From the Argonauts Mythological Sailors to the Argonautes RNA-Silencing Navigators: Their Emerging Roles in Human-Cell Pathologies
Abstract
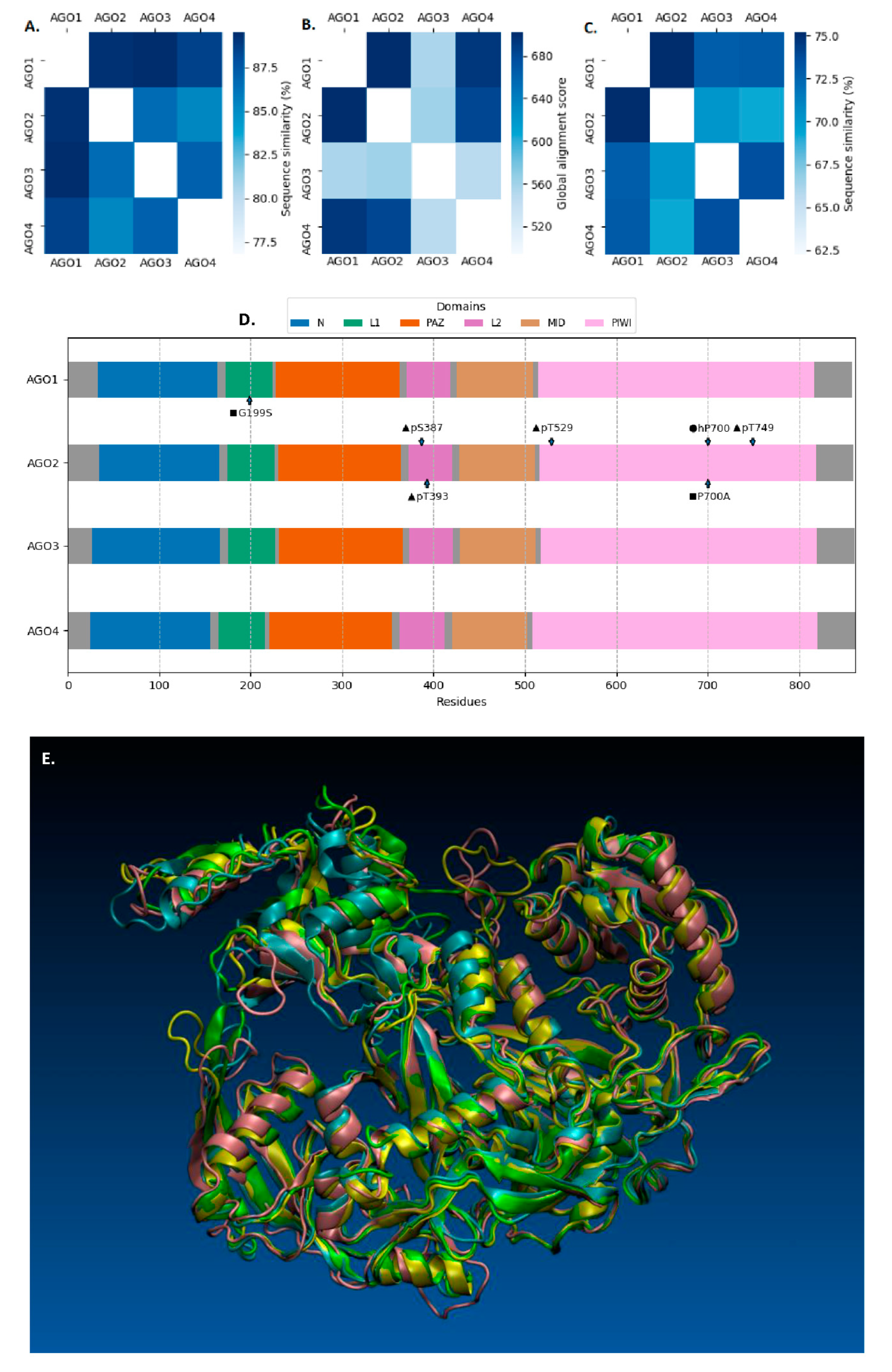
1. Introduction
2. AGOs in Human Diseases
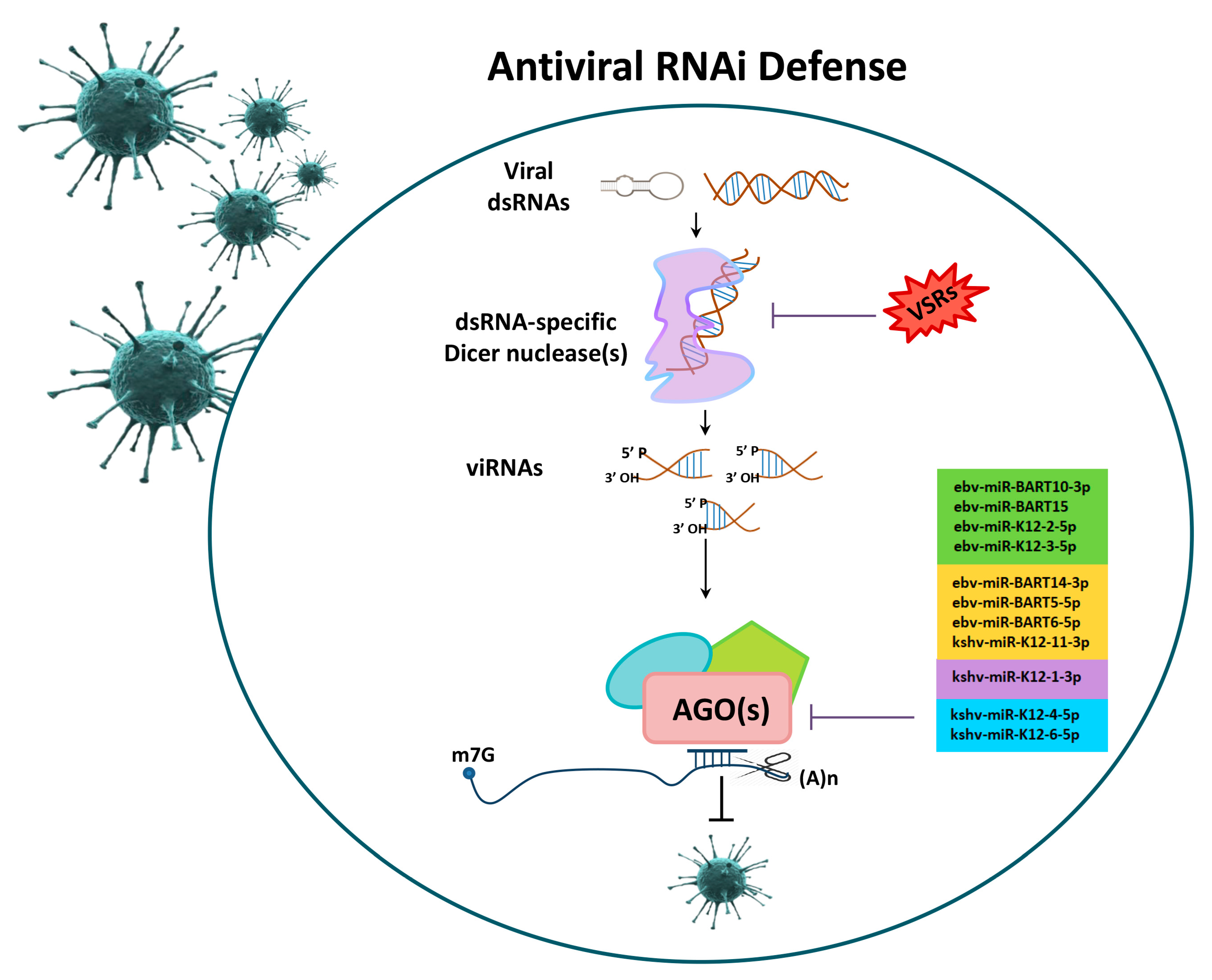
2.1. AGOs in Viral Infections
2.2. AGOs and Autoimmune Diseases
2.3. AGOs in Cancer
2.4. AGOs in Metabolic Deficiencies
2.4.1. Mitochondrial Dysfunctions
2.4.2. Obesity
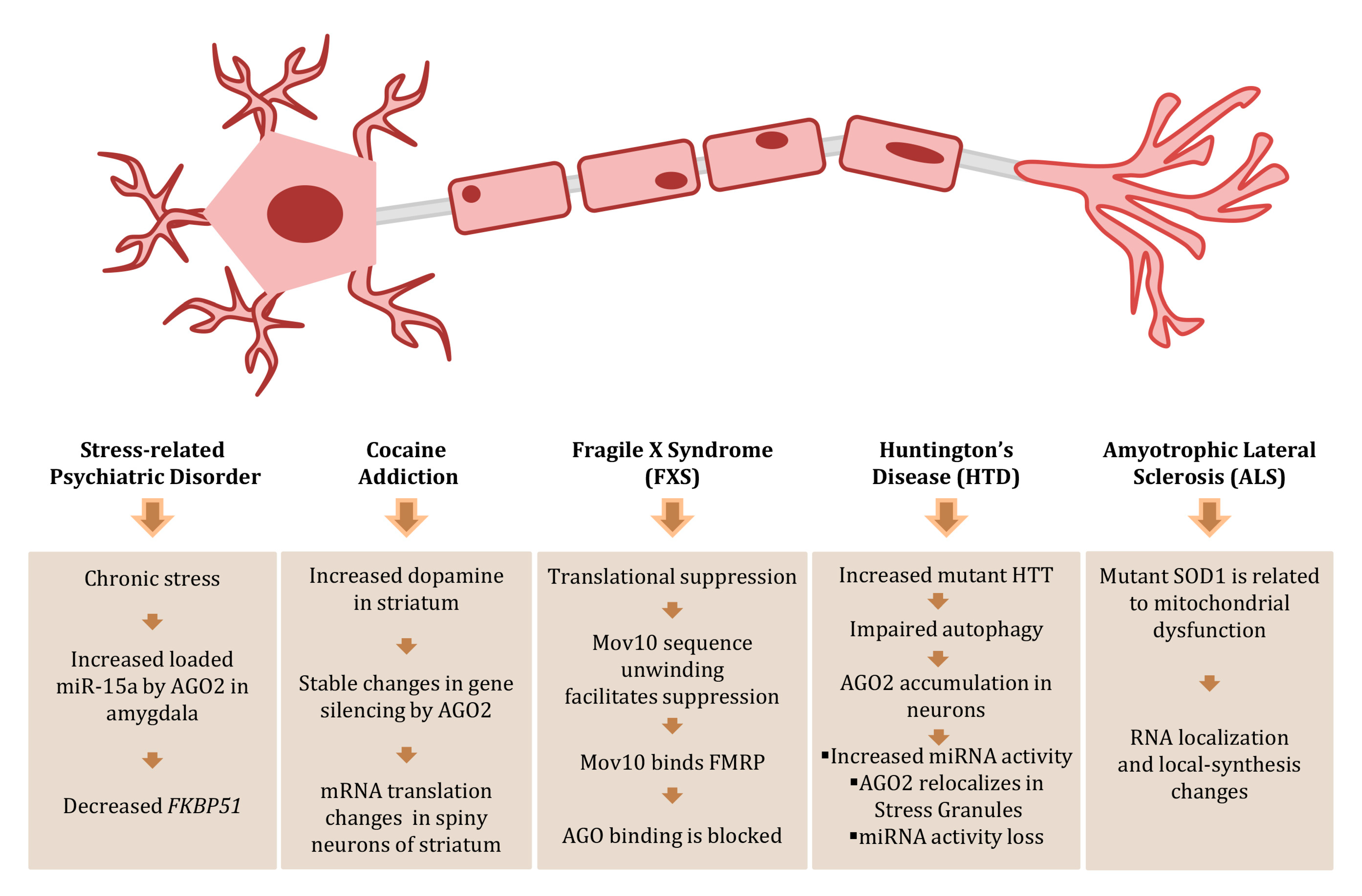
2.5. AGOs in Psychiatric Disorders and Neuronal Diseases
2.6. AGOs and Infertility
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moussian, B.; Schoof, H.; Haecker, A.; Jürgens, G.; Laux, T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998, 17, 1799–1809. [Google Scholar] [CrossRef]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998, 17, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Spradling, A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 1997, 124, 2463–2476. [Google Scholar] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef]
- Hock, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef]
- Schurmann, N.; Trabuco, L.G.; Bender, C.; Russell, R.B.; Grimm, D. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol. 2013, 20, 818–826. [Google Scholar] [CrossRef]
- Joshua-Tor, L. The Argonautes. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 67–72. [Google Scholar] [CrossRef]
- Su, H.; Trombly, M.I.; Chen, J.; Wang, X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009, 23, 304–317. [Google Scholar] [CrossRef]
- Meyer, W.J.; Schreiber, S.; Guo, Y.; Volkmann, T.; Welte, M.A.; Muller, H.A. Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet. 2006, 2, e134. [Google Scholar] [CrossRef]
- Matsui, M.; Li, L.; Janowski, B.A.; Corey, D.R. Reduced Expression of Argonaute 1, Argonaute 2, and TRBP Changes Levels and Intracellular Distribution of RNAi Factors. Sci. Rep. 2015, 5, 12855. [Google Scholar] [CrossRef] [PubMed]
- Hauptmann, J.; Dueck, A.; Harlander, S.; Pfaff, J.; Merkl, R.; Meister, G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. 2013, 20, 814–817. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.A.; Salomon, W.E.; Ryder, S.P.; Aronin, N.; Zamore, P.D. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA 2011, 17, 1858–1869. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute 2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Park, M.S.; Phan, H.D.; Busch, F.; Hinckley, S.H.; Brackbill, J.A.; Wysocki, V.H.; Nakanishi, K. Human Argonaute3 has slicer activity. Nucleic Acids Res. 2017, 45, 11867–11877. [Google Scholar] [CrossRef]
- Hu, Q.; Tanasa, B.; Trabucchi, M.; Li, W.; Zhang, J.; Ohgi, K.A.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat. Struct. Mol. Biol. 2012, 19, 1168–1175. [Google Scholar] [CrossRef]
- Jan, S.Z.; Vormer, T.L. Unraveling transcriptome dynamics in human spermatogenesis. Development 2017, 144, 3659–3673. [Google Scholar] [CrossRef]
- Modzelewski, A.J.; Holmes, R.J.; Hilz, S.; Grimson, A.; Cohen, P.E. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev. Cell 2012, 23, 251–264. [Google Scholar] [CrossRef]
- Faehnle, C.R.; Elkayam, E.; Haase, A.D.; Hannon, G.J.; Joshua-Tor, L. The making of a slicer: Activation of human Argonaute-1. Cell Rep. 2013, 3, 1901–1909. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Sheu-Gruttadauria, J.; Chandradoss, S.D.; Joo, C.; MacRae, I.J. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Araya-Secchi, R.; Brackbill, J.A.; Phan, H.D.; Kehling, A.C.; Abd El-Wahab, E.W.; Dayeh, D.M.; Sotomayor, M.; Nakanishi, K. Multidomain Convergence of Argonaute during RISC Assembly Correlates with the Formation of Internal Water Clusters. Mol. Cell 2019, 75, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinform. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Søndergaard, C.R.; Olsson, M.H.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef]
- Olsson, M.H.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Stagsted, L.V.; Daugaard, I.; Hansen, T.B. The agotrons: Gene regulators or Argonaute protectors? Bioessays News Rev. Mol. Cell. Dev. Biol. 2017, 39. [Google Scholar] [CrossRef]
- Hansen, T.B.; Veno, M.T.; Jensen, T.I.; Schaefer, A.; Damgaard, C.K.; Kjems, J. Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 2016, 7, 11538. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Josa-Prado, F.; Henley, J.M.; Wilkinson, K.A. SUMOylation of Argonaute-2 regulates RNA interference activity. Biochem. Biophys. Res. Commun. 2015, 464, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Dou, J.; Guo, Y.; He, J.; Li, L.; Liu, X.; Chen, R.; Deng, R.; Huang, J.; et al. Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene 2019, 38, 1410–1431. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Hadian, K.; Smirnova, L.; Wulczyn, E.A.; Michel, G.; Nitsch, R.; Krappmann, D.; Wulczyn, F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009, 11, 1411–1420. [Google Scholar] [CrossRef]
- Gibbings, D.; Mostowy, S.; Jay, F.; Schwab, Y.; Cossart, P.; Voinnet, O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat. Cell Biol. 2012, 14, 1314–1321. [Google Scholar] [CrossRef]
- Leung, A.K.; Vyas, S.; Rood, J.E.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 2011, 42, 489–499. [Google Scholar] [CrossRef]
- Butepage, M.; Eckei, L.; Verheugd, P.; Luscher, B. Intracellular Mono-ADP-Ribosylation in Signaling and Disease. Cells 2015, 4, 569–595. [Google Scholar] [CrossRef]
- Quevillon Huberdeau, M.; Zeitler, D.M.; Hauptmann, J.; Bruckmann, A.; Fressigne, L.; Danner, J.; Piquet, S.; Strieder, N.; Engelmann, J.C.; Jannot, G.; et al. Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J. 2017, 36, 2088–2106. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Qi, H.H.; Ongusaha, P.P.; Myllyharju, J.; Cheng, D.; Pakkanen, O.; Shi, Y.; Lee, S.W.; Peng, J.; Shi, Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 2008, 455, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.; Anderson, A.; Bernberg, E.; Kamboj, S.; Huang, E.; Lagasse, G.; Isaacs, G.; Parcells, M.; Meyers, B.C.; Green, P.J.; et al. Sequence conservation and differential expression of Marek’s disease virus microRNAs. J. Virol. 2008, 82, 12213–12220. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef]
- Tang, S.; Bertke, A.S.; Patel, A.; Wang, K.; Cohen, J.I.; Krause, P.R. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. USA 2008, 105, 10931–10936. [Google Scholar] [CrossRef]
