Abstract
The prion protein (PrP) is an enigmatic molecule with a pleiotropic effect on different cell types; it is localized stably in lipid raft microdomains and it is able to recruit downstream signal transduction pathways by its interaction with various biochemical partners. Since its discovery, this lipid raft component has been involved in several functions, although most of the publications focused on the pathological role of the protein. Recent studies report a key role of cellular prion protein (PrPC) in physiological processes, including cellular differentiation. Indeed, the PrPC, whose expression is modulated according to the cell differentiation degree, appears to be part of the multimolecular signaling pathways of the neuronal differentiation process. In this review, we aim to summarize the main findings that report the link between PrPC and stem cells.
Keywords:
prion protein; prions; lipid raft; rafts; raft microdomains; stem cells; mesenchymal stem cells 1. Introduction
The prion protein (PrP), a molecule discovered by Stanley Prusiner, is involved in the transmissible spongiform encephalopathies (TSEs), such as family fatal insomnia (FFI), Creutzfeldt–Jakob disease (CJD), Gerstmann–Sträussler–Scheinker (GSS), and other pathologies [1,2].
Many studies have shown that two possible protein tridimensional conformations may exist: the cellular prion protein (PrPC), normally expressed in all nucleated cells; and the scrapie prion protein (PrPSc), which is involved in the TSEs. The main difference between these isoforms is the content of the β-sheets, which is higher in PrPSc compared to PrPC [3,4,5].
In many conditions, the physiological isoform of PrPC is able to translate into the pathological isoform PrPSc; this transition can be regarded as a post-translational refolding process [6,7], leading to acquiring new physico-chemical properties. In fact, PrPSc has a high β-sheet structure, insolubility, and partial resistance against digestion by proteinase K [8,9,10].
In humans, PrPC is encoded by the PRNP gene, and the protein consists of approximately 250 amino acid residues and a highly conserved membrane-bound glycoprotein anchored by glycosylphosphatidylinositol (GPI) [11].
In reference to the PrPC structure, the N-terminus is distinguished by an octapeptide repeated region able to bind the Cu2+ ions and, for this reason, it is involved in oxidative stress resistance. The middle region contains a cluster of lysine residues and a hydrophobic domain [12], whereas the C-terminal globular domain contains 3 α-helices, 2 short β-sheets, and interconnecting loops [3,13,14]. Moreover, a disulfide bond is found between residues 179 and 214 [15] and, also, some N-linked glycans can be added at residues 181 and 197 [16].
In mammals, PrPC is expressed in all nucleated cells although, it is mainly expressed in neuronal cells [17]. In response to some stimuli, such as proteases or reactive oxygen species (ROS) [18,19], the PrPC, like many other proteins, could be subjected to cleavage in different sites, giving rise to different membrane-bound and soluble fragments of different sizes and features [20].
Since its discovery, it has been well known that PrPC is present on the plasma membrane associated with the lipid raft microdomains [21], structures that represent particular sub-compartments of the plasma membrane enriched in cholesterol and glycosphingolipids, such as GM3, GM1, and GD3 [22,23]. Several studies have shown that lipid rafts are important in the refolding process of the PrPC in PrPSc [24,25]. Indeed, the increasing of the membrane-anchored PrPC local concentration seems to be able to induce a conformational transition accompanied by di- or oligomerization of the PrPC. Elfrink et al. have proposed that membrane anchoring of an excess of prion protein is the structural prerequisite in the development of prion diseases [10,23]. Other authors have suggested that the prion protein–GM1 interaction within lipid rafts at the cell surface could play a significant role in the mechanism predisposing to pathology [26].
Although the PrPSc has been associated with the pathogenesis of TSEs, different reports provided evidences that PrPC is a pivotal molecule with fundamental roles in various physiological and developmental processes [16,27].
Researchers focused on understanding the physiological role of the PrPC [11]. In fact, since its discovery, many functions have been attributed to PrPC, such as the copper-binding ability in brain membrane fractions, which controls the activity of other membrane-associated copper-binding proteins and enhances copper incorporation into superoxide dismutase (SOD) [28,29].
Moreover, the preferential localization of PrPC in the pre- and postsynaptic compartments of the nerve terminals implies that it might be involved in preserving the normal synaptic structure and function by regulating synaptic transmission and plasticity [30]. Recent evidence suggested the possibility that a hypoxia-mediated PrPC increase plays an important role in angiogenesis [31,32].
Additionally, in vitro studies propose that PrPC is involved both in the regulation of neuritogenesis [33,34] as well as axonal growth [35,36], and in tumorigenesis by regulating tumor growth, differentiation, and resistance to conventional therapies. In fact, PrPC overexpression is related to the acquisition of a malignant phenotype of cancer stem cells (CSCs) in different tumors, such as pancreatic ductal adenocarcinoma (PDAC), osteosarcoma, breast cancer, gastric cancer, and glioblastoma multiforme (GBM) [37].
In the last years, several scientists have emphasized a possible role of PrPC in stem cell biology [30,38]. Indeed, as reported in the literature, PrPC is expressed in a wide variety of stem cells, including embryonic and hematopoietic stem cells, and its function has been linked to the modulation of the proliferation and self-renewal capacity of these kind of cells [38,39,40].
Tremblay et al. showed that PrPC is firstly highly expressed during murine embryogenesis, predominantly in post-mitotic neural cells that have undergone neuronal differentiation [41]; subsequently, the PrPC expression expands to non-neuronal tissues. In the human forebrain, PrPC expression starts at the 11th week; it continues until the end of gestation and it occurs predominantly in the axonal tract, suggesting a specific role for this molecule in axonal growth during development [30,36].
A recent study published by Martellucci et al. has furnished evidence of the presence of PrPC in human dental pulp-derived stem cells (hDPSCs) and of its role in the neuronal differentiation process; in addition, it has been shown that the lipid raft’s integrity is essential for the PrPC-induced signal pathways, and that is essential for the neuronal differentiation process of hDPSCs induced by the epidermal growth factor and basic fibroblast growth factor (EGF/bFGF) [42,43]. PrPC plays a key role in the neuronal differentiation process [44], in which the PrPC interacts with EGF-R within lipid rafts, playing a role in the multimolecular signal complexes involved in the hDPSCs neuronal differentiation process [42].
