clear-cell renal cell carcinoma. Medicine (Baltimore) 2015, 94. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Qu, Y.; Xi, W.; Xia, Y.; Bai, Q.; Xiong, Y.; Long, Q.; Xu, J.; Guo, J. Prognostic value of SETD2 expression in patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors. J. Urol. 2016, 196, 1363–1370. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Song, J.; Hu, C.-L.; Chen, S.-B.; Zhang, Q.; Xu, C.-H.; Wu, J.-C.; Hou, D.; Sun, M.; Zhang, Y.-L. SETD2 deficiency predicts poor prognosis in MDS and accelerated MDS-associated leukemogenesis via S100a9. Blood J. 2020, blood-2019001963. [Google Scholar] [CrossRef]
- Parker, H.; Rose-Zerilli, M.J.J.; Larrayoz, M.; Clifford, R.; Edelmann, J.; Blakemore, S.; Gibson, J.; Wang, J.; Ljungström, V.; Wojdacz, T.K. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia 2016, 30, 2179–2186. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martelli, M.; Monaldi, C.; De Santis, S.; Bruno, S.; Mancini, M.; Cavo, M.; Soverini, S. Recent Advances in the Molecular Biology of Systemic Mastocytosis: Implications for Diagnosis, Prognosis, and Therapy. Int. J. Mol. Sci. 2020, 21, 3987. https://doi.org/10.3390/ijms21113987
Martelli M, Monaldi C, De Santis S, Bruno S, Mancini M, Cavo M, Soverini S. Recent Advances in the Molecular Biology of Systemic Mastocytosis: Implications for Diagnosis, Prognosis, and Therapy. International Journal of Molecular Sciences. 2020; 21(11):3987. https://doi.org/10.3390/ijms21113987
Chicago/Turabian StyleMartelli, Margherita, Cecilia Monaldi, Sara De Santis, Samantha Bruno, Manuela Mancini, Michele Cavo, and Simona Soverini. 2020. "Recent Advances in the Molecular Biology of Systemic Mastocytosis: Implications for Diagnosis, Prognosis, and Therapy" International Journal of Molecular Sciences 21, no. 11: 3987. https://doi.org/10.3390/ijms21113987
APA StyleMartelli, M., Monaldi, C., De Santis, S., Bruno, S., Mancini, M., Cavo, M., & Soverini, S. (2020). Recent Advances in the Molecular Biology of Systemic Mastocytosis: Implications for Diagnosis, Prognosis, and Therapy. International Journal of Molecular Sciences, 21(11), 3987. https://doi.org/10.3390/ijms21113987