- Grey, F.; Antoniewicz, A.; Allen, E.; Saugstad, J.; McShea, A.; Carrington, J.C.; Nelson, J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 2005, 79, 12095–12099. [Google Scholar] [CrossRef]
- Cullen, B.R. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011, 25, 1881–1894. [Google Scholar] [CrossRef]
- Goubau, D.; Deddouche, S.; Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Berkhout, B. RNAi-mediated antiviral immunity in mammals. Curr. Opin. Virol. 2018, 32, 9–14. [Google Scholar] [CrossRef]
- Minks, M.A.; West, D.K.; Benvin, S.; Baglioni, C. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem. 1979, 254, 10180–10183. [Google Scholar]
- Schreiber, G. The molecular basis for differential type I interferon signaling. J. Biol. Chem. 2017, 292, 7285–7294. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Seo, G.J.; Kincaid, R.P.; Phanaksri, T.; Burke, J.M.; Pare, J.M.; Cox, J.E.; Hsiang, T.Y.; Krug, R.M.; Sullivan, C.S. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 2013, 14, 435–445. [Google Scholar] [CrossRef]
- Levanova, A.; Poranen, M.M. RNA Interference as a Prospective Tool for the Control of Human Viral Infections. Front. Microbiol. 2018, 9, 2151. [Google Scholar] [CrossRef] [PubMed]
- Goic, B.; Stapleford, K.A.; Frangeul, L.; Doucet, A.J.; Gausson, V.; Blanc, H.; Schemmel-Jofre, N.; Cristofari, G.; Lambrechts, L.; Vignuzzi, M.; et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun. 2016, 7, 12410. [Google Scholar] [CrossRef]
- Wang, X.H.; Aliyari, R.; Li, W.X.; Li, H.W.; Kim, K.; Carthew, R.; Atkinson, P.; Ding, S.W. RNA interference directs innate immunity against viruses in adult Drosophila. Science 2006, 312, 452–454. [Google Scholar] [CrossRef]
- Mahla, R.S.; Reddy, M.C.; Prasad, D.V.; Kumar, H. Sweeten PAMPs: Role of Sugar Complexed PAMPs in Innate Immunity and Vaccine Biology. Front. Immunol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef]
- Takeda, K.; Kaisho, T.; Akira, S. Toll-like receptors. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Webster, J.A.; Madzokere, E.T.; Stephenson, E.B.; Herrero, L.J. Mosquito antiviral defense mechanisms: A delicate balance between innate immunity and persistent viral infection. Parasites Vectors 2019, 12, 165. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Aliyari, R.; Ding, S.W. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 2009, 227, 176–188. [Google Scholar] [CrossRef]
- Li, F.; Ding, S.W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef]
- van Rij, R.P.; Saleh, M.C.; Berry, B.; Foo, C.; Houk, A.; Antoniewski, C.; Andino, R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006, 20, 2985–2995. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, W.X.; Ding, S.W. Induction and suppression of RNA silencing by an animal virus. Science 2002, 296, 1319–1321. [Google Scholar] [CrossRef]
- Aliyari, R.; Wu, Q.; Li, H.W.; Wang, X.H.; Li, F.; Green, L.D.; Han, C.S.; Li, W.X.; Ding, S.W. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 2008, 4, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Nayak, A.; Berry, B.; Tassetto, M.; Kunitomi, M.; Acevedo, A.; Deng, C.; Krutchinsky, A.; Gross, J.; Antoniewski, C.; Andino, R. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Popli, S.; Hari, Y.; Malhotra, P.; Mukherjee, S.; Bhatnagar, R.K. Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 2009, 23, 1845–1857. [Google Scholar] [CrossRef]
- van Mierlo, J.T.; Bronkhorst, A.W.; Overheul, G.J.; Sadanandan, S.A.; Ekstrom, J.O.; Heestermans, M.; Hultmark, D.; Antoniewski, C.; van Rij, R.P. Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog. 2012, 8, e1002872. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Corcoran, D.L.; Gottwein, E.; Frank, C.L.; Kang, D.; Hafner, M.; Nusbaum, J.D.; Feederle, R.; Delecluse, H.J.; Luftig, M.A.; et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012, 8, e1002484. [Google Scholar] [CrossRef] [PubMed]
- Haecker, I.; Gay, L.A.; Yang, Y.; Hu, J.; Morse, A.M.; McIntyre, L.M.; Renne, R. Ago HITS-CLIP expands understanding of Kaposi’s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012, 8, e1002884. [Google Scholar] [CrossRef] [PubMed]
- Moon, I.Y.; Choi, J.H.; Chung, J.W.; Jang, E.S.; Jeong, S.H.; Kim, J.W. MicroRNA20 induces methylation of hepatitis B virus covalently closed circular DNA in human hepatoma cells. Mol. Med. Rep. 2019, 20, 2285–2293. [Google Scholar] [CrossRef]
- Eckenfelder, A.; Segeral, E.; Pinzon, N.; Ulveling, D.; Amadori, C.; Charpentier, M.; Nidelet, S.; Concordet, J.P.; Zagury, J.F.; Paillart, J.C.; et al. Argonaute proteins regulate HIV-1 multiply spliced RNA and viral production in a Dicer independent manner. Nucleic Acids Res. 2017, 45, 4158–4173. [Google Scholar] [CrossRef]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Parameswaran, P.; Sklan, E.; Wilkins, C.; Burgon, T.; Samuel, M.A.; Lu, R.; Ansel, K.M.; Heissmeyer, V.; Einav, S.; Jackson, W.; et al. Six RNA viruses and forty-one hosts: Viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010, 6, e1000764. [Google Scholar] [CrossRef]
- Weng, K.F.; Hung, C.T.; Hsieh, P.T.; Li, M.L.; Chen, G.W.; Kung, Y.A.; Huang, P.N.; Kuo, R.L.; Chen, L.L.; Lin, J.Y.; et al. A cytoplasmic RNA virus generates functional viral small RNAs and regulates viral IRES activity in mammalian cells. Nucleic Acids Res. 2014, 42, 12789–12805. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Whisnant, A.W.; Kornepati, A.V.; Marshall, J.B.; Bogerd, H.P.; Cullen, B.R. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc. Natl. Acad. Sci. USA 2015, 112, E6945–E6954. [Google Scholar] [CrossRef]
- Umbach, J.L.; Cullen, B.R. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009, 23, 1151–1164. [Google Scholar] [CrossRef] [PubMed]
- Samir, M.; Vaas, L.A.; Pessler, F. MicroRNAs in the Host Response to Viral Infections of Veterinary Importance. Front. Vet. Sci. 2016, 3, 86. [Google Scholar] [CrossRef] [PubMed]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar] [CrossRef] [PubMed]
- Billy, E.; Brondani, V.; Zhang, H.; Muller, U.; Filipowicz, W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 2001, 98, 14428–14433. [Google Scholar] [CrossRef]
- Maillard, P.V.; Ciaudo, C.; Marchais, A.; Li, Y.; Jay, F.; Ding, S.W.; Voinnet, O. Antiviral RNA interference in mammalian cells. Science 2013, 342, 235–238. [Google Scholar] [CrossRef]
- Eggenberger, J.; Blanco-Melo, D.; Panis, M.; Brennand, K.J.; tenOever, B.R. Type I interferon response impairs differentiation potential of pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2019, 116, 1384–1393. [Google Scholar] [CrossRef]