PrPC is constitutively present in lipid rafts [45] and in a wide variety of stem cells [39,40]; in fact, it is involved in stemness modulation and, mostly, in the self-renewal and proliferation of tissue-resident stem cells and the neuronal differentiation of neural stem cells [46].
The purpose of this review is to highlight the role of PrPC as a lipid raft component during neuronal and neuronal differentiation processes.
2. Prion Protein as Raft Component
Since its discovery, various authors have showed that PrPC is present on the cell plasma membrane—associated with different gangliosides and cholesterol—within particular structures named “lipid raft microdomains” [21,47]. These structures represent domains of plasma membrane that contain high concentrations of cholesterol and glycosphingolipids, such as GM3, GM1, GD3, and proteins; lipid rafts can dissociate and associate rapidly, forming functional clusters in cell membranes [22,48]. They are distinct regions of the plasma membrane where they may represent a large fraction. Rafts are characterized by a distinctive protein and lipid composition, depending on the cell type or tissue [49].
A lot of proteins are present within lipid rafts; in particular, those involved in cell signaling. In fact, a number of proteins involved in the signal transduction pathways have been copurified with lipid rafts on a sucrose gradient as ZAP-70, Fyn, p56lck [50,51]. For this reason, lipid rafts are thought to be involved in the regulation of signal transduction pathways [52,53].
These clusters, present in the outer monolayer of the plasma membrane, provide highly efficient lipid–protein modules, which operate in membrane trafficking and cell signaling [52]. Several authors theorized lipid rafts as sequestering platforms of specific proteins, thus modulating cell signaling [21,53,54].
Different evidence suggests that there are probably different mechanisms through which rafts may control cell signaling. For example, rafts may contain incomplete signaling pathways that are activated when a receptor is recruited within the raft or by suppressing the intrinsic activity of the signaling proteins present within rafts [55].
It was also demonstrated that some proteins are mainly distributed within lipid rafts or are recruited in response to specific stimuli [35].
Like other glycosylphosphatidylinositol (GPI)-anchored proteins, most PrPC molecules are found in lipid rafts from neural and non-neural cells [56]. PrPC is localized stably in the lipid raft microdomains and able to recruit the downstream signal transduction pathways by interaction with various partners [57].
In a recent study, it was reported that GM2 lipid rafts are present on the plasma membrane of hDPSCs. Besides, the PrPC is able to associate specifically with GM2 in non-treated hDPSCs while, after the neuronal differentiation process induced by EGF/bFGF, the PrPC associates preferentially with GD3 lipid rafts. Moreover, this study suggests that all the main lipid constitutive components of the rafts, such as the gangliosides and cholesterol, are essential for the hDPSCs neuronal differentiation process [35]. Thus, the results point out the functional role of the lipid rafts and PrPC in the hDPSCs neuronal differentiation process, suggesting that these structures may represent specific chambers, where multimolecular signaling complexes, including the lipids (i.e., gangliosides, cholesterol) and proteins (i.e., PrPC, EGF-R), play a role in neuronal differentiation [22,35]. It was found that, following neuronal differentiation of the hDPSCs, EGF-R is recruited within the lipid raft where it interacts with PrPC (Figure 1).
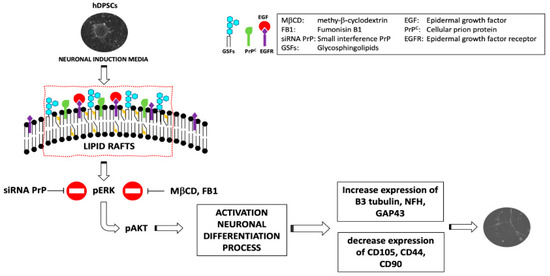
Figure 1.
Role of lipid raft inhibitors and siRNA PrP in the neurodifferentiation process of hDPSCs.
In fact, lipid raft inhibitors, such as fumonisin B1 and methyl-β-cyclodextrin (MβCD), significantly prevented ERK 1/2 and Akt phosphorylation and neuronal differentiation process induced by EGF/bFGF [35]. It was also observed that silencing of PrPC by the usage of a specific small interference RNA (siRNA PrP), in order to ablate its function, affected neuronal differentiation process mediated by EGF/bFGF [35].
In agreement with this data, other authors highlighted the association of PrPC with two components of the EGF-R macromolecular complex, such as Grb-2 [58] and Src [59]. This indicates that PrPC may be part of the cell membrane complexes that regulate EGF/EGF-R signaling [42]. It was hypothesized that, after neuronal induction, EGF-R is recruited within the lipid rafts, where, interacting with PrPC triggers the signal transduction, starting the neuronal differentiation process [42].
3. Prion Protein and Signaling Pathway
PrPC is a highly conserved cell surface GPI-anchored glycoprotein that, as reported above, was first identified as a molecule able to bind Cu++ [28]. Cross-linkage of GPI-anchored proteins usually results in protein sorting, shedding, and cell signaling, which is greatly influenced by the GPI-anchor signal sequence [60]. In particular, crosslink of PrPC on the surface of T-lymphocytes has been associated with various cellular responses, such as the intracellular Ca++ mobilization [61], Src, and extracellular-signal-regulated kinase (ERK) activation or capping of the lipid raft microdomains with Fyn phosphorylation [62]. Indeed, anti-PrPC antibody coimmunoprecipitated Fyn; as a downstream target of Fyn activation, PrPC was shown to activate the ERK1/2, which takes part in the mitogen-activated protein kinase (MAPK) cascades [63]. Recently, Martellucci et al. confirmed and extended the role of PrPC in this signaling pathway, demonstrating that recombinant prion protein 23–231 (recPrPC) is involved in the neuronal differentiation process, by activating ERK 1/2 and Akt [35,42]. Interestingly, this activity required an endogenous PrPC to mediate the way of the signal triggered by recPrPC. Furthermore, PrPC was shown to activate the Lyn- and Syk-dependent signal transduction pathways in different cell types [51,64]. It leads to a transient release of Ca++ that induces activation of protein kinase C- and Ca++-dependent tyrosine kinases. In addition, a direct interaction of PrPC with synapsin Ib and the adapter protein Grb-2 was also reported [58].
