RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Results
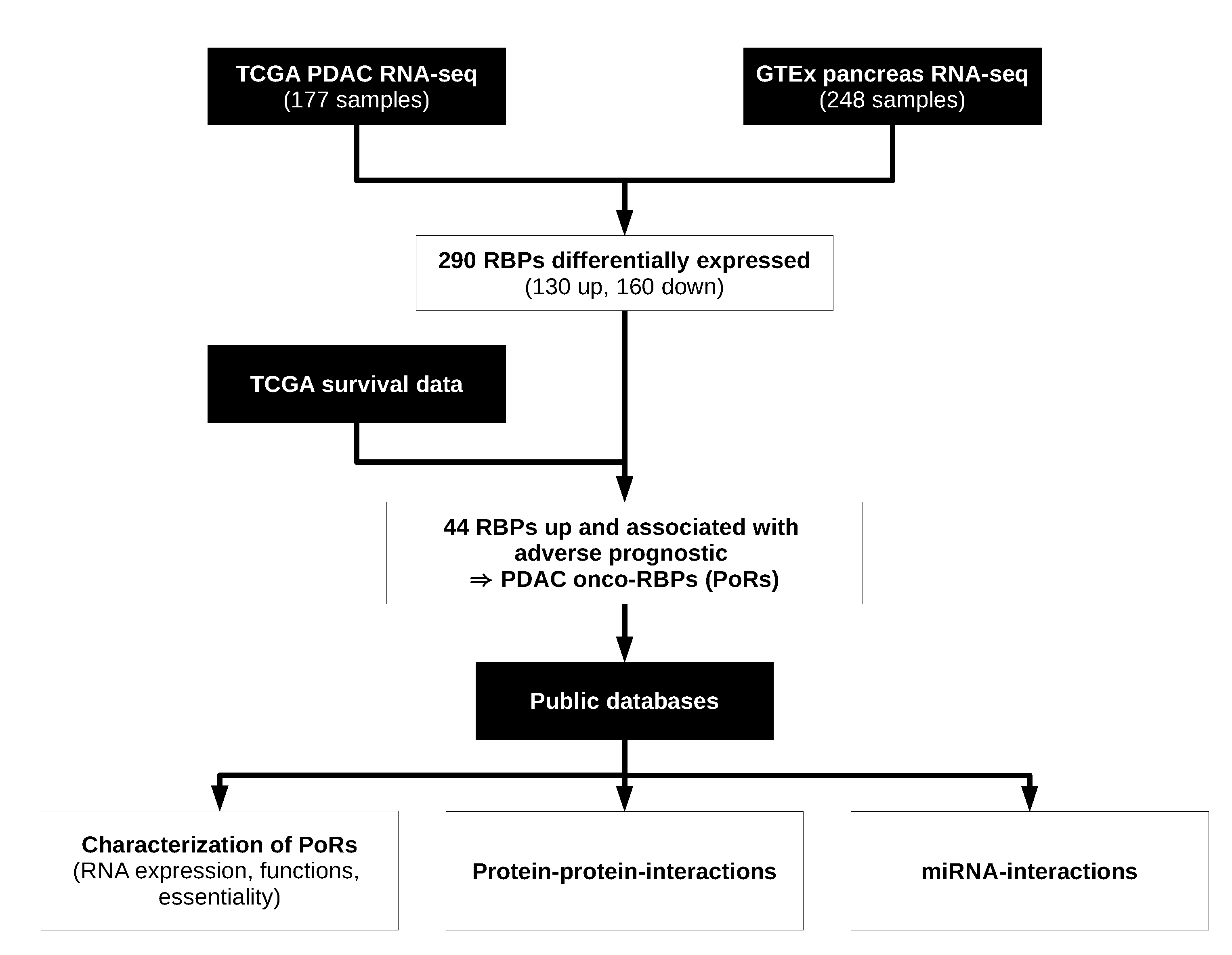
2.1. Workflow for the Identification and Characterization of Oncogenic RBPs in PDAC
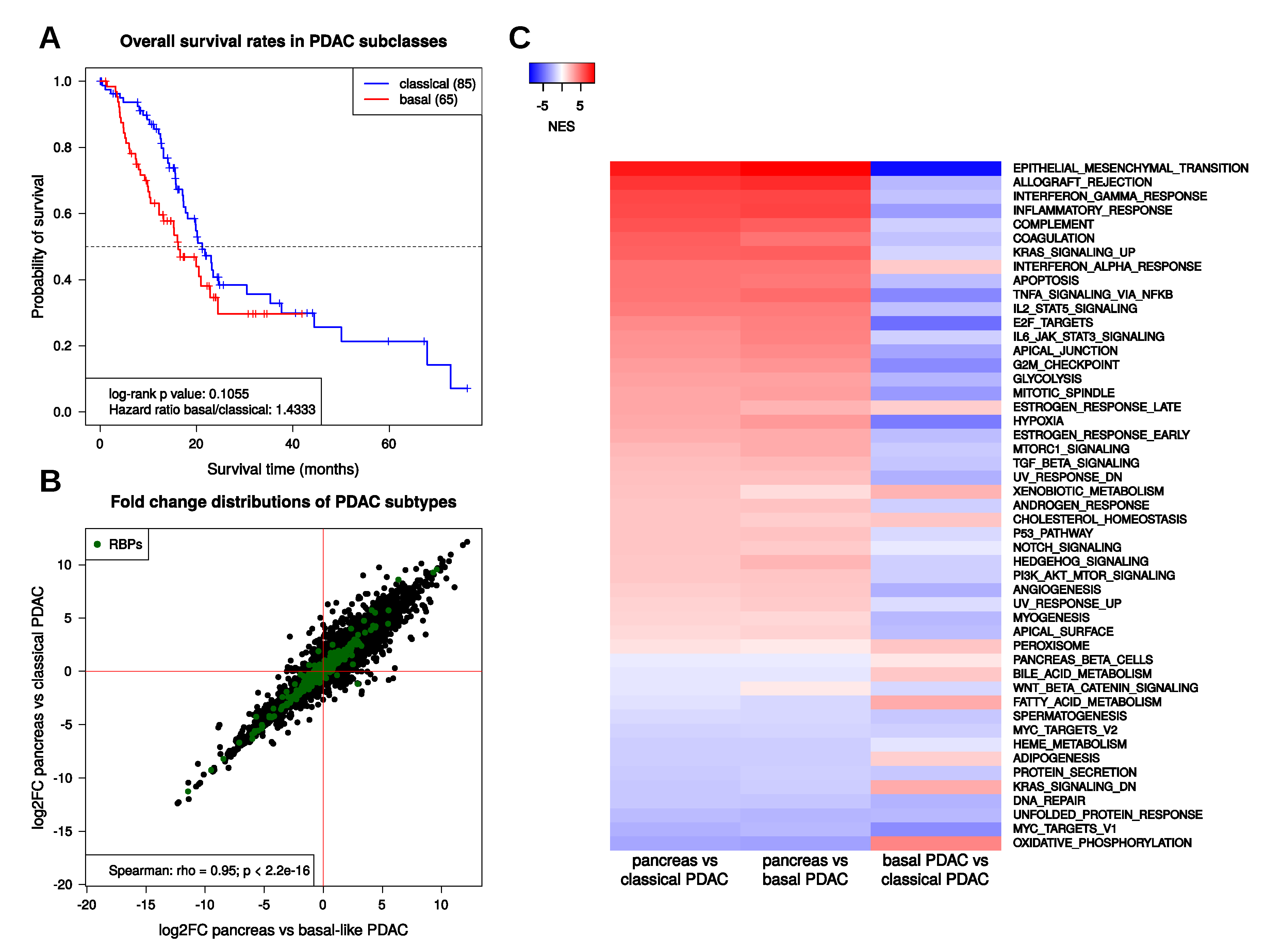
2.2. Transcriptional Regulation in PDAC
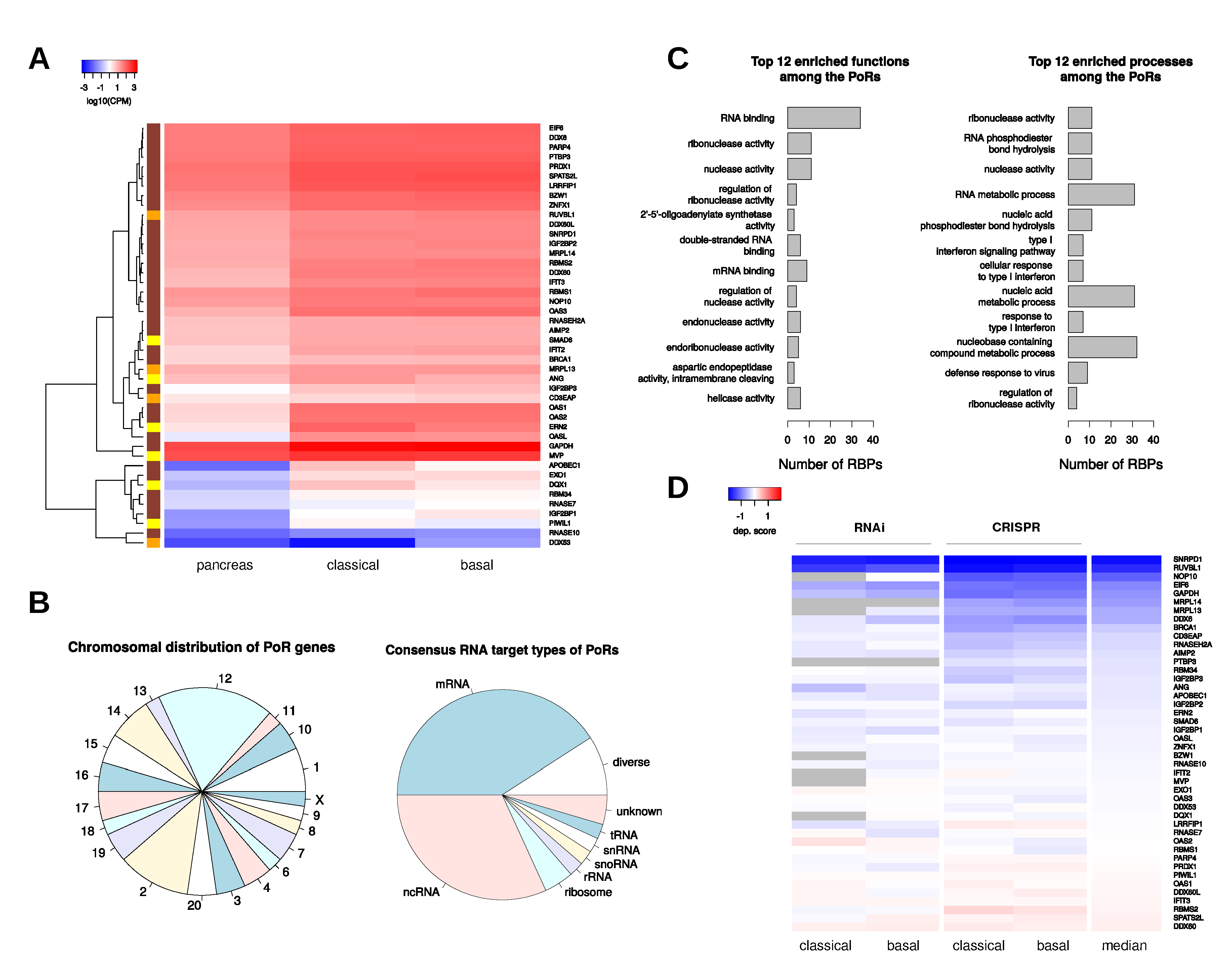
2.3. Transcriptional Regulation of RBPs in PDAC
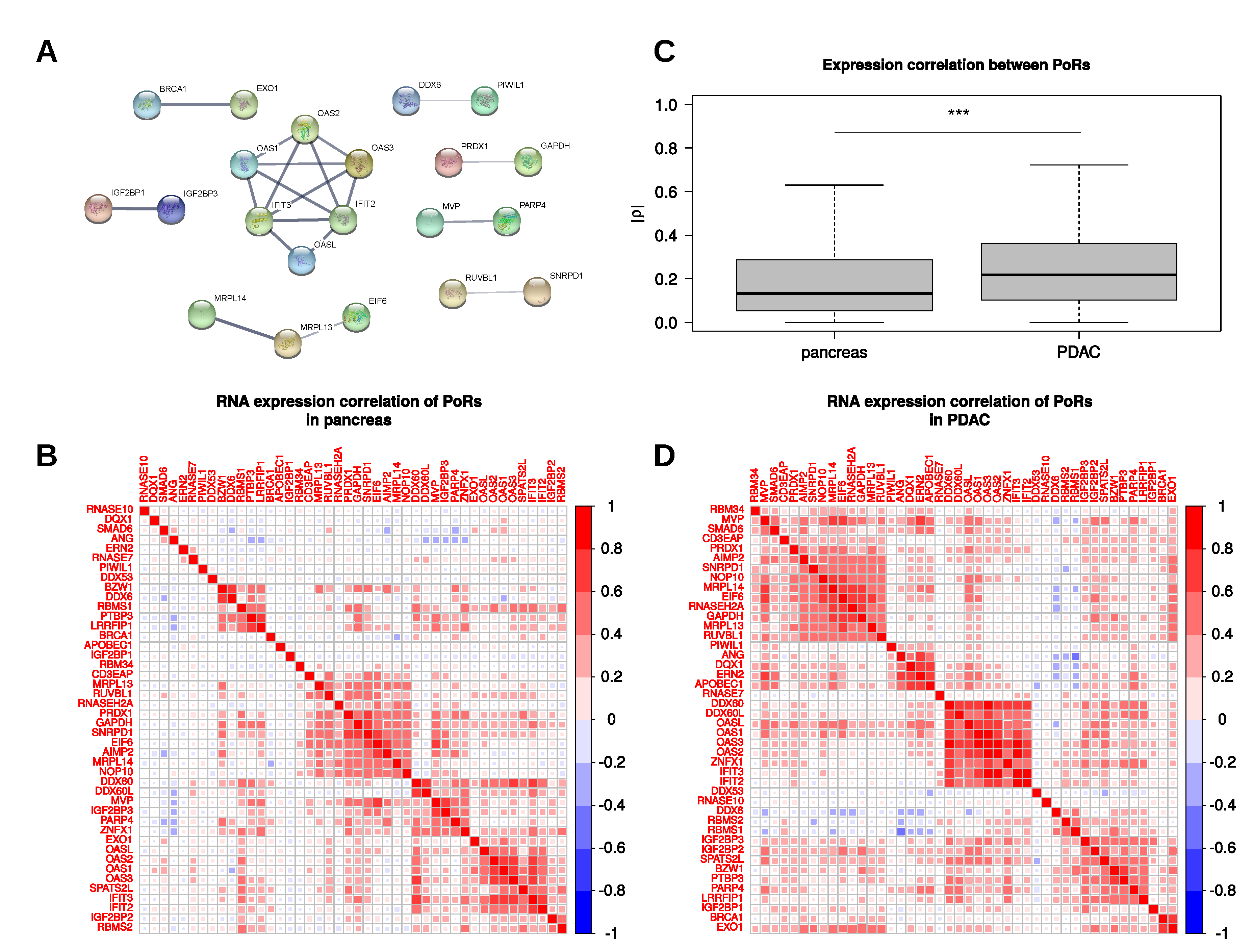
2.4. Characterization and Interactions of the PoRs
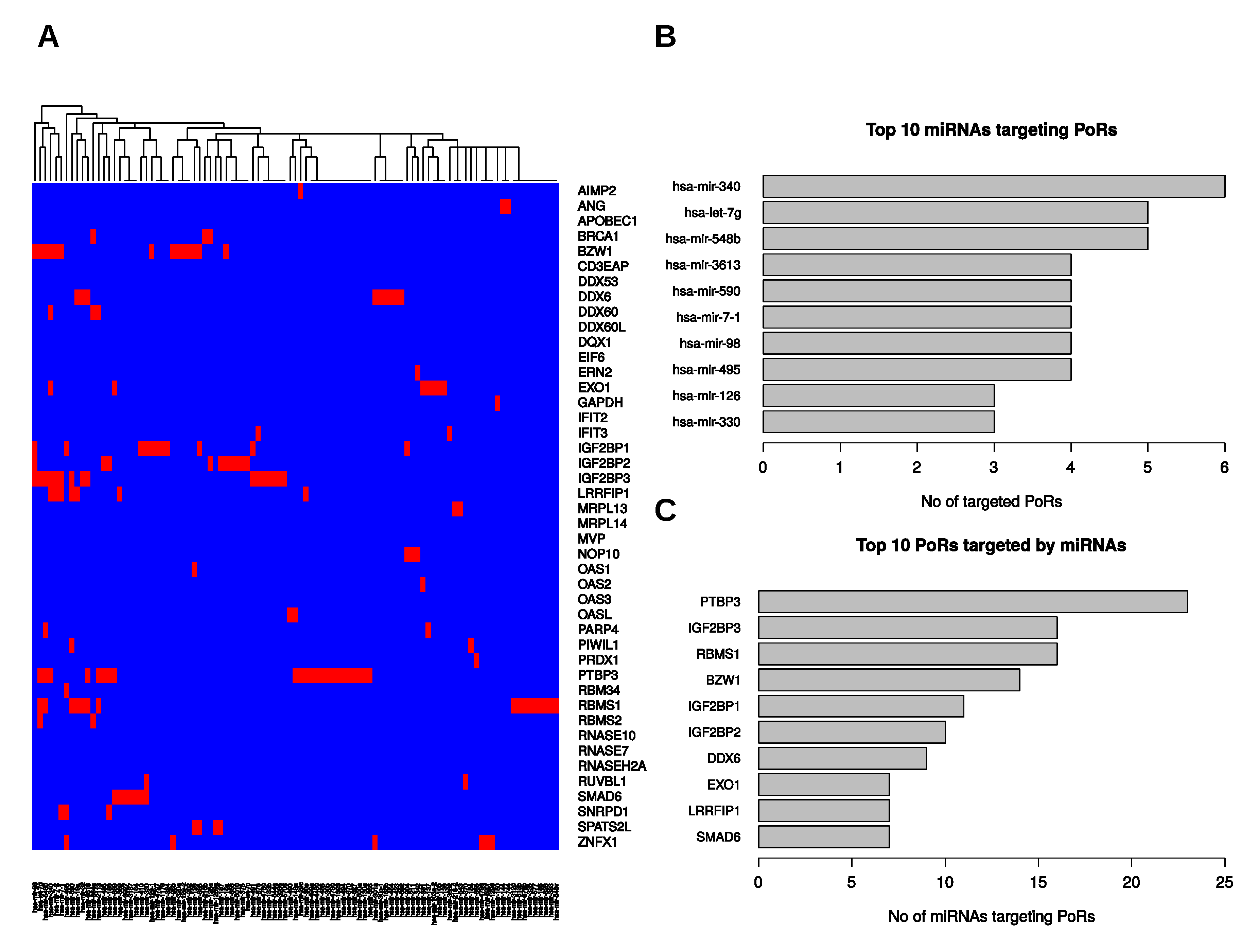
2.5. Post-Transcriptional Regulation of the PoRs by microRNAs
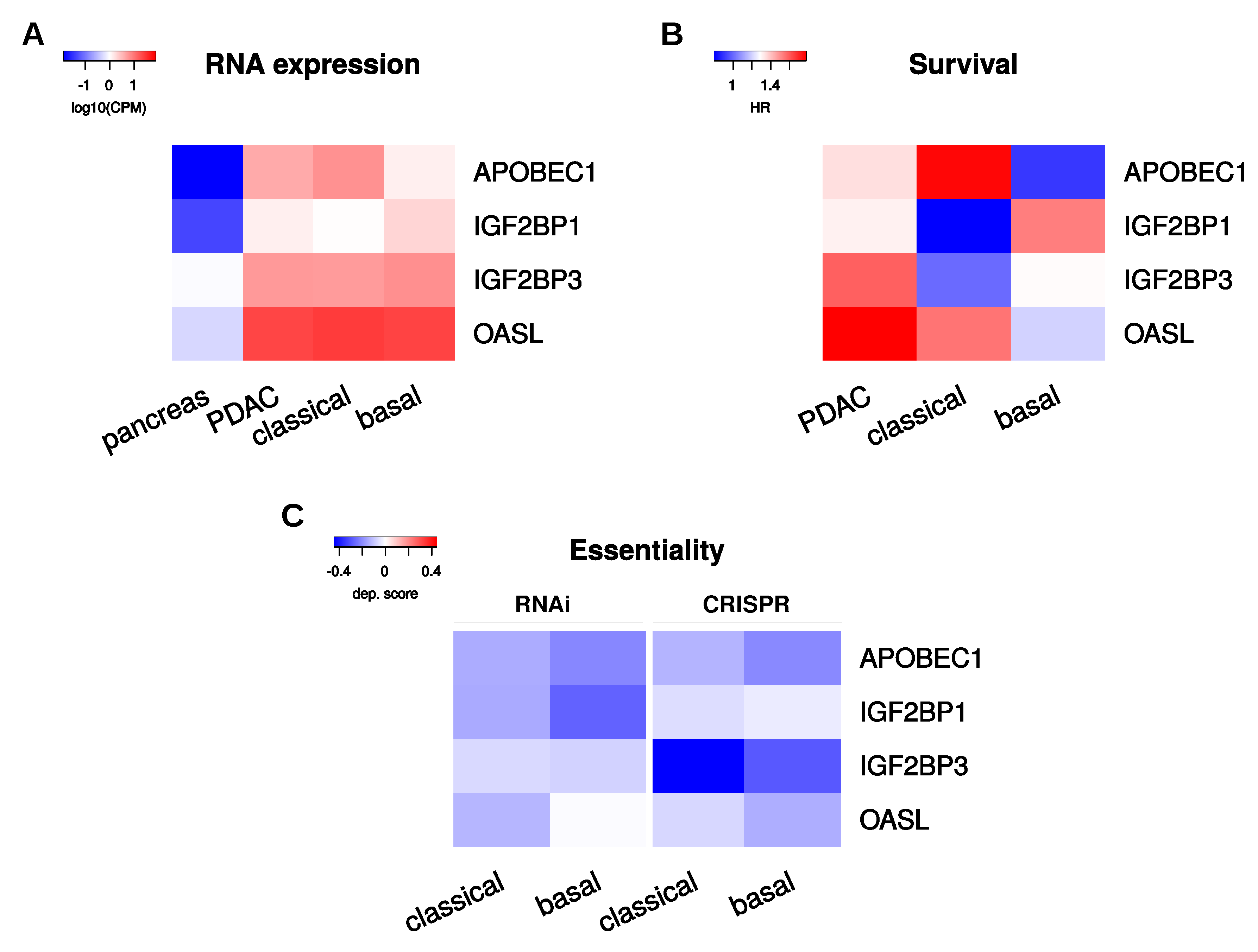
2.6. Selection of Candidate PoRs for Therapeutic Targeting
3. Discussion
4. Materials and Methods
4.1. RNA-seq Data Processing, Differential Expression, and Survival Analysis
4.2. Functional Enrichment Analyses
4.3. RNA Expression and Dependency Scores in PDAC Cell Lines
4.4. Protein–Protein Interaction Analysis
4.5. MicroRNA Binding Prediction
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Chen, C.C.; Chen, L.L.; Li, C.P.; Hsu, Y.T.; Jiang, S.S.; Fan, C.S.; Chua, K.V.; Huang, S.X.; Shyr, Y.M.; Chen, L.T.; et al. Myeloid-derived macrophages and secreted HSP90α induce pancreatic ductal adenocarcinoma development. Oncoimmunology 2018, 7, e1424612. [Google Scholar] [CrossRef]