- Li, Y.; Lu, J.; Han, Y.; Fan, X.; Ding, S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013, 342, 231–234. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, Y.; Zhang, Y.; Zhou, H.; Deng, Y.Q.; Li, X.F.; Miao, M.; Zhang, Q.; Zhong, B.; Hu, Y.; et al. Human Virus-Derived Small RNAs Can Confer Antiviral Immunity in Mammals. Immunity 2017, 46, 992–1004. [Google Scholar] [CrossRef]
- Wang, S.; Sun, X.; Yi, C.; Zhang, D.; Lin, X.; Sun, X.; Chen, H.; Jin, M. AGO2 Negatively Regulates Type I Interferon Signaling Pathway by Competition Binding IRF3 with CBP/p300. Front. Cell. Infect. Microbiol. 2017, 7, 195. [Google Scholar] [CrossRef]
- Matskevich, A.A.; Moelling, K. Dicer is involved in protection against influenza A virus infection. J. Gen. Virol. 2007, 88, 2627–2635. [Google Scholar] [CrossRef]
- Casseb, S.M.; Simith, D.B.; Melo, K.F.; Mendonca, M.H.; Santos, A.C.; Carvalho, V.L.; Cruz, A.C.; Vasconcelos, P.F. Drosha, DGCR8, and Dicer mRNAs are down-regulated in human cells infected with dengue virus 4, and play a role in viral pathogenesis. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Hassan, T.; McKiernan, P.J.; McElvaney, N.G.; Cryan, S.A.; Greene, C.M. Therapeutic modulation of miRNA for the treatment of proinflammatory lung diseases. Expert Rev. Anti-Infect. Ther. 2012, 10, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Foster, P.S.; Plank, M.; Collison, A.; Tay, H.L.; Kaiko, G.E.; Li, J.; Johnston, S.L.; Hansbro, P.M.; Kumar, R.K.; Yang, M.; et al. The emerging role of microRNAs in regulating immune and inflammatory responses in the lung. Immunol. Rev. 2013, 253, 198–215. [Google Scholar] [CrossRef] [PubMed]
- Globinska, A.; Pawelczyk, M.; Kowalski, M.L. MicroRNAs and the immune response to respiratory virus infections. Expert Rev. Clin. Immunol. 2014, 10, 963–971. [Google Scholar] [CrossRef]
- Tahamtan, A.; Tavakoli-Yaraki, M.; Rygiel, T.P.; Mokhtari-Azad, T.; Salimi, V. Effects of cannabinoids and their receptors on viral infections. J. Med. Virol. 2016, 88, 1–12. [Google Scholar] [CrossRef]
- Salimi, V.; Hennus, M.P.; Mokhtari-Azad, T.; Shokri, F.; Janssen, R.; Hodemaekers, H.M.; Rygiel, T.P.; Coenjaerts, F.E.; Meyaard, L.; Bont, L. Opioid receptors control viral replication in the airways. Crit. Care Med. 2013, 41, 205–214. [Google Scholar] [CrossRef]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells. J. Virol. 2015, 89, 9029–9043. [Google Scholar] [CrossRef]
- Adiliaghdam, F.; Basavappa, M.; Saunders, T.L.; Harjanto, D.; Prior, J.T.; Cronkite, D.A.; Papavasiliou, N.; Jeffrey, K.L. A Requirement for Argonaute 4 in Mammalian Antiviral Defense. Cell Rep. 2020, 30, 1690–1701. [Google Scholar] [CrossRef]
- Tokiyoshi, E.; Watanabe, M.; Inoue, N.; Hidaka, Y.; Iwatani, Y. Polymorphisms and expression of genes encoding Argonautes 1 and 2 in autoimmune thyroid diseases. Autoimmunity 2018, 51, 35–42. [Google Scholar] [CrossRef]
- Li, G.; You, D.; Ma, J. The Role of Autoimmunity in the Pathogenesis of Sudden Sensorineural Hearing Loss. Neural Plast. 2018, 2018, 7691473. [Google Scholar] [CrossRef]
- Han, S.Y.; Kim, S.; Shin, D.H.; Cho, J.H.; Nam, S.I. The Expression of AGO2 and DGCR8 in Idiopathic Sudden Sensorineural Hearing Loss. Clin. Exp. Otorhinolaryngol. 2014, 7, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Sibony, M.; Abdullah, M.; Greenfield, L.; Raju, D.; Wu, T.; Rodrigues, D.M.; Galindo-Mata, E.; Mascarenhas, H.; Philpott, D.J.; Silverberg, M.S.; et al. Microbial Disruption of Autophagy Alters Expression of the RISC Component AGO2, a Critical Regulator of the miRNA Silencing Pathway. Inflamm. Bowel Dis. 2015, 21, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Sethi, A.; Kulkarni, N.; Sonar, S.; Lal, G. Role of miRNAs in CD4 T cell plasticity during inflammation and tolerance. Front. Genet. 2013, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Lewkowicz, P.; Cwiklinska, H.; Mycko, M.P.; Cichalewska, M.; Domowicz, M.; Lewkowicz, N.; Jurewicz, A.; Selmaj, K.W. Dysregulated RNA-Induced Silencing Complex (RISC) Assembly within CNS Corresponds with Abnormal miRNA Expression during Autoimmune Demyelination. J. Neurosci. Off. J. Soc. Neurosci. 2015, 35, 7521–7537. [Google Scholar] [CrossRef]
- Smibert, P.; Yang, J.S.; Azzam, G.; Liu, J.L.; Lai, E.C. Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 2013, 20, 789–795. [Google Scholar] [CrossRef]
- Treadwell, E.L.; Alspaugh, M.A.; Sharp, G.C. Characterization of a new antigen-antibody system (Su) in patients with systemic lupus erythematosus. Arthritis Rheum. 1984, 27, 1263–1271. [Google Scholar] [CrossRef]
- Jakymiw, A.; Lian, S.; Eystathioy, T.; Li, S.; Satoh, M.; Hamel, J.C.; Fritzler, M.J.; Chan, E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005, 7, 1267–1274. [Google Scholar] [CrossRef]
- Jakymiw, A.; Ikeda, K.; Fritzler, M.J.; Reeves, W.H.; Satoh, M.; Chan, E.K. Autoimmune targeting of key components of RNA interference. Arthritis Res. Ther. 2006, 8, R87. [Google Scholar] [CrossRef]
- Satoh, M.; Reeves, W.H. Induction of lupus-associated autoantibodies in BALB/c mice by intraperitoneal injection of pristane. J. Exp. Med. 1994, 180, 2341–2346. [Google Scholar] [CrossRef]
- Bloch, D.B.; Nobre, R.A.; Yang, W.H. GW/P-bodies and autoimmune disease. Adv. Exp. Med. Biol. 2013, 768, 61–70. [Google Scholar] [CrossRef]
- Wei, K.; Wu, L.; Chen, Y.; Lin, Y.; Wang, Y.; Liu, X.; Xie, D. Argonaute protein as a linker to command center of physiological processes. Chin. J. Cancer Res. 2013, 25, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Fritzler, M.J.; Pauls, J.D.; Kinsella, T.D.; Bowen, T.J. Antinuclear, anticytoplasmic, and anti-Sjogren’s syndrome antigen A (SS-A/Ro) antibodies in female blood donors. Clin. Immunol. Immunopathol. 1985, 36, 120–128. [Google Scholar] [CrossRef]
- Ceribelli, A.; Yao, B.; Dominguez-Gutierrez, P.R.; Nahid, M.A.; Satoh, M.; Chan, E.K. MicroRNAs in systemic rheumatic diseases. Arthritis Res. Ther. 2011, 13, 229. [Google Scholar] [CrossRef] [PubMed]
- Onodera, H. A new precipitating antibody to the microsomal fraction in patients with connective tissue diseases. Okayama Igakkai Zasshi (J. Okayama Med Assoc.) 1986, 98, 747–758. [Google Scholar] [CrossRef][Green Version]
- Vazquez-Del Mercado, M.; Sanchez-Orozco, L.V.; Pauley, B.A.; Chan, J.Y.; Chan, E.K.; Panduro, A.; Maldonado Gonzalez, M.; Jimenez-Luevanos, M.A.; Martin-Marquez, B.T.; Palafox-Sanchez, C.A.; et al. Autoantibodies to a miRNA-binding protein Argonaute2 (Su antigen) in patients with hepatitis C virus infection. Clin. Exp. Rheumatol. 2010, 28, 842–848. [Google Scholar] [PubMed]