PrPC may also activate the neuroprotective signaling pathway(s). In fact, dimerization of PrPC leads to clustering in multimolecular complexes and serves to regulate different aspects of neuronal homeostasis, whereas intracellular dimerization appears to be the most relevant event in neuroprotection, via N1 and C1 prion metabolites [23]. Indeed, the dimerization stimulates α-cleavage and thus the production of the neuroprotective fragments.
The neuroprotective functions of PrPC may be attributed to its BCL-2-like properties [65]. PrPC protects against cell death by preventing the conformational change of BAX occurring during BAX activation [66]. A neuroprotective activity of PrPC was reported by Mitteregger et al. [67], who revealed that both the C-terminal GPI anchor and the N-terminal domain are required for this physiological activity. In particular, the cAMP-dependent protein kinase A (PKA) seems to mediate the neuroprotective signals, as demonstrated by Chiarini et al. [68].
In addition, the phosphatidylinositol 3-kinase/AKT (PI3K/AKT) pathway is involved in the regulation of PrPC-induced neuroprotection. Thus, PrPC might act as a signaling molecule at the cell surface to promote stress-protective signaling under physiological conditions, which can be switched to toxic signaling through the interaction with β-sheet-rich conformers. In this concern, the role of PrPC may depend on its intracellular localization [61]. Indeed, PrPC translocation from lipid rafts to non-lipid rafts prevents p38 and caspase-3 activation with consequent inhibition of cell apoptosis. PrPC is generally reported as a plasma membrane protein; however, studies revealed the presence of endogenous PrPC as an interacting protein with the membrane/organelles [69], as well as with the cytoskeleton network. In fact, lipid microdomains are similarly formed at subcellular organelles, including the endoplasmic reticulum, Golgi complex, and mitochondria, named lipid raft-like microdomains [70]. In the last few years, Mattei et al. identified PrPC as a new component of mitochondrial raft-like microdomains in T cells undergoing CD95/Fas-mediated apoptosis, indicating that PrPC could undergo intracellular re-localization via the ER–mitochondria-associated membranes (MAM) and microtubular network [69].
A new and innovative point of view suggests that PrPC increases the calcineurin activity, resulting in decreased AMPK phosphorylation that induces autophagic cell death [71]. Interestingly, this study demonstrated that the prion protein–calcineurin activation was involved not only in prion protein-mediated neuronal cell death but also in the AMPK and autophagy signaling pathways. It regulates metabolic homeostasis [72] by controlling autophagy [73]. However, the overexpression of PrPC inhibited the autophagic flux signals, lipid accumulation, as well as the PPAR-γ and C/EBP-α mRNA and protein expression levels in comparison to the control cells [74].
In conclusion, the modular structure, the variety of binding partners, and the typical localization within the lipid rafts, suggest that PrPC may be a key component of the dynamic platforms on the cell surface, with the capability to assemble multicomponent complexes through different domains, triggering different signaling pathways that regulate differentiation and cell fate.
4. Prion Protein and Stem Cells
Different studies demonstrated a strong relationship between the cellular form of prion protein and stem cells, considering the PrPC as an element of the pluripotency and self-renewal matrix [38,75]. Furthermore, the expression of PrPC is modulated according to the degree of stem cell differentiation and it is involved in the molecular signaling that underlies the differentiation process of several cell lineages [42,76].
In human embryonic stem cell (hESC) differentiation, it was observed that PrPC is involved in controlling the cell cycle dynamics, self-renewal, and the fate of this kind of cell. Furthermore, silencing PrPC in hESCs undergoing spontaneous differentiation altered the dynamics of the cell cycle and changed the balance between the lineages of the three germ layers, where differentiation toward ectodermal lineages was suppressed. Moreover, over-expression of PrPC in hESCs undergoing spontaneous differentiation inhibited differentiation toward lineages of all three germ layers and helped to preserve high proliferation activity. These results illustrate that PrPC is involved in key activities that dictate the status of the hESCs, including regulation of the cell cycle dynamics, controlling the switch between self-renewal and differentiation, and determining the fate of hESCs differentiation. Thus, PrPC is at the crossroads of several signaling pathways that regulate the switch between preservation of or departure from the self-renewal state, control cell proliferation activity, and define the stem cell fate [39].
In human mesenchymal stem cells (hMSCs), the PrPC has been shown to enhance proliferation and promote self-renewal of this kind of cell. In fact, the expression of PrPC decreased in hMSCs following ex vivo expansion. When PrPC expression was knocked down, the hMSCs showed a significant reduction in proliferation and differentiation. In contrast, the hMSCs expanded in the presence of small molecule 3/689, a modulator of PrPC expression, showing retention of PrPC expression with ex vivo expansion and an extended lifespan of up to 10 population doublings [76]. On the basis of this findings, a lot of preclinical and clinical studies indicated PrPC as a potential target for therapeutic strategy.
Indeed, as reported by Lee et al., hMSCs are promising candidates for stem cell-based therapy in ischemic diseases that induce pathophysiological conditions, such as oxidative stress and inflammation. The authors demonstrated how melatonin promotes hMSCs functionality and enhances MSC-mediated neovascularization in ischemic tissues through the upregulation of PrPC expression. So, melatonin-treated hMSCs could provide a therapeutic strategy for vessel regeneration in ischemic disease, and the targeting of PrPC levels may prove instrumental for MSC-based therapies [77]. In reference to melatonin, another work team showed that this hormone inhibits colon cancer stem cells (CSCs) by regulating the PrPC–Oct4 axis. Indeed, in specimens from patients with colorectal cancer, the expressions of PrPC and Oct4 were significantly correlated with metastasis and tumor stages. Co-treatment with 5-fluorouracil (5-FU) and melatonin inhibited the stem cell markers Oct4, Nanog, Sox2, and ALDH1A1 by downregulating PrPC. In this way, tumor growth, proliferation, and tumor-mediated angiogenesis were suppressed. In colorectal CSCs, PRNP overexpression protects Oct4 against inhibition by 5-FU and melatonin. So, the authors suggest that the co-treatment with anticancer drugs and melatonin is a potential therapy for colorectal cancer and PrPC maintains cancer stemness during tumor progression. Therefore, targeting the PrPC–Oct4 axis may prove instrumental in colorectal cancer therapy [78].
