Impaired Expression of Chloroplast HSP90C Chaperone Activates Plant Defense Responses with a Possible Link to a Disease-Symptom-Like Phenotype
Abstract
1. Introduction
2. Results
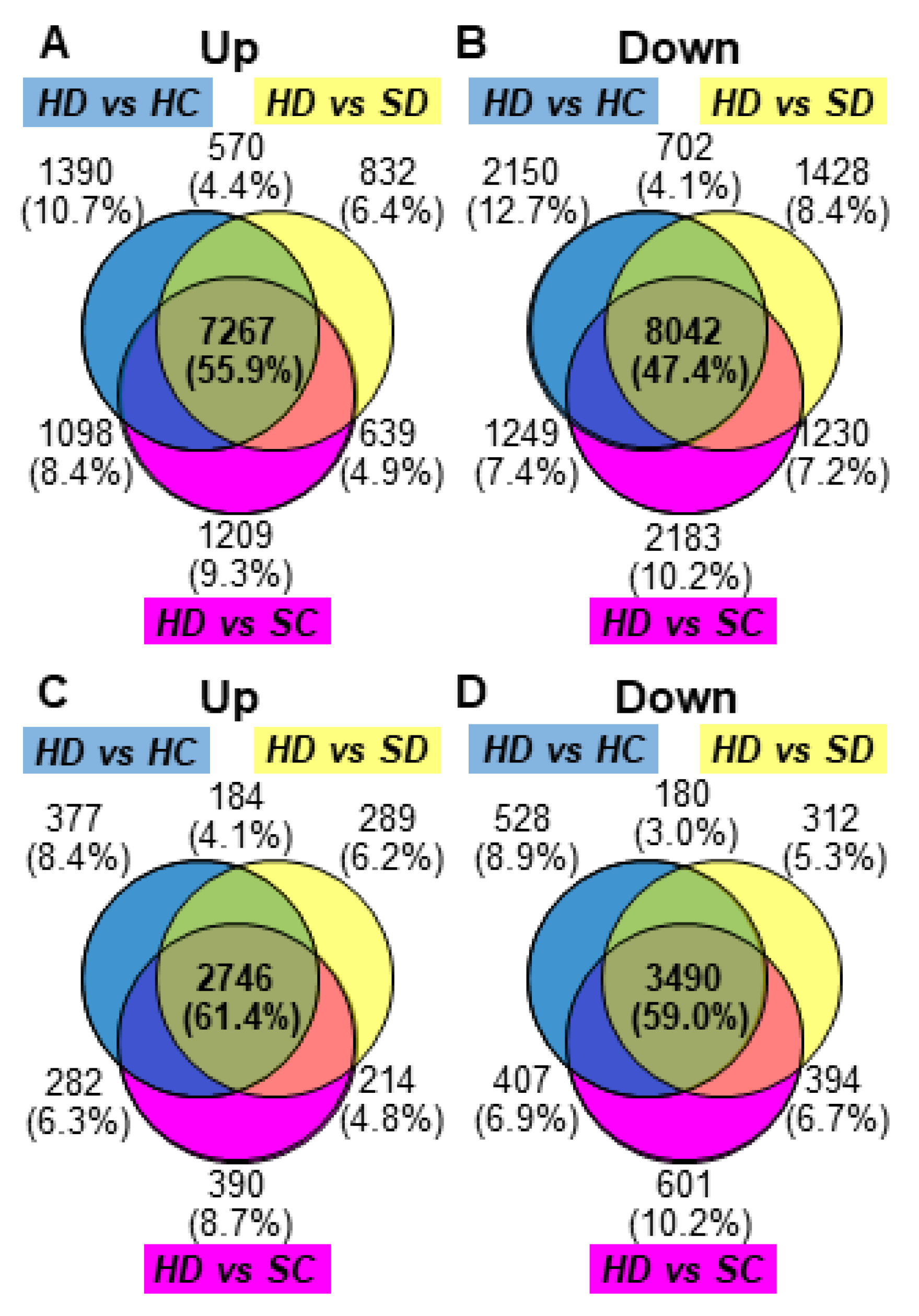
2.1. RNA Sequencing, Mapping, and Identification of Differentially Expressed Genes (DEGs)
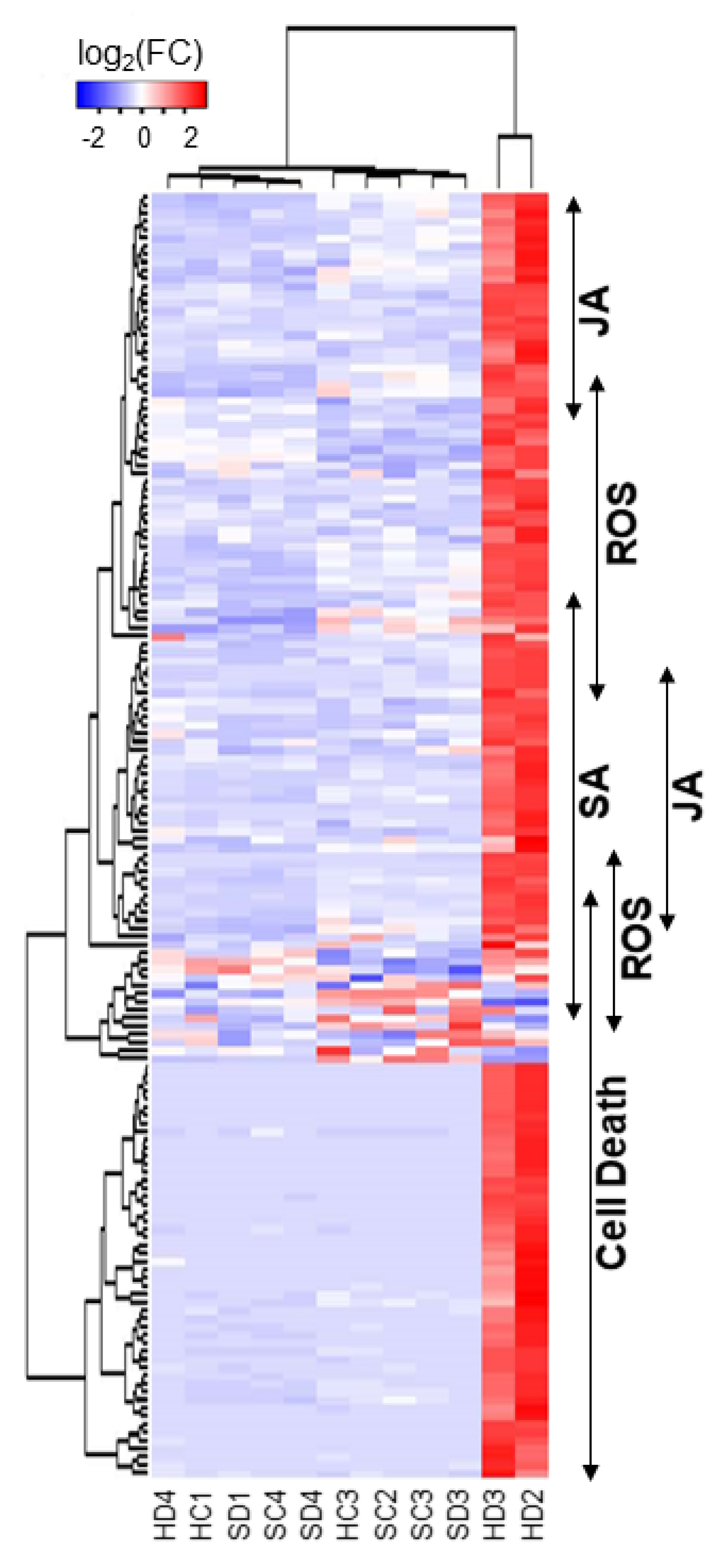
2.2. Functional Classification of DEGs
2.3. Detection of Cell Death and Reactive Oxygen Species in HPS90C-Silenced Plants
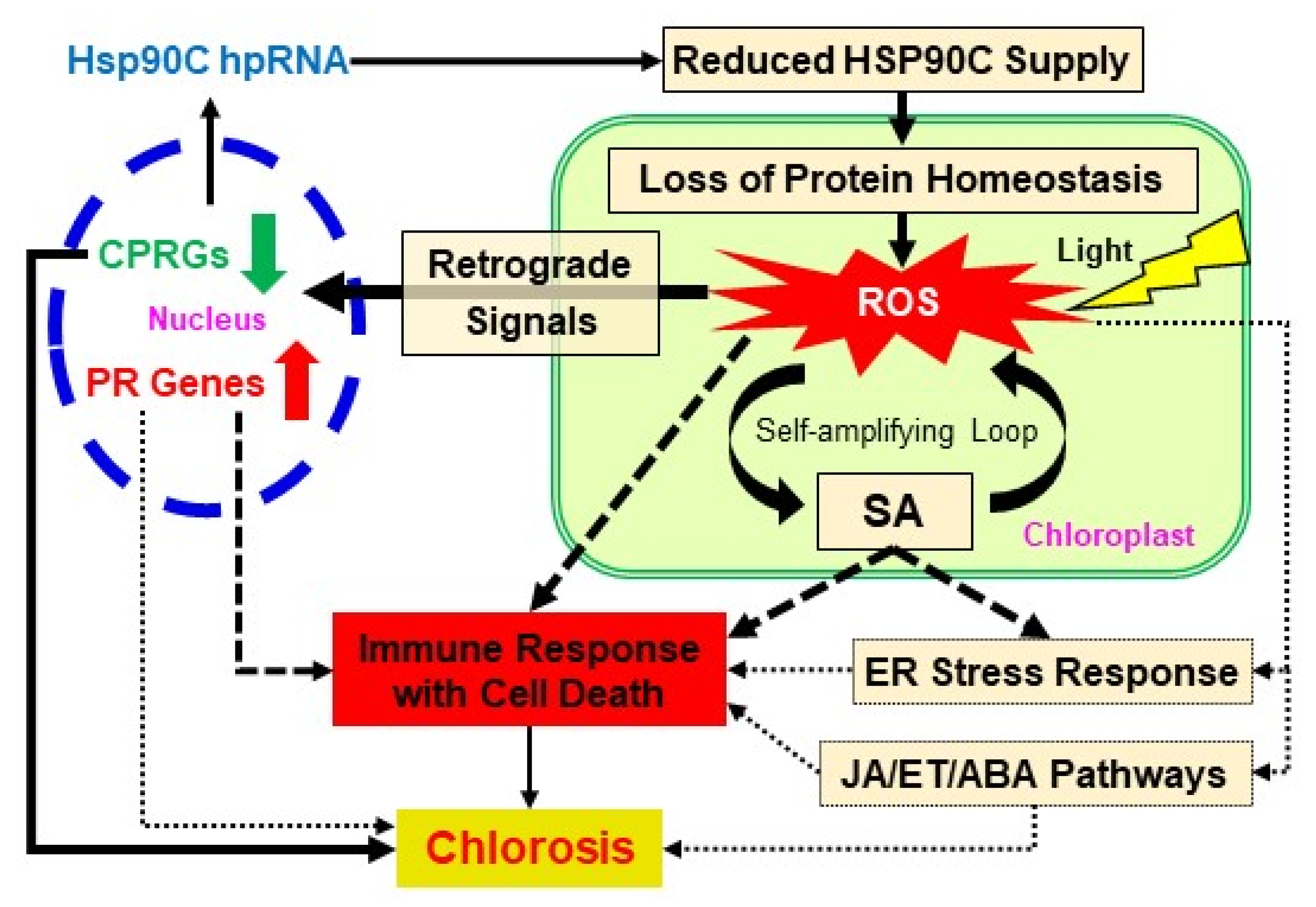
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Extraction and Sequencing of RNA
4.3. RNA-seq Data Analysis
4.4. Quantitative RT-PCR
4.5. Determination of Cell Death
4.6. DAB (3,3’-Diaminobenzidine) Staining for Hydrogen Peroxide Detection
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Manfre, A.; Glenn, M.; Nuñez, A.; Moreau, R.A.; Dardick, C. Light quantity and photosystem function mediate host susceptibility to Turnip mosaic virus via a salicylic acid-independent mechanism. Mol. Plant. Microbe Interact. 2011, 24, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Ohki, S.T. Single amino acid substitutions at residue 129 in the coat protein of cucumber mosaic virus affect symptom expression and thylakoid structure. Arch. Virol. 2011, 156, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Ogata, Y.; Hirata, Y.; Ohki, S.T. Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with cucumber mosaic virus. Mol. Plant. Pathol. 2014, 15, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zhang, Y.; Wang, C.; Lei, R.; Wu, Y.; Li, X.; Zhu, S. Cucumber mosaic virus coat protein induces the development of chlorotic symptoms through interacting with the chloroplast ferredoxin i protein. Sci. Rep. 2018, 8, 1205. [Google Scholar] [CrossRef] [PubMed]