- Conger, A.K.; Martin, E.C.; Yan, T.J.; Rhodes, L.V.; Hoang, V.T.; La, J.; Anbalagan, M.; Burks, H.E.; Rowan, B.G.; Nephew, K.P.; et al. Argonaute 2 Expression Correlates with a Luminal B Breast Cancer Subtype and Induces Estrogen Receptor Alpha Isoform Variation. Non-Coding RNA 2016, 2, 8. [Google Scholar] [CrossRef]
- Voller, D.; Reinders, J.; Meister, G.; Bosserhoff, A.K. Strong reduction of AGO2 expression in melanoma and cellular consequences. Br. J. Cancer 2013, 109, 3116–3124. [Google Scholar] [CrossRef]
- Di Leva, G.; Croce, C.M. The Role of microRNAs in the Tumorigenesis of Ovarian Cancer. Front. Oncol. 2013, 3, 153. [Google Scholar] [CrossRef]
- Yang, F.Q.; Huang, J.H.; Liu, M.; Yang, F.P.; Li, W.; Wang, G.C.; Che, J.P.; Zheng, J.H. Argonaute 2 is up-regulated in tissues of urothelial carcinoma of bladder. Int. J. Clin. Exp. Pathol. 2014, 7, 340–347. [Google Scholar]
- Bian, X.J.; Zhang, G.M.; Gu, C.Y.; Cai, Y.; Wang, C.F.; Shen, Y.J.; Zhu, Y.; Zhang, H.L.; Dai, B.; Ye, D.W. Down-regulation of Dicer and Ago2 is associated with cell proliferation and apoptosis in prostate cancer. Tumour Biol. 2014, 35, 11571–11578. [Google Scholar] [CrossRef]
- Kim, M.S.; Oh, J.E.; Kim, Y.R.; Park, S.W.; Kang, M.R.; Kim, S.S.; Ahn, C.H.; Yoo, N.J.; Lee, S.H. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J. Pathol. 2010, 221, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Min, H.; Ha, J.Y.; Kim, B.H.; Choi, M.S.; Kim, S. Dysregulation of the miRNA biogenesis components DICER1, DROSHA, DGCR8 and AGO2 in clear cell renal cell carcinoma in both a Korean cohort and the cancer genome atlas kidney clear cell carcinoma cohort. Oncol. Lett. 2019, 18, 4337–4345. [Google Scholar] [CrossRef] [PubMed]
- Masciarelli, S.; Quaranta, R.; Iosue, I.; Colotti, G.; Padula, F.; Varchi, G.; Fazi, F.; Del Rio, A. A small-molecule targeting the microRNA binding domain of argonaute 2 improves the retinoic acid differentiation response of the acute promyelocytic leukemia cell line NB4. ACS Chem. Biol. 2014, 9, 1674–1679. [Google Scholar] [CrossRef] [PubMed]
- Iosue, I.; Quaranta, R.; Masciarelli, S.; Fontemaggi, G.; Batassa, E.M.; Bertolami, C.; Ottone, T.; Divona, M.; Salvatori, B.; Padula, F.; et al. Argonaute 2 sustains the gene expression program driving human monocytic differentiation of acute myeloid leukemia cells. Cell Death Dis. 2013, 4, e926. [Google Scholar] [CrossRef] [PubMed]
- De Santa, F.; Iosue, I.; Del Rio, A.; Fazi, F. microRNA biogenesis pathway as a therapeutic target for human disease and cancer. Curr. Pharm. Des. 2013, 19, 745–764. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Hou, Y.X.; Langlais, P.; Erickson, P.; Zhu, J.; Shi, C.X.; Luo, M.; Zhu, Y.; Xu, Y.; Mandarino, L.J.; et al. Expression of the cereblon binding protein argonaute 2 plays an important role for multiple myeloma cell growth and survival. BMC Cancer 2016, 16, 297. [Google Scholar] [CrossRef]
- Mostafavi, S.; Ray, D.; Warde-Farley, D.; Grouios, C.; Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008, 9 (Suppl. 1), S4. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, H.; Fang, J.; Yang, Z.; Chen, R.; Wang, Y.; Zhao, X.; Ge, S.; Yu, J.; Huang, J. AGO2 phosphorylation by c-Src kinase promotes tumorigenesis. Neoplasia 2020, 22, 129–141. [Google Scholar] [CrossRef]
- Zhang, X.; Graves, P.; Zeng, Y. Overexpression of human Argonaute2 inhibits cell and tumor growth. Biochim. Biophys. Acta 2013, 1830, 2553–2561. [Google Scholar] [CrossRef]
- Sharma, N.R.; Wang, X.; Majerciak, V.; Ajiro, M.; Kruhlak, M.; Meyers, C.; Zheng, Z.M. Cell Type- and Tissue Context-dependent Nuclear Distribution of Human Ago2. J. Biol. Chem. 2016, 291, 2302–2309. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, X.S.; Wang, C.X.; Liu, B.; Li, Q.; Zhou, X.J. Up-regulation of Ago2 expression in gastric carcinoma. Med. Oncol. 2013, 30, 628. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, F.; Fang, E.; Xiao, W.; Mei, H.; Li, H.; Li, D.; Song, H.; Wang, J.; Hong, M.; et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019, 26, 1346–1364. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ren, D.; Guo, W.; Wang, Z.; Huang, S.; Du, H.; Song, L.; Peng, X. Loss of miR-100 enhances migration, invasion, epithelial-mesenchymal transition and stemness properties in prostate cancer cells through targeting Argonaute 2. Int. J. Oncol. 2014, 45, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Yoo, N.J.; Hur, S.Y.; Kim, M.S.; Lee, J.Y.; Lee, S.H. Immunohistochemical analysis of RNA-induced silencing complex-related proteins AGO2 and TNRC6A in prostate and esophageal cancers. APMIS 2010, 118, 271–276. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, G.; Kong, C.; Bi, J.; Gong, D.; Yu, X.; Shi, D.; Zhan, B.; Ye, P. EIF2C, Dicer, and Drosha are up-regulated along tumor progression and associated with poor prognosis in bladder carcinoma. Tumour Biol. 2015, 36, 5071–5079. [Google Scholar] [CrossRef]
- Rabien, A.; Ratert, N.; Hogner, A.; Erbersdobler, A.; Jung, K.; Ecke, T.H. Diagnostic and Prognostic Potential of MicroRNA Maturation Regulators Drosha, AGO1 and AGO2 in Urothelial Carcinomas of the Bladder. Int. J. Mol. Sci. 2018, 19, 1622. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, L.; Barlogie, B.; Stephens, O.; Wu, X.; Williams, D.R.; Cartron, M.A.; van Rhee, F.; Nair, B.; Waheed, S.; et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc. Natl. Acad. Sci. USA 2010, 107, 7904–7909. [Google Scholar] [CrossRef]
- Kwon, S.Y.; Lee, J.H.; Kim, B.; Park, J.W.; Kwon, T.K.; Kang, S.H.; Kim, S. Complexity in regulation of microRNA machinery components in invasive breast carcinoma. Pathol. Oncol. Res. 2014, 20, 697–705. [Google Scholar] [CrossRef]
- Casey, M.C.; Prakash, A.; Holian, E.; McGuire, A.; Kalinina, O.; Shalaby, A.; Curran, C.; Webber, M.; Callagy, G.; Bourke, E.; et al. Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer. BMC Cancer 2019, 19, 712. [Google Scholar] [CrossRef]
- Cheng, C.; Fu, X.; Alves, P.; Gerstein, M. mRNA expression profiles show differential regulatory effects of microRNAs between estrogen receptor-positive and estrogen receptor-negative breast cancer. Genome Biol. 2009, 10, R90. [Google Scholar] [CrossRef]