In the same direction of Lee et al., many studies demonstrated that MSCs promote regeneration of injured tissues, interacting with the PrPC that plays an active role in neuronal survival and angioneurogenesis [77,78,79,80]. In fact, hypoxia enhanced the proliferative potential of MSCs by promoting the expression of normal PrPC, suggesting that hypo-MSCs offer a therapeutic strategy for accelerated neovasculogenesis in ischemic diseases, and that PrPC comprises a potential target for MSC-based therapies [81]. Corsaro et al. also showed that PrPC regulates different biological functions in human tumors, including glioblastoma (GBM). The authors analyzed the role of PrPC in GBM cell pathogenicity, focusing on tumor-initiating cells (TICs or CSCs), the subpopulation responsible for development, progression, and recurrence of most malignancies. Analyzing four GBM CSC-enriched cultures, they showed that PrPC expression is directly correlated with the proliferation rate of the cells. To better define its role in CSCs biology, they knocked-down PrPC expression in two of these GBM-derived CSCs cultures by specific lentiviral-delivered shRNAs. The work provided evidence that the CSC proliferation rate, spherogenesis, and in vivo tumorigenicity are significantly inhibited in PrPC downregulated cells. Moreover, PrPC downregulation caused loss of expression of the stemness and self-renewal markers (NANOG, Sox2) as well as the activation of differentiation pathways (i.e. increased GFAP expression). The authors suggested that PrPC controls the stemness properties of human GBM CSCs and that its downregulation induces the acquisition of a more differentiated and less oncogenic phenotype [82].
5. Prion Protein in Neural and Neuronal Differentiation Processes
The spectrum of proposed biological functions of PrPC has been expanded rapidly over the last decade. Extensive experimental works disclosed multiple physiological roles of PrPC at the molecular, cellular, and systemic levels, affecting the homeostasis of copper, neuroprotection, stem cell renewal, and memory mechanisms, among others. Various authors proposed that the biological function of the PrPC is that of a cell surface scaffold protein, based on the striking similarities of its functional properties with those of scaffold proteins involved in the organization of intracellular signal transduction pathways [57,83]. However, PrPC is highly conserved in mammals and is present on all nucleated cells, although it is mainly expressed in the central and peripheral nervous system. So, an increasing number of authors investigated the role of PrPC as a key component of multimolecular complexes during the neuronal differentiation process [43]. As reported by Lee et al., PrPC is a glycoprotein that is expressed on the cell surface beginning with the early stages of embryonic stem cell differentiation. The ectopic expression of PrPC in ESCs triggers differentiation toward endodermal, mesodermal, and ectodermal lineages, whereas silencing of PrPC suppresses the differentiation toward ectodermal but not endodermal or mesodermal lineages [39]. Starting with the role of PrPC in controlling the balance between cells of different lineages, the authors also tested whether PrPC controls the differentiation of hESCs into cells of the neuron-, oligodendrocyte-, and astrocyte-committed lineages. They found that silencing of PrPC suppressed the differentiation toward all three lineages. Moreover, switching PrPC expression during a differentiation time course revealed that silencing PrPC expression during the very initial stage that corresponds to embryonic bodies has a more significant impact than silencing it at the later stages of differentiation. Their work illustrated that PrPC controls differentiation of hESCs toward the neuron-, oligodendrocyte-, and astrocyte-committed lineages, and is likely involved at the stage of uncommitted neural progenitor cells rather than lineage-committed neural progenitors [46]. The importance of PrPC at the early stages of neural differentiation is represented by different studies.
Prodromidou et al. showed that PrPC is essential for proper neural stem/precursor cells (NPCs) proliferation, neuronal differentiation, and, moreover, PrPC is required for the NPC response to the neural cell adhesion molecule. In the absence of PrPC, NCAM not only fails to promote neuronal differentiation but also induces an accumulation of doublecortin-positive neuronal progenitors at the proliferation stage. So, the authors demonstrated that PrPC plays a critical role in neuronal differentiation of the NPCs and suggest that this function is, at least in part, NCAM-dependent [84]. Steele et al. investigated the role of PrPC in neural development in adult neurogenesis, which occurs constitutively in the dentate gyrus of the hippocampus and in the olfactory bulb from precursors in the subventricular zone rostral migratory stream. Loss and gain-of-function experiments demonstrate that the PrPC levels correlate with the differentiation of multipotent neural precursors into mature neurons in vitro, and that the PrPC levels positively influence neuronal differentiation in a dose-dependent manner [85].
Taken together, this data suggests that PrPC plays a common and important role in the commission of NPCs toward the neuron-, oligodendrocyte-, and astrocyte lineages; indeed, silencing of the PRNP gene prevents both the neural and neuronal differentiation process.
Recent studies show that both the recombinant form [35] and cleavage products of PrPC [86] are involved in the neuronal differentiation process and neuritogenesis. Martellucci et al. demonstrated that recPrPC was able to activate the neuronal differentiation process and induce the expression of the typical neuronal markers, such as β3-Tubulin, NFH, and GAP-43; in fact, the authors reported that, when the PrPC was silenced by siRNA, the neuro-induction was blocked. Furthermore, lipid raft inhibitors, such as fumonisin B1 and MβCD, significantly prevented the neuronal differentiation process, suggesting that lipid raft integrity plays a key role in recPrP activity [34]
Furthermore, Collins et al. identified a switch between neural stem cell (NSC) proliferation and quiescence through the changing intracellular redox signaling, showing that N-terminal post-translational cleavage products of the PrPC induce a quiescent state, halting the NSCs’ cellular growth, migration, and neurite outgrowth. Quiescence is initiated by the PrP cleavage products through reducing the intracellular levels of reactive oxygen species. First, inhibition of redox signaling results in increased mitochondrial fission, which rapidly signals quiescence. Thereafter, quiescence is maintained through downstream increases in the expression and activity of superoxide dismutase-2, which reduces mitochondrial superoxide. Besides, the authors observed that PrP is predominantly cleaved in quiescent NSCs, indicating a homeostatic role for this cascade [86].