- Shimura, H.; Pantaleo, V.; Ishihara, T.; Myojo, N.; Inaba, J.-I.; Sueda, K.; Burgyán, J.; Masuta, C. A Viral Satellite RNA Induces Yellow Symptoms on Tobacco By Targeting a Gene Involved in Chlorophyll Biosynthesis Using the RNA Silencing Machinery. PLoS Pathog. 2011, 7, e1002021. [Google Scholar] [CrossRef]
- Smith, N.A.; Eamens, A.L.; Wang, M.B. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011, 7, e1002022. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Small RNAs containing the pathogenic determinant of a chloroplast- replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant. J. 2012, 70, 991–1003. [Google Scholar] [CrossRef]
- Delgado, S.; Navarro, B.; Serra, P.; Gentit, P.; Cambra, M.Á.; Chiumenti, M.; De Stradis, A.; Di Serio, F.; Flores, R. How sequence variants of a plastid-replicating viroid with one single nucleotide change initiate disease in its natural host. RNA Biol. 2019, 16, 906–917. [Google Scholar] [CrossRef]
- Bhor, S.A.; Tateda, C.; Mochizuki, T.; Sekine, K.T.; Yaeno, T.; Yamaoka, N.; Nishiguchi, M.; Kobayashi, K. Inducible expression of magnesium protoporphyrin chelatase subunit I (CHLI)-amiRNA provides insights into cucumber mosaic virus Y satellite RNA-induced chlorosis symptoms. VirusDisease 2017, 28, 69–80. [Google Scholar] [CrossRef]
- Bhor, S.A.; Tateda, C.; Mochizuki, T.; Sekine, K.T.; Yaeno, T.; Yamaoka, N.; Nishiguchi, M.; Kobayashi, K. Inducible transgenic tobacco system to study the mechanisms underlying chlorosis mediated by the silencing of chloroplast heat shock protein 90. VirusDisease 2017, 28, 81–92. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, A.S.; Pooggin, M.M. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses 2012, 4, 2578–2597. [Google Scholar] [CrossRef] [PubMed]