- Vaksman, O.; Hetland, T.E.; Trope, C.G.; Reich, R.; Davidson, B. Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum. Pathol. 2012, 43, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ivan, M.; Hawkins, S.M. The role of MicroRNA molecules and MicroRNA-regulating machinery in the pathogenesis and progression of epithelial ovarian cancer. Gynecol. Oncol. 2017, 147, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.C.; Ho, M.Y.; Liang, S.M.; Liang, C.M. Autophagic degradation of SQSTM1 inhibits ovarian cancer motility by decreasing DICER1 and AGO2 to induce MIRLET7A-3P. Autophagy 2018, 14, 2065–2082. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Jin, X.; Ham, S.W.; Lee, S.Y.; Seo, S.; Kim, S.C.; Kim, S.H.; Kim, H. IRF7 promotes glioma cell invasion by inhibiting AGO2 expression. Tumour Biol. 2015, 36, 5561–5569. [Google Scholar] [CrossRef]
- Feng, B.; Hu, P.; Lu, S.J.; Chen, J.B.; Ge, R.L. Increased argonaute 2 expression in gliomas and its association with tumor progression and poor prognosis. Asian Pac. J. Cancer Prev. 2014, 15, 4079–4083. [Google Scholar] [CrossRef]
- Cheng, N.; Li, Y.; Han, Z.G. Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology 2013, 57, 1906–1918. [Google Scholar] [CrossRef]
- Ye, Z.; Jin, H.; Qian, Q. Argonaute 2: A Novel Rising Star in Cancer Research. J. Cancer 2015, 6, 877–882. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, H.; Liu, H.; Lv, S.; Wang, B.; Wang, R.; Liu, H.; Ding, M.; Yang, Y.; Li, L.; et al. MiRNA-99a directly regulates AGO2 through translational repression in hepatocellular carcinoma. Oncogenesis 2014, 3, e97. [Google Scholar] [CrossRef]
- Zhang, H.; Sheng, C.; Yin, Y.; Wen, S.; Yang, G.; Cheng, Z.; Zhu, Q. PABPC1 interacts with AGO2 and is responsible for the microRNA mediated gene silencing in high grade hepatocellular carcinoma. Cancer Lett. 2015, 367, 49–57. [Google Scholar] [CrossRef]
- Völler, D.; Linck, L.; Bruckmann, A.; Hauptmann, J.; Deutzmann, R.; Meister, G.; Bosserhoff, A.K. Argonaute Family Protein Expression in Normal Tissue and Cancer Entities. PLoS ONE 2016, 11, e0161165. [Google Scholar] [CrossRef]
- Sand, M.; Skrygan, M.; Georgas, D.; Arenz, C.; Gambichler, T.; Sand, D.; Altmeyer, P.; Bechara, F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012, 51, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Lv, J.; Liu, M.; Tang, H. miR-346 Up-regulates Argonaute 2 (AGO2) Protein Expression to Augment the Activity of Other MicroRNAs (miRNAs) and Contributes to Cervical Cancer Cell Malignancy. J. Biol. Chem. 2015, 290, 30342–30350. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, M.; Feng, Y.; Xu, Y.F.; Che, J.P.; Wang, G.C.; Zheng, J.H.; Gao, H.J. Evaluation of Argonaute protein as a predictive marker for human clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2013, 6, 1086–1094. [Google Scholar] [PubMed]
- Li, L.; Yu, C.; Gao, H.; Li, Y. Argonaute proteins: Potential biomarkers for human colon cancer. BMC Cancer 2010, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Papachristou, D.J.; Korpetinou, A.; Giannopoulou, E.; Antonacopoulou, A.G.; Papadaki, H.; Grivas, P.; Scopa, C.D.; Kalofonos, H.P. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. Int. J. Pathol. 2011, 459, 431–440. [Google Scholar] [CrossRef]
- Li, P.; Meng, J.; Zhai, Y.; Zhang, H.; Yu, L.; Wang, Z.; Zhang, X.; Cao, P.; Chen, X.; Han, Y.; et al. Argonaute 2 and nasopharyngeal carcinoma: A genetic association study and functional analysis. BMC Cancer 2015, 15, 862. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Y.; Wang, B.; Chen, X.; Li, W.; Dong, P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-derived FaDu cells. Oncotarget 2017, 8, 54735–54746. [Google Scholar] [CrossRef]
- Piroozian, F.; Bagheri Varkiyani, H.; Koolivand, M.; Ansari, M.; Afsa, M.; AtashAbParvar, A.; MalekZadeh, K. The impact of variations in transcription of DICER and AGO2 on exacerbation of childhood B-cell lineage acute lymphoblastic leukaemia. Int. J. Exp. Pathol. 2019, 100, 184–191. [Google Scholar] [CrossRef]
- Chang, S.S.; Smith, I.; Glazer, C.; Hennessey, P.; Califano, J.A. EIF2C is overexpressed and amplified in head and neck squamous cell carcinoma. ORL J. Otorhinolaryngol. Relat. Spec. 2010, 72, 337–343. [Google Scholar] [CrossRef]
- Papachristou, D.J.; Sklirou, E.; Corradi, D.; Grassani, C.; Kontogeorgakos, V.; Rao, U.N. Immunohistochemical analysis of the endoribonucleases Drosha, Dicer and Ago2 in smooth muscle tumours of soft tissues. Histopathology 2012, 60, E28–E36. [Google Scholar] [CrossRef]
- Zhao, X.; Guan, J.L. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 2011, 63, 610–615. [Google Scholar] [CrossRef]
- Winter, J.; Diederichs, S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011, 8, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Mihaly, Z.; Kormos, M.; Lanczky, A.; Dank, M.; Budczies, J.; Szasz, M.A.; Gyorffy, B. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res. Treat. 2013, 140, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Jeon, S.; Lee, K.M.; Han, S.; Song, M.; Choi, J.Y.; Park, S.K.; Yoo, K.Y.; Noh, D.Y.; Ahn, S.H.; et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and breast cancer survival. BMC Cancer 2012, 12, 195. [Google Scholar] [CrossRef]
- Horikawa, Y.; Wood, C.G.; Yang, H.; Zhao, H.; Ye, Y.; Gu, J.; Lin, J.; Habuchi, T.; Wu, X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 7956–7962. [Google Scholar] [CrossRef]
- Guo, Z.; Shu, Y.; Zhou, H.; Zhang, W. Identification of diagnostic and prognostic biomarkers for cancer: Focusing on genetic variations in microRNA regulatory pathways (Review). Mol. Med. Rep. 2016, 13, 1943–1952. [Google Scholar] [CrossRef]
- Bermisheva, M.A.; Takhirova, Z.R.; Gilyazova, I.R.; Khusnutdinova, E.K. MicroRNA Biogenesis Pathway Gene Polymorphisms Are Associated with Breast Cancer Risk. Russ. J. Genet. 2018, 54, 568–575. [Google Scholar] [CrossRef]
- Song, X.; Zhong, H.; Wu, Q.; Wang, M.; Zhou, J.; Zhou, Y.; Lu, X.; Ying, B. Association between SNPs in microRNA machinery genes and gastric cancer susceptibility, invasion, and metastasis in Chinese Han population. Oncotarget 2017, 8, 86435–86446. [Google Scholar] [CrossRef]
- Kershaw, M.H.; Westwood, J.A.; Parker, L.L.; Wang, G.; Eshhar, Z.; Mavroukakis, S.A.; White, D.E.; Wunderlich, J.R.; Canevari, S.; Rogers-Freezer, L.; et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 6106–6115. [Google Scholar] [CrossRef]
- Fang, X.; Yin, Z.; Li, X.; Xia, L.; Zhou, B. Polymorphisms in GEMIN4 and AGO1 Genes Are Associated with the Risk of Lung Cancer: A Case-Control Study in Chinese Female Non-Smokers. Int. J. Environ. Res. Public Health 2016, 13, 939. [Google Scholar] [CrossRef] [PubMed]