- Knudsen, E.S.; Balaji, U.; Mannakee, B.; Vail, P.; Eslinger, C.; Moxom, C.; Mansour, J.; Witkiewicz, A.K. Pancreatic cancer cell lines as patient-derived avatars: Genetic characterisation and functional utility. Gut 2018, 67, 508–520. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Juiz, N.A.; Iovanna, J.; Dusetti, N. Pancreatic Cancer Heterogeneity Can Be Explained Beyond the Genome. Front. Oncol. 2019, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Abreu, F.B.; Liu, X.; Tsongalis, G.J. miRNA analysis in pancreatic cancer: The Dartmouth experience. Clin. Chem. Lab. Med. 2017, 55, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Lu, D.; Lin, Y.; Lv, Y.; He, L. A seven-miRNA expression-based prognostic signature and its corresponding potential competing endogenous RNA network in early pancreatic cancer. Exp. Ther. Med. 2019, 18, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, B.; Chen, D.; Liu, L.; Huang, C.; Lu, Z.; Lun, L.; Wan, X. miR-139 and miR-200c regulate pancreatic cancer endothelial cell migration and angiogenesis. Oncol. Rep. 2015, 34, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wan, B.S.; Cheng, M.; Zhang, L. Insulin-like growth factor 2 mRNA-binding protein 1 promotes cell proliferation via activation of AKT and is directly targeted by microRNA-494 in pancreatic cancer. World J. Gastroenterol. 2019, 25, 6063–6076. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wu, Z.; Cheng, Y.; Wei, W.; Hao, L. Insulin-like growth factor 2 mRNA binding protein 2 promotes aerobic glycolysis and cell proliferation in pancreatic ductal adenocarcinoma via stabilizing GLUT1 mRNA. Acta Biochim. Biophys. Sin. (Shanghai) 2019, 51, 743–752. [Google Scholar] [CrossRef]