Thus, the shed PrPC and its cleavage products are biologically active fragments that may potentially participate with other biological processes, such as the differentiation process. In fact, proteolytic cleavage events may alter either the biological functions of PrPC or produce protein fragments harboring specific intrinsic properties, thus contributing to a higher biological complexity.
6. Conclusions Remarks
The prion protein is an enigmatic protein with a pleiotropic effect on different cell types. It was previously shown to play a key role in some physiological processes, including cellular activation, apoptosis, and differentiation, by functional interaction with a multimolecular signaling complex through lipid rafts. Recent evidence pointed out its role in stem cell differentiation, where it appears to be involved in the molecular signaling of neuronal differentiation.
Author Contributions
S.M., C.S., M.S., V.M., writing, review, and editing; F.S., V.M., bibliographic research, editing; S.M., M.S., V.M., critical discussion, editing. All authors have read and approved published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors want to thank Amy Heath Butt for English editing contribution.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef]
- Asher, D.M.; Gregori, L. Human transmissible spongiform encephalopathies: Historic view. Handb. Clin. Neurol. 2018, 153, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Requena, J.R.; Wille, H. The structure of the infectious prion protein and its propagation. Prog. Mol. Biol. Transl. Sci. 2017, 150, 341–359. [Google Scholar] [CrossRef]
- Lima, A.N.; Oliveira, R.J.; Braz, A.S.K.; Costa, M.G.S.; Perahia, D.; Scott, A.L. Effects of pH and aggregation in the human prion conversion into scrapie form: A study using molecular dynamics with excited normal modes. Eur. Biophys. J. 2018, 47, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, C.J.; Zhang, K.; Munn, A.L.; Wiegmans, A.; Wei, M.Q. Prion protein scrapie and the normal cellular prion protein. Prion 2015, 10, 63–82. [Google Scholar] [CrossRef] [PubMed]
- Priola, S.A. Cell biology approaches to studying prion diseases. Methods Mol. Biol. 2017, 1658, 83–94. [Google Scholar] [CrossRef]
- Aguilar-Calvo, P.; Xiao, X.; Bett, C.; Eraña, H.; Soldau, K.; Castilla, J.; Nilsson, K.P.R.; Surewicz, W.K.; Sigurdson, C.J. Post-translational modifications in PrP expand the conformational diversity of prions in vivo. Sci. Rep. 2017, 7, 43295. [Google Scholar] [CrossRef] [PubMed]
- Abskharon, R.; Wang, F.; Wohlkonig, A.; Ruan, J.; Soror, S.H.; Giachin, G.; Pardon, E.; Zou, W.; Legname, G.; Ma, J.; et al. Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion. PLoS Pathog. 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Šafář, J.; Roller, P.P.; Gajdusek, D.C.; Gibbs, C.J. Thermal stability and conformational transitions of scrapie amyloid (prion) protein correlate with infectivity. Protein Sci. 1993, 2, 2206–2216. [Google Scholar] [CrossRef]
- Elfrink, K.; Ollesch, J.; Stöhr, J.; Willbold, D.; Riesner, D.; Gerwert, K. Structural changes of membrane-anchored native PrP(C). Proc. Natl. Acad. Sci. USA 2008, 105, 10815–10819. [Google Scholar] [CrossRef]
- Zhang, B.; Shen, P.; Yin, X.; Dai, Y.; Ding, M.; Cui, L. Expression and functions of cellular prion proteins in immunocytes. Scand. J. Immunol. 2019, 91. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yin, S.; Wang, X.; Zha, L.; Sy, M.-S.; Ma, J. Role of the highly conserved middle region of prion protein (PrP) in PrP−lipid interaction. Biochemistry 2010, 49, 8169–8176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Riek, R.; Hornemann, S.; Wider, G.; Billeter, M.; Glockshuber, R.; Wüthrich, K. NMR structure of the mouse prion protein domain PrP(121–231). Nature 1996, 382, 180–182. [Google Scholar] [CrossRef] [PubMed]
- Haire, L.; Whyte, S.; Vasisht, N.; Gill, A.; Verma, C.; Dodson, E.; Dodson, G.; Bayley, P.M. The crystal structure of the globular domain of sheep prion protein. J. Mol. Biol. 2004, 336, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Biljan, I.; Ilc, G.; Plavec, J. Analysis of prion protein structure using nuclear magnetic resonance spectroscopy. Methods Mol. Biol. 2017, 1658, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Castle, A.R.; Gill, A. Physiological functions of the cellular prion protein. Front. Mol. Biosci. 2017, 4, 698. [Google Scholar] [CrossRef]
- Watts, J.C.; Bourkas, M.E.C.; Arshad, H. The function of the cellular prion protein in health and disease. Acta Neuropathol. 2017, 135, 159–178. [Google Scholar] [CrossRef]
- Parkin, E.; Watt, N.T.; Turner, A.J.; Hooper, N.M.; Hong, M.; Luo, S.; Baumeister, P.; Huang, J.-M.; Gogia, R.K.; Li, M.; et al. Dual mechanisms for shedding of the cellular prion protein. J. Biol. Chem. 2004, 279, 11170–11178. [Google Scholar] [CrossRef]
- Taylor, D.R.; Parkin, E.; Cocklin, S.L.; Ault, J.R.; Ashcroft, A.E.; Turner, A.J.; Hooper, N.M. Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J. Biol. Chem. 2009, 284, 22590–22600. [Google Scholar] [CrossRef]
- Lewis, V.; Johanssen, V.A.; Crouch, P.J.; Klug, G.M.; Hooper, N.M.; Collins, S.J. Prion protein “gamma-cleavage”: Characterizing a novel endoproteolytic processing event. Cell. Mol. Life Sci. 2015, 73, 667–683. [Google Scholar] [CrossRef]