- Dobrijevic, Z.; Matijasevic, S.; Savic-Pavicevic, D.; Brajuskovic, G. Association between genetic variants in genes encoding Argonaute proteins and cancer risk: A meta-analysis. Pathol. Res. Pract. 2020, 216, 152906. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.O.; Du, Y.; Wang, Y.; Chang, W.C.; Chen, C.H.; et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013, 497, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; So, J.; Davis-Dusenbery, B.N.; Qi, H.H.; Bloch, D.B.; Shi, Y.; Lagna, G.; Hata, A. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol. Cell. Biol. 2011, 31, 4760–4774. [Google Scholar] [CrossRef]
- Chen, Z.; Lai, T.C.; Jan, Y.H.; Lin, F.M.; Wang, W.C.; Xiao, H.; Wang, Y.T.; Sun, W.; Cui, X.; Li, Y.S.; et al. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. J. Clin. Investig. 2013, 123, 1057–1067. [Google Scholar] [CrossRef]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Rendtlew Danielsen, J.M.; Yang, Y.G.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef]
- Gao, M.; Wei, W.; Li, M.M.; Wu, Y.S.; Ba, Z.; Jin, K.X.; Li, M.M.; Liao, Y.Q.; Adhikari, S.; Chong, Z.; et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014, 24, 532–541. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Shay, J.W.; Bacchetti, S. A survey of telomerase activity in human cancer. Eur. J. Cancer 1997, 33, 787–791. [Google Scholar] [CrossRef]
- Laudadio, I.; Orso, F.; Azzalin, G.; Calabro, C.; Berardinelli, F.; Coluzzi, E.; Gioiosa, S.; Taverna, D.; Sgura, A.; Carissimi, C.; et al. AGO2 promotes telomerase activity and interaction between the telomerase components TERT and TERC. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Ruberg, S.; Girard, M.; Cagnard, N.; Hanein, S.; Chretien, D.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE 2011, 6, e20746. [Google Scholar] [CrossRef] [PubMed]
- Hock, J.; Weinmann, L.; Ender, C.; Rudel, S.; Kremmer, E.; Raabe, M.; Urlaub, H.; Meister, G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007, 8, 1052–1060. [Google Scholar] [CrossRef]
- Jagannathan, R.; Thapa, D.; Nichols, C.E.; Shepherd, D.L.; Stricker, J.C.; Croston, T.L.; Baseler, W.A.; Lewis, S.E.; Martinez, I.; Hollander, J.M. Translational Regulation of the Mitochondrial Genome Following Redistribution of Mitochondrial MicroRNA in the Diabetic Heart. Circ. Cardiovasc. Genet. 2015, 8, 785–802. [Google Scholar] [CrossRef]
- Barrey, E.; Saint-Auret, G.; Bonnamy, B.; Damas, D.; Boyer, O.; Gidrol, X. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE 2011, 6, e20220. [Google Scholar] [CrossRef]
- Bian, Z.; Li, L.M.; Tang, R.; Hou, D.X.; Chen, X.; Zhang, C.Y.; Zen, K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010, 20, 1076–1078. [Google Scholar] [CrossRef]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef]
- Kren, B.T.; Wong, P.Y.; Sarver, A.; Zhang, X.; Zeng, Y.; Steer, C.J. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009, 6, 65–72. [Google Scholar] [CrossRef]
- Sripada, L.; Tomar, D.; Prajapati, P.; Singh, R.; Singh, A.K.; Singh, R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: Detailed analysis of mitochondrial associated miRNA. PLoS ONE 2012, 7, e44873. [Google Scholar] [CrossRef]
- Jeandard, D.; Smirnova, A.; Tarassov, I.; Barrey, E.; Smirnov, A. Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells 2019, 8, 286. [Google Scholar] [CrossRef] [PubMed]
- Pare, J.M.; Tahbaz, N.; Lopez-Orozco, J.; LaPointe, P.; Lasko, P.; Hobman, T.C. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol. Biol. Cell 2009, 20, 3273–3284. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, S.; Navarro-Gonzalez, C.; Panadero, J.; Villarroya, M.; Boutoual, R.; Sanchez-Alcazar, J.A.; Armengod, M.E. The MELAS mutation m.3243A > G alters the expression of mitochondrial tRNA fragments. Biochim. Biophys. Acta. Mol. Cell Res. 2019, 1866, 1433–1449. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Kumar, P.; Kiran, M.; Su, Z.; Malik, A.; Dutta, A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 2018, 24, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.; Phan, T.; Deng, X.; Zhou, J.; Lee, I.; Lee, Y.S.; et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. J. Am. Soc. Gene Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef]
- Kanai, A. Molecular Biology of the Transfer RNA Revisited; Frontiers E-books: Lausanne, Switzerland, 2014. [Google Scholar]
- Pederson, T. Regulatory RNAs derived from transfer RNA? RNA 2010, 16, 1865–1869. [Google Scholar] [CrossRef]
- Madiraju, A.K.; Erion, D.M.; Rahimi, Y.; Zhang, X.M.; Braddock, D.T.; Albright, R.A.; Prigaro, B.J.; Wood, J.L.; Bhanot, S.; MacDonald, M.J.; et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 2014, 510, 542–546. [Google Scholar] [CrossRef]
- Yang, L.; Li, P.; Fu, S.; Calay, E.S.; Hotamisligil, G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010, 11, 467–478. [Google Scholar] [CrossRef]
- Zhang, C.; Seo, J.; Murakami, K.; Salem, E.S.B.; Bernhard, E.; Borra, V.J.; Choi, K.; Yuan, C.L.; Chan, C.C.; Chen, X.; et al. Hepatic Ago2-mediated RNA silencing controls energy metabolism linked to AMPK activation and obesity-associated pathophysiology. Nat. Commun. 2018, 9, 3658. [Google Scholar] [CrossRef] [PubMed]
- Soufi, F.G.; Zonouzi, A.A.P.; Eftekhar, E.; Kamali, K.; Chegeni, S.A.; Zonouzi, A.P. The expression of Drosha, DGCR8, Dicer and Ago-2 genes are upregulated in human umbilical vein endothelial cells under hyperglycemic condition. Endocr. Regul. 2018, 52, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wang, Z.; Bishop, C.A.; Weitkunat, K.; Feng, X.; Tarbier, M.; Luo, J.; Friedlander, M.R.; Burkhardt, R.; Klaus, S.; et al. Control of hepatic gluconeogenesis by Argonaute2. Mol. Metab. 2018, 18, 15–24. [Google Scholar] [CrossRef]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef]
- Tattikota, S.G.; Rathjen, T.; McAnulty, S.J.; Wessels, H.H.; Akerman, I.; van de Bunt, M.; Hausser, J.; Esguerra, J.L.; Musahl, A.; Pandey, A.K.; et al. Argonaute2 mediates compensatory expansion of the pancreatic beta cell. Cell Metab. 2014, 19, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Tattikota, S.G.; Rathjen, T.; Hausser, J.; Khedkar, A.; Kabra, U.D.; Pandey, V.; Sury, M.; Wessels, H.H.; Mollet, I.G.; Eliasson, L.; et al. miR-184 Regulates Pancreatic beta-Cell Function According to Glucose Metabolism. J. Biol. Chem. 2015, 290, 20284–20294. [Google Scholar] [CrossRef] [PubMed]
- Volk, N.; Pape, J.C.; Engel, M.; Zannas, A.S.; Cattane, N.; Cattaneo, A.; Binder, E.B.; Chen, A. Amygdalar MicroRNA-15a Is Essential for Coping with Chronic Stress. Cell Rep. 2016, 17, 1882–1891. [Google Scholar] [CrossRef]