- Krishna, P.; Gloor, G. The Hsp90 family of proteins in arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 238–246. [Google Scholar] [CrossRef]
- Xu, Z.S.; Li, Z.Y.; Chen, Y.; Chen, M.; Li, L.C.; Ma, Y.Z. Heat shock protein 90 in plants: Molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 2012, 13, 15706–15723. [Google Scholar] [CrossRef]
- Cao, D.; Froehlich, J.E.; Zhang, H.; Cheng, C.L. The chlorate-resistant and photomorphogenesis-defective mutant cr88 encodes a chloroplast-targeted HSP90. Plant. J. 2003, 33, 107–118. [Google Scholar] [CrossRef]
- Altieri, D.C.; Stein, G.S.; Lian, J.B.; Languino, L.R. TRAP-1, the mitochondrial Hsp90. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 767–773. [Google Scholar] [CrossRef]
- Altieri, D.C. Mitochondrial HSP90s and tumor cell metabolism. Autophagy 2013, 9, 244–245. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Watanabe, Y.; Ito, N.; Nonaka, H.; Takeda, N.; Sakai, T.; Kanaya, H.; Okada, K. SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 2002, 21, 898–908. [Google Scholar] [CrossRef]
- Marzec, M.; Eletto, D.; Argon, Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 774–787. [Google Scholar] [CrossRef]
- Schroda, M.; Mühlhaus, T. A “foldosome” in the chloroplast? Plant. Signal. Behav. 2009, 4, 301–303. [Google Scholar] [CrossRef]
- Inoue, H.; Li, M.; Schnell, D.J. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Fan, P.; Jiang, P.; Lv, S.; Chen, X.; Li, Y. Chloroplast-targeted Hsp90 plays essential roles in plastid development and embryogenesis in Arabidopsis possibly linking with VIPP1. Physiol. Plant. 