- Dahlem, C.; Barghash, A.; Puchas, P.; Haybaeck, J.; Kessler, S.M. The Insulin-Like Growth Factor 2 mRNA Binding Protein IMP2/IGF2BP2 is Overexpressed and Correlates with Poor Survival in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 3204. [Google Scholar] [CrossRef] [PubMed]
- Ennajdaoui, H.; Howard, J.M.; Sterne-Weiler, T.; Jahanbani, F.; Coyne, D.J.; Uren, P.J.; Dargyte, M.; Katzman, S.; Draper, J.M.; Wallace, A.; et al. IGF2BP3 Modulates the Interaction of Invasion-Associated Transcripts with RISC. Cell Rep. 2016, 15, 1876–1883. [Google Scholar] [CrossRef]
- Schaeffer, D.F.; Owen, D.R.; Lim, H.J.; Buczkowski, A.K.; Chung, S.W.; Scudamore, C.H.; Huntsman, D.G.; Ng, S.S.; Owen, D.A. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer 2010, 10, 59. [Google Scholar] [CrossRef]
- Kugel, S.; Sebastián, C.; Fitamant, J.; Ross, K.N.; Saha, S.K.; Jain, E.; Gladden, A.; Arora, K.S.; Kato, Y.; Rivera, M.N.; et al. SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell 2016, 165, 1401–1415. [Google Scholar] [CrossRef]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; LaPierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R.I. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef]
- Busch, B.; Bley, N.; Müller, S.; Glaß, M.; Misiak, D.; Lederer, M.; Vetter, M.; Strauß, H.G.; Thomssen, C.; Hüttelmaier, S. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016, 44, 3845–3864. [Google Scholar] [CrossRef]