- Blake, D.D.; Weathers, F.W.; Nagy, L.M.; Kaloupek, D.G.; Gusman, F.D.; Charney, D.S.; Keane, T.M. The development of a Clinician-Administered PTSD Scale. J. Trauma. Stress 1995, 8, 75–90. [Google Scholar] [CrossRef]
- Heinzelmann, M.; Gill, J. Epigenetic Mechanisms Shape the Biological Response to Trauma and Risk for PTSD: A Critical Review. Nurs. Res. Pract. 2013, 2013, 417010. [Google Scholar] [CrossRef]
- Bam, M.; Yang, X.; Zumbrun, E.E.; Ginsberg, J.P.; Leyden, Q.; Zhang, J.; Nagarkatti, P.S.; Nagarkatti, M. Decreased AGO2 and DCR1 in PBMCs from War Veterans with PTSD leads to diminished miRNA resulting in elevated inflammation. Transl. Psychiatry 2017, 7, e1222. [Google Scholar] [CrossRef]
- Schaefer, A.; Im, H.I.; Veno, M.T.; Fowler, C.D.; Min, A.; Intrator, A.; Kjems, J.; Kenny, P.J.; O′Carroll, D.; Greengard, P. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J. Exp. Med. 2010, 207, 1843–1851. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, F.V.; Bell, K.; Cox, H.; Broadie, K.S.; Tully, T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat. Neurosci. 2008, 11, 1143–1145. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, R.J., 3rd; Bear, M.F. The autistic neuron: Troubled translation? Cell 2008, 135, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Liu-Yesucevitz, L.; Bassell, G.J.; Gitler, A.D.; Hart, A.C.; Klann, E.; Richter, J.D.; Warren, S.T.; Wolozin, B. Local RNA translation at the synapse and in disease. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 16086–16093. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Berry-Kravis, E.; Kaufmann, W.E.; Ono, M.Y.; Tartaglia, N.; Lachiewicz, A.; Kronk, R.; Delahunty, C.; Hessl, D.; Visootsak, J.; et al. Advances in the treatment of fragile X syndrome. Pediatrics 2009, 123, 378–390. [Google Scholar] [CrossRef]
- van Kouwenhove, M.; Kedde, M.; Agami, R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 2011, 11, 644–656. [Google Scholar] [CrossRef]
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol. Cell 2014, 54, 573–585. [Google Scholar] [CrossRef]
- Kenny, P.J.; Zhou, H.; Kim, M.; Skariah, G.; Khetani, R.S.; Drnevich, J.; Arcila, M.L.; Kosik, K.S.; Ceman, S. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014, 9, 1729–1741. [Google Scholar] [CrossRef]
- Sakaguchi, A.; Yamashita, Y.; Ishii, T.; Uehara, T.; Kosaki, K.; Takahashi, T.; Takenouchi, T. Further evidence of a causal association between AGO1, a critical regulator of microRNA formation, and intellectual disability/autism spectrum disorder. Eur. J. Med. Genet. 2019, 62, 103537. [Google Scholar] [CrossRef]
- Pircs, K.; Petri, R.; Madsen, S.; Brattas, P.L.; Vuono, R.; Ottosson, D.R.; St-Amour, I.; Hersbach, B.A.; Matusiak-Bruckner, M.; Lundh, S.H.; et al. Huntingtin Aggregation Impairs Autophagy, Leading to Argonaute-2 Accumulation and Global MicroRNA Dysregulation. Cell Rep. 2018, 24, 1397–1406. [Google Scholar] [CrossRef]
- Vandoorne, T.; De Bock, K.; Van Den Bosch, L. Energy metabolism in ALS: An underappreciated opportunity? Acta Neuropathol. 2018, 135, 489–509. [Google Scholar] [CrossRef] [PubMed]
- Maniataki, E.; Mourelatos, Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA 2005, 11, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.N.; Kar, A.N.; Kowalak, J.A.; Gale, J.R.; Aschrafi, A.; Chen, C.Y.; Gioio, A.E.; Kaplan, B.B. Axonal localization and mitochondrial association of precursor microRNA 338. Cell. Mol. Life Sci. 2016, 73, 4327–4340. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Visavadiya, N.P.; Pandya, J.D.; Nelson, P.T.; Sullivan, P.G.; Springer, J.E. Mitochondria-associated microRNAs in rat hippocampus following traumatic brain injury. Exp. Neurol. 2015, 265, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Gershoni-Emek, N.; Altman, T.; Ionescu, A.; Costa, C.J.; Gradus-Pery, T.; Willis, D.E.; Perlson, E. Localization of RNAi Machinery to Axonal Branch Points and Growth Cones Is Facilitated by Mitochondria and Is Disrupted in ALS. Front. Mol. Neurosci. 2018, 11, 311. [Google Scholar] [CrossRef]
- Kanekura, K.; Yagi, T.; Cammack, A.J.; Mahadevan, J.; Kuroda, M.; Harms, M.B.; Miller, T.M.; Urano, F. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum. Mol. Genet. 2016, 25, 1803–1813. [Google Scholar] [CrossRef]
- Russo, A.; Scardigli, R.; La Regina, F.; Murray, M.E.; Romano, N.; Dickson, D.W.; Wolozin, B.; Cattaneo, A.; Ceci, M. Increased cytoplasmic TDP-43 reduces global protein synthesis by interacting with RACK1 on polyribosomes. Hum. Mol. Genet. 2017, 26, 1407–1418. [Google Scholar] [CrossRef]
- Cestra, G.; Rossi, S.; Di Salvio, M.; Cozzolino, M. Control of mRNA Translation in ALS Proteinopathy. Front. Mol. Neurosci. 2017, 10, 85. [Google Scholar] [CrossRef]
- Tom Dieck, S.; Hanus, C.; Schuman, E.M. SnapShot: Local protein translation in dendrites. Neuron 2014, 81, 958. [Google Scholar] [CrossRef][Green Version]
- Rotem, N.; Magen, I.; Ionescu, A.; Gershoni-Emek, N.; Altman, T.; Costa, C.J.; Gradus, T.; Pasmanik-Chor, M.; Willis, D.E.; Ben-Dov, I.Z.; et al. ALS Along the Axons—Expression of Coding and Noncoding RNA Differs in Axons of ALS models. Sci. Rep. 2017, 7, 44500. [Google Scholar] [CrossRef]
- Zappulo, A.; van den Bruck, D.; Ciolli Mattioli, C. RNA localization is a key determinant of neurite-enriched proteome. Nat. Commun. 2017, 8, 583. [Google Scholar] [CrossRef] [PubMed]
- Peredo, J.; Villace, P.; Ortin, J.; de Lucas, S. Human Staufen1 associates to miRNAs involved in neuronal cell differentiation and is required for correct dendritic formation. PLoS ONE 2014, 9, e113704. [Google Scholar] [CrossRef] [PubMed]
- Gershoni-Emek, N.; Chein, M.; Gluska, S.; Perlson, E. Amyotrophic lateral sclerosis as a spatiotemporal mislocalization disease: Location, location, location. Int. Rev. Cell Mol. Biol. 2015, 315, 23–71. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.J.; Cooper-Knock, J.; Dodd, J.E.; Stopford, M.J.; Mihaylov, S.R.; Kirby, J.; Shaw, P.J.; Hautbergue, G.M. Invited review: Decoding the pathophysiological mechanisms that underlie RNA dysregulation in neurodegenerative disorders: A review of the current state of the art. Neuropathol. Appl. Neurobiol. 2015, 41, 109–134. [Google Scholar] [CrossRef]
- Kapur, M.; Monaghan, C.E.; Ackerman, S.L. Regulation of mRNA Translation in Neurons-A Matter of Life and Death. Neuron 2017, 96, 616–637. [Google Scholar] [CrossRef]
- Lotti, F.; Maggi, M. Ultrasound of the male genital tract in relation to male reproductive health. Hum. Reprod. Update 2015, 21, 56–83. [Google Scholar] [CrossRef]