- Mattei, V.; Manganelli, V.; Martellucci, S.; Capozzi, A.; Mantuano, E.; Longo, A.; Ferri, A.; Garofalo, T.; Sorice, M.; Misasi, R.; et al. A multimolecular signaling complex including PrP C and LRP1 is strictly dependent on lipid rafts and is essential for the function of tissue plasminogen activator. J. Neurochem. 2019, 152, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Mattei, V.; Santacroce, C.; Tasciotti, V.; Martellucci, S.; Santilli, F.; Manganelli, V.; Piccoli, L.; Misasi, R.; Sorice, M.; Garofalo, T.; et al. Role of lipid rafts in neuronal differentiation of dental pulp-derived stem cells. Exp. Cell Res. 2015, 339, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Mattei, V.; Martellucci, S.; Santilli, F.; Manganelli, V.; Garofalo, T.; Candelise, N.; Caruso, A.; Sorice, M.; Scaccianoce, S.; Misasi, R.; et al. Morphine withdrawal modifies prion protein expression in rat hippocampus. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Kazlauskaite, J.; Sanghera, N.; Sylvester, I.; Venien-Bryan, C.; Pinheiro, T.J. Structural changes of the prion protein in lipid membranes leading to aggregation and fibrillization. Biochemistry 2003, 42, 3295–3304. [Google Scholar] [CrossRef] [PubMed]
- Sanghera, N.; Pinheiro, T.J. Binding of prion protein to lipid membranes and implications for prion conversion. J. Mol. Biol. 2002, 315, 1241–1256. [Google Scholar] [CrossRef] [PubMed]
- Botto, L.; Cunati, D.; Coco, S.; Sesana, S.; Bulbarelli, A.; Biasini, E.; Colombo, L.; Negro, A.; Chiesa, R.; Masserini, M.; et al. Role of lipid rafts and GM1 in the segregation and processing of prion protein. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Lopes, M.; Santos, T.G. Prion potency in stem cells biology. Prion 2012, 6, 142–146. [Google Scholar] [CrossRef]
- Arcos-López, T.; Qayyum, M.; Rivillas-Acevedo, L.; Miotto, M.C.; Grande-Aztatzi, R.; Fernández, C.O.; Hedman, B.; Hodgson, K.O.; Vela, A.; Solomon, E.I.; et al. Spectroscopic and theoretical study of CuIBinding to His111 in the human prion protein fragment 106–115. Inorg. Chem. 2016, 55, 2909–2922. [Google Scholar] [CrossRef]
- Brown, D.R.; Wong, B.S.; Hafiz, F.; Clive, C.; Haswell, S.J.; Jones, I.M. Normal prion protein has an activity like that of superoxide dismutase. Biochem. J. 1999, 344, 1–5. [Google Scholar] [CrossRef]
- Wulf, M.-A.; Senatore, A.; Aguzzi, A. The biological function of the cellular prion protein: An update. BMC Biol. 2017, 15, 34. [Google Scholar] [CrossRef]
- Yun, S.P.; Han, Y.-S.; Lee, J.H.; Yoon, Y.M.; Yun, C.W.; Rhee, P.; Lee, S.H. Role of hypoxia-mediated cellular prion protein functional change in stem cells and potential application in angiogenesis (Review). Mol. Med. Rep. 2017, 16, 5747–5751. [Google Scholar] [CrossRef] [PubMed]
- Monache, S.D.; Martellucci, S.; Clementi, L.; Pulcini, F.; Santilli, F.; Mei, C.; Piccoli, L.; Angelucci, A.; Mattei, V. In vitro conditioning determines the capacity of dental pulp stem cells to function as pericyte-like cells. Stem Cells Dev. 2019, 28, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, X.T.A.; Tran, T.H.; Cojoc, D.; Legname, G. Copper binding regulates cellular prion protein function. Mol. Neurobiol. 2019, 56, 6121–6133. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, S.; Zurzolo, C.; Paladino6S. Organization of GPI-anchored proteins at the cell surface and its physiopathological relevance. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 403–419. [Google Scholar] [CrossRef]
- Martellucci, S.; Santacroce, C.; Santilli, F.; Piccoli, L.; Monache, S.D.; Angelucci, A.; Misasi, R.; Sorice, M.; Mattei, V. Cellular and molecular mechanisms mediated by recPrPC involved in the neuronal differentiation process of mesenchymal stem cells. Int. J. Mol. Sci. 2019, 20, 345. [Google Scholar] [CrossRef] [PubMed]
- Parrie, L.E.; Crowell, J.A.; Telling, G.C.; Bessen, R.A. The cellular prion protein promotes olfactory sensory neuron survival and axon targeting during adult neurogenesis. Dev. Biol. 2018, 438, 23–32. [Google Scholar] [CrossRef]
- Ryskalin, L.; Busceti, C.; Biagioni, F.; Limanaqi, F.; Familiari, P.; Frati, A.; Fornai, F. Prion protein in glioblastoma multiforme. Int. J. Mol. Sci. 2019, 20, 5107. [Google Scholar] [CrossRef]
- Mediano, D.R.; Ranera, B.; Bolea, R.; Sanz-Rubio, D.; Martín-Burriel, I. The potential of mesenchymal stem cell in prion research. Zoonoses Public Health 2014, 62, 165–178. [Google Scholar] [CrossRef]
- Zhang, C.C.; Steele, A.D.; Lindquist, S.; Lodish, H.F. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc. Natl. Acad. Sci. USA 2006, 103, 2184–2189. [Google Scholar] [CrossRef]
- Lee, Y.J.; Baskakov, I.V. The cellular form of the prion protein is involved in controlling cell cycle dynamics, self-renewal and the fate of human embryonic stem cell differentiation. J. Neurochem. 2012, 124, 310–322. [Google Scholar] [CrossRef]