- Dong, X.; Li, Y.; Hess, K.R.; Abbruzzese, J.L.; Li, D. DNA mismatch repair gene polymorphisms affect survival in pancreatic cancer. Oncologist 2011, 16, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.F.; Jimbo, M.; Wulfkuhle, J.; Gallagher, I.; Deng, J.; Enyenihi, L.; Meisner-Kober, N.; Londin, E.; Rigoutsos, I.; Sawicki, J.A.; et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene 2016, 35, 2529–2541. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Weng, L.; Jia, Y.; Liu, B.; Wu, S.; Xue, L.; Yin, X.; Mao, A.; Wang, Z.; Shang, M. PTBP3 promotes malignancy and hypoxia-induced chemoresistance in pancreatic cancer cells by ATG12 up-regulation. J. Cell. Mol. Med. 2020, 24, 2917–2930. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdȯttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Wang, L.; Rowe, R.G.; Jaimes, A.; Yu, C.; Nam, Y.; Pearson, D.S.; Zhang, J.; Xie, X.; Marion, W.; Heffron, G.J.; et al. Small-Molecule Inhibitors Disrupt let-7 Oligouridylation and Release the Selective Blockade of let-7 Processing by LIN28. Cell Rep. 2018, 23, 3091–3101. [Google Scholar] [CrossRef]
- Zhao, Y.; Altendorf-Hofmann, A.; Pozios, I.; Camaj, P.; Däberitz, T.; Wang, X.; Niess, H.; Seeliger, H.; Popp, F.; Betzler, C.; et al. Elevated interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) is a poor prognostic marker in pancreatic ductal adenocarcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 1061–1068. [Google Scholar] [CrossRef]