2014, 150, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.E.; Yeung, C.; Babaei-Rad, R.; Zhao, R. Cosuppression of the chloroplast localized molecular chaperone HSP90.5 impairs plant development and chloroplast biogenesis in Arabidopsis. BMC Res. Notes 2014, 7, 643. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, Y.; Iwata, Y.; Ashida, M.; Mishiba, K.I.; Koizumi, N. Exogenous salicylic acid activates two signaling arms of the unfolded protein response in arabidopsis. Plant. Cell Physiol. 2014, 55, 1772–1778. [Google Scholar] [CrossRef]
- Kørner, C.J.; Du, X.; Vollmer, M.E.; Pajerowska-Mukhtar, K.M. Endoplasmic reticulum stress signaling in plant immunity—At the crossroad of life and death. Int. J. Mol. Sci. 2015, 16, 26582–26598. [Google Scholar] [CrossRef]
- Xia, C.; Li, S.; Hou, W.; Fan, Z.; Xiao, H.; Lu, M.; Sano, T.; Zhang, Z. Global transcriptomic changes induced by infection of cucumber (Cucumis sativus L.) with mild and severe variants of hop stunt viroid. Front. Microbiol. 2017, 8, 2427. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Ding, B.; Fei, Z. Comprehensive Transcriptome Analyses Reveal that Potato Spindle Tuber Viroid Triggers Genome-Wide Changes in Alternative Splicing, Inducible trans -Acting Activity of Phased Secondary Small Interfering RNAs, and Immune Responses. J. Virol. 2017, 91, e00247-17. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- Satoh, K.; Kondoh, H.; Sasaya, T.; Shimizu, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Selective modification of rice (Oryza sativa) gene expression by rice stripe virus infection. J. Gen. Virol. 2010, 91, 294–305. [Google Scholar] [CrossRef]
- Postnikova, O.A.; Nemchinov, L.G. Comparative analysis of microarray data in Arabidopsis transcriptome during compatible interactions with plant viruses. Virol. J. 2012, 9, 101. [Google Scholar] [CrossRef]