- Tremblay, P.; Bouzamondo-Bernstein, E.; Heinrich, C.; Prusiner, S.B.; DeArmond, S.J. Developmental expression of PrP in the post-implantation embryo. Brain Res. 2007, 1139, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Martellucci, S.; Manganelli, V.; Santacroce, C.; Santilli, F.; Piccoli, L.; Sorice, M.; Mattei, V. Role of prion protein-EGFR multimolecular complex during neuronal differentiation of human dental pulp-derived stem cells. Prion 2018, 12, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Martellucci, S.; Santacroce, C.; Manganelli, V.; Santilli, F.; Piccoli, L.; Cassetta, M.; Misasi, R.; Sorice, M.; Mattei, V. Isolation, propagation, and prion protein expression during neuronal differentiation of human dental pulp stem cells. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed]
- Martin-Lannerée, S.; Halliez, S.; Hirsch, T.; Hernandez-Rapp, J.; Passet, B.; Tomkiewicz, C.; Díaz, A.V.; Torres, J.M.; Launay, J.-M.; Béringue, V.; et al. The cellular prion protein controls notch signaling in neural stem/progenitor cells. Stem Cells 2016, 35, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Caputo, A.; Sarnataro, D.; Campana, V.; Costanzo, M.; Negro, A.; Sorgato, M.C.; Zurzolo, C. Doppel and PrPC co-immunoprecipitate in detergent-resistant membrane domains of epithelial FRT cells. Biochem. J. 2009, 425, 341–351. [Google Scholar] [CrossRef]
- Lee, Y.J.; Baskakov, I.V. The cellular form of the prion protein guides the differentiation of human embryonic stem cells into neuron-, oligodendrocyte-, and astrocyte-committed lineages. Prion 2014, 8, 266–275. [Google Scholar] [CrossRef]
- Iwabuchi, K. Gangliosides in the immune system: Role of Glycosphingolipids and Glycosphingolipid-enriched lipid rafts in immunological functions. Methods Mol. Biol. 2018, 83–95. [Google Scholar] [CrossRef]
- Kraft, M.L. Sphingolipid organization in the plasma membrane and the mechanisms that influence it. Front. Cell Dev. Biol. 2017, 4, 677. [Google Scholar] [CrossRef]
- Bieberich, E. Sphingolipids and lipid rafts: Novel concepts and methods of analysis. Chem. Phys. Lipids 2018, 216, 114–131. [Google Scholar] [CrossRef]
- Barbat, C.; Trucy, M.; Sorice, M.; Garofalo, T.; Manganelli, V.; Fischer, A.; Mazerolles, F. p56lck, LFA-1 and PI3K but not SHP-2 interact with GM1- or GM3-enriched microdomains in a CD4–p56lck association-dependent manner. Biochem. J. 2007, 402, 471–481. [Google Scholar] [CrossRef]
- Mattei, V.; Garofalo, T.; Misasi, R.; Circella, A.; Manganelli, V.; Lucania, G.; Pavan, A.; Sorice, M. Prion protein is a component of the multimolecular signaling complex involved in T cell activation. FEBS Lett. 2004, 560, 14–18. [Google Scholar] [CrossRef]
- Sorice, M.; Matarrese, P.; Manganelli, V.; Tinari, A.; Giammarioli, A.M.; Mattei, V.; Misasi, R.; Garofalo, T.; Malorni, W. Role of GD3-CLIPR-59 association in lymphoblastoid T cell apoptosis triggered by CD95/Fas. PLoS ONE 2010, 5, e8567. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F.; Gajate, C. Lipid rafts as major platforms for signaling regulation in cancer. Adv. Biol. Regul. 2015, 57, 130–146. [Google Scholar] [CrossRef]
- Mollinedo, F.; Gajate, C. Lipid rafts as signaling hubs in cancer cell survival/death and invasion: Implications in tumor progression and therapy. J. Lipid Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J. Lipid rafts: Bringing order to chaos. J. Lipid Res. 2003, 44, 655–667. [Google Scholar] [CrossRef]
- Vey, M.; Pilkuhn, S.; Wille, H.; Nixon, R.; De Armond, S.J.; Smart, E.J.; Anderson, R.G.W.; Taraboulos, A.; Prusiner, S.B. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl. Acad. Sci. USA 1996, 93, 14945–14949. [Google Scholar] [CrossRef]
- Linden, R. The biological function of the prion protein: A cell surface scaffold of signaling modules. Front. Mol. Neurosci. 2017, 10, 336. [Google Scholar] [CrossRef]
- Spielhaupter, C.; Schätzl, H.M. PrPC directly interacts with proteins involved in signaling pathways. J. Biol. Chem. 2001, 276, 44604–44612. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.R.; Hooper, N.M. The prion protein and lipid rafts. Mol. Membr. Biol. 2006, 23, 89–99. [Google Scholar] [CrossRef]
- Puig, B.; Altmeppen, H.C.; Linsenmeier, L.; Chakroun, K.; Wegwitz, F.; Piontek, U.K.; Tatzelt, J.; Bate, C.; Magnus, T.; Glatzel, M.; et al. GPI-anchor signal sequence influences PrPC sorting, shedding and signalling, and impacts on different pathomechanistic aspects of prion disease in mice. PLoS Pathog. 2019, 15. [Google Scholar] [CrossRef]
- Didonna, A. Prion protein and its role in signal transduction. Cell. Mol. Biol. Lett. 2013, 18, 209–230. [Google Scholar] [CrossRef]
- Hirsch, T.; Martin-Lannerée, S.; Mouillet-Richard, S. Functions of the prion protein. Prog. Mol. Biol. Transl. Sci. 2017, 150, 1–34. [Google Scholar] [CrossRef]
- Schneider, B.; Mutel, V.; Pietri, M.; Ermonval, M.; Mouillet-Richard, S.; Kellermann, O. NADPH oxidase and extracellular regulated kinases 1/2 are targets of prion protein signaling in neuronal and nonneuronal cells. Proc. Natl. Acad. Sci. USA 2003, 100, 13326–13331. [Google Scholar] [CrossRef] [PubMed]
- Combs, C.K.; Johnson, D.E.; Cannady, S.B.; Lehman, T.M.; Landreth, G.E. Identification of microglial signal transduction pathways mediating a neurotoxic response to amyloidogenic fragments of beta-amyloid and prion proteins. J. Neurosci. 1999, 19, 928–939. [Google Scholar] [CrossRef]
- Kuwahara, C.; Takeuchi, A.M.; Nishimura, T.; Haraguchi, K.; Kubosaki, A.; Matsumoto, Y.; Saeki, K.; Matsumoto, Y.; Yokoyama, T.; Itohara, S.; et al. Prions prevent neuronal cell-line death. Nature 1999, 400, 225–226. [Google Scholar] [CrossRef]