- Müller, S.; Bley, N.; Glaß, M.; Busch, B.; Rousseau, V.; Misiak, D.; Fuchs, T.; Lederer, M.; Hüttelmaier, S. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 2018, 46, 6285–6303. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Chen, B.; Aran, D.; Charalel, J.; Yau, C.; Wolf, D.M.; van ’t Veer, L.J.; Butte, A.J.; Goldstein, T.; Sirota, M. Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat. Commun. 2019, 10, 3574. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.Y.; Kang, J.S.; Hwang, Y.S.; Kim, Y.J. Oligoadenylate synthase-like (OASL) proteins: Dual functions and associations with diseases. Exp. Mol. Med. 2015, 47, e144. [Google Scholar] [CrossRef] [PubMed]
- Ishidate, T.; Ozturk, A.R.; Durning, D.J.; Sharma, R.; Shen, E.Z.; Chen, H.; Seth, M.; Shirayama, M.; Mello, C.C. ZNFX-1 Functions within Perinuclear Nuage to Balance Epigenetic Signals. Mol. Cell 2018, 70, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, S.; Jia, X.; Ge, Y.; Ling, T.; Nie, M.; Lan, X.; Chen, S.; Xu, A. Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat. Cell Biol. 2019, 21, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Grünvogel, O.; Esser-Nobis, K.; Reustle, A.; Schult, P.; Müller, B.; Metz, P.; Trippler, M.; Windisch, M.P.; Frese, M.; Binder, M.; et al. DDX60L Is an Interferon-Stimulated Gene Product Restricting Hepatitis C Virus Replication in Cell Culture. J. Virol. 2015, 89, 10548–10568. [Google Scholar] [CrossRef] [PubMed]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576. [Google Scholar] [CrossRef]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Tomimatsu, N.; Mukherjee, B.; Catherine Hardebeck, M.; Ilcheva, M.; Vanessa Camacho, C.; Louise Harris, J.; Porteus, M.; Llorente, B.; Khanna, K.K.; Burma, S. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat. Commun. 2014, 5, 3561. [Google Scholar] [CrossRef]
- Bell, J.L.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 2013, 70, 2657–2675. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Ren, P.; Zhang, Y.; Jiang, J.; Zhang, H. MicroRNAs-491-5p suppresses cell proliferation and invasion by inhibiting IGF2BP1 in non-small cell lung cancer. Am. J. Transl. Res. 2016, 8, 485–495. [Google Scholar]
- Lu, L.; Cai, M.; Peng, M.; Wang, F.; Zhai, X. miR-491-5p functions as a tumor suppressor by targeting IGF2 in colorectal cancer. Cancer Manag. Res. 2019, 11, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lü, Y.; Jiang, Q.; Yang, Y.; Dang, C.; Sun, R. miR-491 inhibits BGC-823 cell migration via targeting HMGA2. Int. J. Biol. Markers 2019, 34, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479–480, 131–145. [Google Scholar] [CrossRef]
- Olson, M.E.; Harris, R.S.; Harki, D.A. APOBEC Enzymes as Targets for Virus and Cancer Therapy. Cell Chem. Biol. 2018, 25, 36–49. [Google Scholar] [CrossRef]
- Yamanaka, S.; Balestra, M.E.; Ferrell, L.D.; Fan, J.; Arnold, K.S.; Taylor, S.; Taylor, J.M.; Innerarity, T.L. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. USA 1995, 92, 8483–8487. [Google Scholar] [CrossRef]