- Conine, C.C.; Batista, P.J.; Gu, W.; Claycomb, J.M.; Chaves, D.A.; Shirayama, M.; Mello, C.C. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2010, 107, 3588–3593. [Google Scholar] [CrossRef]
- Han, T.; Manoharan, A.P.; Harkins, T.T.; Bouffard, P.; Fitzpatrick, C.; Chu, D.S.; Thierry-Mieg, D.; Thierry-Mieg, J.; Kim, J.K. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 18674–18679. [Google Scholar] [CrossRef]
- Conine, C.C.; Moresco, J.J.; Gu, W.; Shirayama, M.; Conte, D., Jr.; Yates, J.R., 3rd; Mello, C.C. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 2013, 155, 1532–1544. [Google Scholar] [CrossRef]
- van Wolfswinkel, J.C.; Ketting, R.F. The role of small non-coding RNAs in genome stability and chromatin organization. J. Cell Sci. 2010, 123, 1825–1839. [Google Scholar] [CrossRef]
- Modzelewski, A.J.; Hilz, S.; Crate, E.A.; Schweidenback, C.T.; Fogarty, E.A.; Grenier, J.K.; Freire, R.; Cohen, P.E.; Grimson, A. Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. J. Cell Sci. 2015, 128, 2314–2327. [Google Scholar] [CrossRef] [PubMed]
- Maine, E.M. Meiotic silencing in Caenorhabditis elegans. Int. Rev. Cell Mol. Biol. 2010, 282, 91–134. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, H.; Liu, Y.; Zhao, F.; Zhu, S.; Xie, C.; Tang, T.S.; Guo, C. Germline deletion of huntingtin causes male infertility and arrested spermiogenesis in mice. J. Cell Sci. 2016, 129, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Savas, J.N.; Makusky, A.; Ottosen, S.; Baillat, D.; Then, F.; Krainc, D.; Shiekhattar, R.; Markey, S.P.; Tanese, N. Huntington′s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc. Natl. Acad. Sci. USA 2008, 105, 10820–10825. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.; Tang, F.; O′Carroll, D.; Lao, K.; Surani, M.A. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin 2009, 2, 9. [Google Scholar] [CrossRef]
- Stein, P.; Rozhkov, N.V.; Li, F.; Cardenas, F.L.; Davydenko, O.; Vandivier, L.E.; Gregory, B.D.; Hannon, G.J.; Schultz, R.M. Essential Role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 2015, 11, e1005013. [Google Scholar] [CrossRef]
- Claycomb, J.M.; Batista, P.J.; Pang, K.M.; Gu, W.; Vasale, J.J.; van Wolfswinkel, J.C.; Chaves, D.A.; Shirayama, M.; Mitani, S.; Ketting, R.F.; et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 2009, 139, 123–134. [Google Scholar] [CrossRef]
- Yigit, E.; Batista, P.J.; Bei, Y.; Pang, K.M.; Chen, C.C.; Tolia, N.H.; Joshua-Tor, L.; Mitani, S.; Simard, M.J.; Mello, C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 2006, 127, 747–757. [Google Scholar] [CrossRef]
- Gerson-Gurwitz, A.; Wang, S.; Sathe, S.; Green, R.; Yeo, G.W.; Oegema, K.; Desai, A. A Small RNA-Catalytic Argonaute Pathway Tunes Germline Transcript Levels to Ensure Embryonic Divisions. Cell 2016, 165, 396–409. [Google Scholar] [CrossRef]
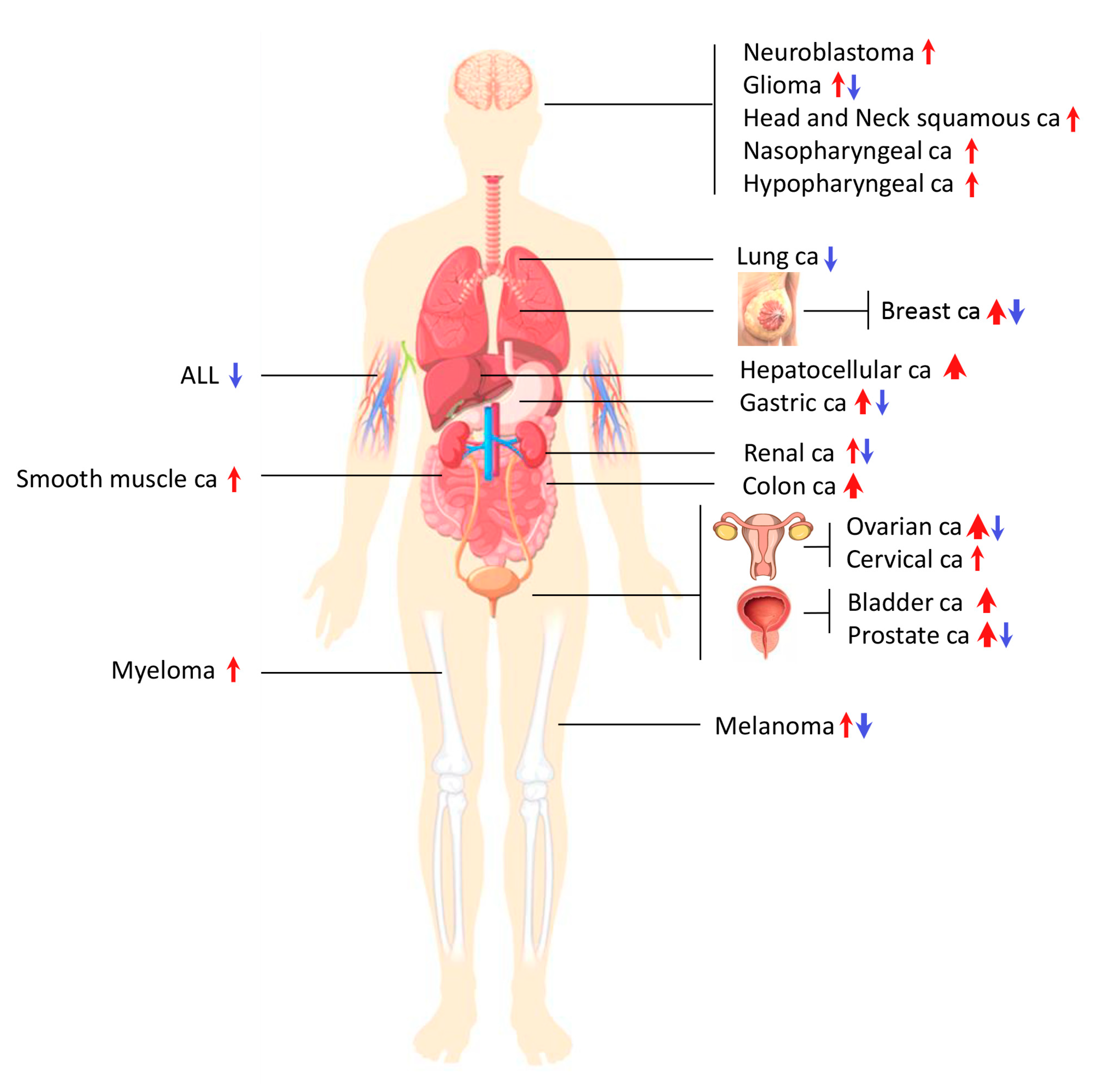
| Cancer Type | AGO Type per Citation | Deregulation (Up or Down) | References |
|---|---|---|---|
| Gastric cancer | AGO2 | upregulated | [141] |
| AGO2 | downregulated | [131] | |
| AGO2 | upregulated | [142] | |
| Prostate cancer | AGO2 | upregulated | [141] |
| AGO2 | upregulated | [143] | |
| AGO2 | downregulated | [130] | |
| AGO2 | upregulated | [144] | |
| Neuroblastoma | AGO2 | upregulated | [141] |
| Bladder cancer | AGO2 | upregulated | [129] |
| AGO2 | upregulated | [145] | |
| AGO1 | upregulated | [146] | |
| AGO2 | upregulated | ||
| Myeloma | AGO2 | upregulated | [147] |
| Breast cancer | AGO2 | downregulated | [148] |
| AGO2 | upregulated | [126] | |
| AGO2 | upregulated | [149] | |
| AGO2 | downregulated | [148] | |
| AGO1 AGO2 | upregulated | [150] | |
| Ovarian cancer | AGO1 AGO2 | upregulated | [151] |
| AGO1 AGO2 | upregulated | [152] | |
| AGO2 | upregulated | [153] | |
| AGO2 | downregulated | [154] | |
| AGO2 | upregulated | [155] | |
| Hepatocellular carcinoma | AGO2 | upregulated | [156] |
| AGO2 | upregulated | [157] | |
| AGO2 | upregulated | [158] | |
| AGO2 | upregulated | [159] | |
| Melanoma | AGO1 | downregulated | [160] |
| AGO2 | downregulated | ||
| AGO3 | downregulated | ||
| AGO4 | downregulated | ||
| AGO2 | downregulated | [127] | |
| AGO1 AGO2 | upregulated | [161] | |
| Cervical cancer | AGO2 | upregulated | [162] |
| Renal carcinoma | AGO1 AGO2 | upregulated | [163] |
| AGO2 | downregulated | [132] | |
| Colon cancer | AGO2 | upregulated | [142] |
| AGO1 | upregulated | [164] | |
| AGO2 | upregulated | ||
| AGO3 | upregulated | ||
| AGO4 | upregulated | ||
| AGO2 | upregulated | [165] | |
| Nasopharyngeal carcinoma (NPC) | AGO2 | upregulated (specific genetic variants) | [166] |
| Hypopharyngeal cancer | AGO2 | upregulated | [167] |
| Lung cancer | AGO2 | downregulated | [139] |
| Acute lymphoblastic leukemia (ALL) | AGO2 | downregulated | [168] |
| Head and neck squamous cell carcinoma | AGO2 | upregulated | [169] |
| Smooth muscle tumors | AGO2 | upregulated | [170] |
| Glioma | AGO2 | upregulated | [155] |
| AGO2 | downregulated | [154] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantazopoulou, V.I.; Georgiou, S.; Kakoulidis, P.; Giannakopoulou, S.N.; Tseleni, S.; Stravopodis, D.J.; Anastasiadou, E. From the Argonauts Mythological Sailors to the Argonautes RNA-Silencing Navigators: Their Emerging Roles in Human-Cell Pathologies. Int. J. Mol. Sci. 2020, 21, 4007. https://doi.org/10.3390/ijms21114007
Pantazopoulou VI, Georgiou S, Kakoulidis P, Giannakopoulou SN, Tseleni S, Stravopodis DJ, Anastasiadou E. From the Argonauts Mythological Sailors to the Argonautes RNA-Silencing Navigators: Their Emerging Roles in Human-Cell Pathologies. International Journal of Molecular Sciences. 2020; 21(11):4007. https://doi.org/10.3390/ijms21114007
Chicago/Turabian StylePantazopoulou, Vasiliki I., Stella Georgiou, Panos Kakoulidis, Stavroula N. Giannakopoulou, Sofia Tseleni, Dimitrios J. Stravopodis, and Ema Anastasiadou. 2020. "From the Argonauts Mythological Sailors to the Argonautes RNA-Silencing Navigators: Their Emerging Roles in Human-Cell Pathologies" International Journal of Molecular Sciences 21, no. 11: 4007. https://doi.org/10.3390/ijms21114007
APA StylePantazopoulou, V. I., Georgiou, S., Kakoulidis, P., Giannakopoulou, S. N., Tseleni, S., Stravopodis, D. J., & Anastasiadou, E. (2020). From the Argonauts Mythological Sailors to the Argonautes RNA-Silencing Navigators: Their Emerging Roles in Human-Cell Pathologies. International Journal of Molecular Sciences, 21(11), 4007. https://doi.org/10.3390/ijms21114007