- Roucou, X.; Giannopoulos, P.N.; Zhang, Y.; Jodoin, J.; Goodyer, C.G.; Leblanc, A.C. Cellular prion protein inhibits proapoptotic Bax conformational change in human neurons and in breast carcinoma MCF-7 cells. Cell Death Differ. 2005, 12, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Mitteregger, G.; Vosko, M.; Krebs, B.; Xiang, W.; Kohlmannsperger, V.; Nölting, S.; Hamann, G.F.; Kretzschmar, H.A. The role of the octarepeat region in neuroprotective function of the cellular prion protein. Brain Pathol. 2007, 17, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, L.B.; Freitas, A.R.O.; Zanata, S.; Brentani, R.R.; Martins, V.R.; Linden, R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002, 21, 3317–3326. [Google Scholar] [CrossRef]
- Mattei, V.; Matarrese, P.; Garofalo, T.; Tinari, A.; Gambardella, L.; Ciarlo, L.; Manganelli, V.; Tasciotti, V.; Misasi, R.; Malorni, W.; et al. Recruitment of cellular prion protein to mitochondrial raft-like microdomains contributes to apoptosis execution. Mol. Biol. Cell 2011, 22, 4842–4853. [Google Scholar] [CrossRef]
- Garofalo, T.; Ferri, A.; Sorice, M.; Azmoon, P.; Grasso, M.; Mattei, V.; Capozzi, A.; Manganelli, V.; Misasi, R. Neuroglobin overexpression plays a pivotal role in neuroprotection through mitochondrial raft-like microdomains in neuroblastoma SK-N-BE2 cells. Mol. Cell. Neurosci. 2018, 88, 167–176. [Google Scholar] [CrossRef]
- Hong, J.-M.; Moon, J.-H.; Park, S.-Y. Human prion protein-mediated calcineurin activation induces neuron cell death via AMPK and autophagy pathway. Int. J. Biochem. Cell Biol. 2020, 119, 105680. [Google Scholar] [CrossRef]
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Vingtdeux, V.; Chandakkar, P.; Zhao, H.; D’Abramo, C.; Davies, P.; Marambaud, P. Novel synthetic small-molecule activators of AMPK as enhancers of autophagy and amyloid-β peptide degradation. FASEB J. 2010, 25, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-K.; Lee, J.-H.; Kim, S.-W.; Hong, J.-M.; Seol, J.-W.; Park, S.-Y. Cellular prion protein regulates the differentiation and function of adipocytes through autophagy flux. Mol. Cell. Endocrinol. 2019, 481, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.; Ramos-Ibeas, P.; Pericuesta, E.; Ramirez, M.A.; Gutierrez-Adan, A. The role of prion protein in stem cell regulation. Reproduction 2013, 146, R91–R99. [Google Scholar] [CrossRef]
- Mohanty, S.T.; Cairney, C.; Chantry, A.; Madan, S.; Fernandes, J.A.; Howe, S.J.; Moore, H.D.; Thompson, M.J.; Chen, B.; Thrasher, A.; et al. A small molecule modulator of prion protein increases human mesenchymal stem cell lifespan, ex vivo expansion, and engraftment to bone marrow in NOD/SCID mice. Stem Cells 2012, 30, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Han, Y.-S.; Lee, S.H. Potentiation of biological effects of mesenchymal stem cells in ischemic conditions by melatonin via upregulation of cellular prion protein expression. J. Pineal Res. 2017, 62. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yun, C.W.; Han, Y.-S.; Kim, S.; Jeong, D.; Kwon, H.Y.; Kim, H.; Baek, M.-J.; Lee, S.H. Melatonin and 5-fluorouracil co-suppress colon cancer stem cells by regulating cellular prion protein-Oct4 axis. J. Pineal Res. 2018, 65, e12519. [Google Scholar] [CrossRef] [PubMed]
- Doeppner, T.R.; Kaltwasser, B.; Schlechter, J.; Jäschke, J.; Kilic, E.; Bähr, M.; Hermann, D.M.; Weiße, J. Cellular prion protein promotes post-ischemic neuronal survival, angioneurogenesis and enhances neural progenitor cell homing via proteasome inhibition. Cell Death Dis. 2015, 6. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoon, Y.M.; Song, K.; Noh, H.; Lee, S.H. Melatonin suppresses senescence-derived mitochondrial dysfunction in mesenchymal stem cells via the HSPA1L–mitophagy pathway. Aging Cell 2020, 19. [Google Scholar] [CrossRef]
- Han, Y.-S.; Lee, J.H.; Yoon, Y.M.; Yun, C.W.; Noh, H.; Lee, S.H. Hypoxia-induced expression of cellular prion protein improves the therapeutic potential of mesenchymal stem cells. Cell Death Dis. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, A.; Bajetto, A.; Thellung, S.; Begani, G.; Villa, V.; Nizzari, M.; Pattarozzi, A.; Solari, A.; Gatti, M.; Pagano, A.; et al. Cellular prion protein controls stem cell-like properties of human glioblastoma tumor-initiating cells. Oncotarget 2016, 7, 38638–38657. [Google Scholar] [CrossRef] [PubMed]
- Langeberg, L.K.; Scott, J.D. Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 2015, 16, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Prodromidou, K.; Papastefanaki, F.; Sklaviadis, T.; Matsas, R. Functional cross-talk between the cellular prion protein and the neural cell adhesion molecule is critical for neuronal differentiation of neural stem/precursor cells. Stem Cells 2014, 32, 1674–1687. [Google Scholar] [CrossRef] [PubMed]
- Steele, A.D.; Emsley, J.G.; Özdinler, P.H.; Lindquist, S.; Macklis, J.D. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 3416–3421. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.J.; Tumpach, C.; Groveman, B.R.; Drew, S.C.; Haigh, C.L. Prion protein cleavage fragments regulate adult neural stem cell quiescence through redox modulation of mitochondrial fission and SOD2 expression. Cell. Mol. Life Sci. 2018, 75, 3231–3249. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).