- Yamanaka, S.; Poksay, K.S.; Arnold, K.S.; Innerarity, T.L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997, 11, 321–333. [Google Scholar] [CrossRef]
- Chester, A.; Scott, J.; Anant, S.; Navaratnam, N. RNA editing: Cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta 2000, 1494, 1–13. [Google Scholar] [CrossRef]
- Niavarani, A.; Shahrabi Farahani, A.; Sharafkhah, M.; Rassoulzadegan, M. Pancancer analysis identifies prognostic high-APOBEC1 expression level implicated in cancer in-frame insertions and deletions. Carcinogenesis 2018, 39, 327–335. [Google Scholar] [CrossRef]
- Lv, J.; Wang, L.; Shen, H.; Wang, X. Regulatory roles of OASL in lung cancer cell sensitivity to Actinidia chinensis Planch root extract (acRoots). Cell Biol. Toxicol. 2018, 34, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Lederer, M.; Bley, N.; Schleifer, C.; Hüttelmaier, S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin. Cancer Biol. 2014, 29, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Söhr, N.; Hüttelmaier, S. IGF2BP1: A post-transcriptional “driver” of tumor cell migration. Cell Adh. Migr. 2012, 6, 312–318. [Google Scholar]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Müller, S.; Glaß, M.; Singh, A.K.; Haase, J.; Bley, N.; Fuchs, T.; Lederer, M.; Dahl, A.; Huang, H.; Chen, J.; et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019, 47, 375–390. [Google Scholar] [CrossRef]
- Mahapatra, L.; Andruska, N.; Mao, C.; Le, J.; Shapiro, D.J. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl. Oncol. 2017, 10, 818–827. [Google Scholar] [CrossRef]
- Gonzalez, K.J.; Moncada-Giraldo, D.M.; Gutierrez, J.B. In silico identification of potential inhibitors against human 2’-5’- oligoadenylate synthetase (OAS) proteins. Comput. Biol. Chem. 2020, 85, 107211. [Google Scholar] [CrossRef]
- Hartmann, R.; Walko, G.; Justesen, J. Inhibition of 2′-5′ oligoadenylate synthetase by divalent metal ions. FEBS Lett. 2001, 507, 54–58. [Google Scholar] [CrossRef][Green Version]
- Li, L.; Gai, X. The association between dietary zinc intake and risk of pancreatic cancer: A meta-analysis. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Bewick, V.; Cheek, L.; Ball, J. Statistics review 12: Survival analysis. Crit. Care 2004, 8, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glaß, M.; Michl, P.; Hüttelmaier, S. RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 4190. https://doi.org/10.3390/ijms21114190
Glaß M, Michl P, Hüttelmaier S. RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma. International Journal of Molecular Sciences. 2020; 21(11):4190. https://doi.org/10.3390/ijms21114190
Chicago/Turabian StyleGlaß, Markus, Patrick Michl, and Stefan Hüttelmaier. 2020. "RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma" International Journal of Molecular Sciences 21, no. 11: 4190. https://doi.org/10.3390/ijms21114190
APA StyleGlaß, M., Michl, P., & Hüttelmaier, S. (2020). RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma. International Journal of Molecular Sciences, 21(11), 4190. https://doi.org/10.3390/ijms21114190