- Zanardo, L.G.; de Souza, G.B.; Alves, M.S. Transcriptomics of plant–virus interactions: A review. Theor. Exp. Plant. Physiol. 2019, 31, 103–125. [Google Scholar] [CrossRef]
- Waliullah, S.; Mochizuki, T.; Sekine, K.T.; Atsumi, G.; Ali, M.E.; Yaeno, T.; Yamaoka, N.; Nishiguchi, M.; Kobayashi, K. Artificial induction of a plant virus protein in transgenic tobacco provides a synchronous system for analyzing the process of leaf chlorosis. Physiol. Mol. Plant. Pathol. 2014, 88, 43–51. [Google Scholar] [CrossRef]
- Waliullah, S.; Kosaka, N.; Yaeno, T.; Ali, M.E.; Sekine, K.T.; Atsumi, G.; Yamaoka, N.; Nishiguchi, M.; Takahashi, H.; Kobayashi, K. Cauliflower mosaic virus Tav protein induces leaf chlorosis in transgenic tobacco through a host response to virulence function of Tav. J. Gen. Plant. Pathol. 2015, 81, 261–270. [Google Scholar] [CrossRef]
- Crawford, T.; Lehotai, N.; Strand, Å. The role of retrograde signals during plant stress responses. J. Exp. Bot. 2018, 69, 2783–2795. [Google Scholar] [CrossRef] [PubMed]
- Leister, D.; Romani, I.; Mittermayr, L.; Paieri, F.; Fenino, E.; Kleine, T. Identification of target genes and transcription factors implicated in translation-dependent retrograde signaling in Arabidopsis. Mol. Plant. 2014, 7, 1228–1247. [Google Scholar] [CrossRef]
- Henzler, T.; Steudle, E. Transport and metabolic degradation of hydrogen peroxide in chara corallina: Model calculations and measurements with the pressure probe suggest transport of h202 across water channels. J. Exp. Bot. 2000, 51, 2053–2066. [Google Scholar] [CrossRef]
- Saito, M.; Watanabe, S.; Yoshikawa, H.; Nakamoto, H. Interaction of the molecular chaperone HtpG with uroporphyrinogen decarboxylase in the cyanobacterium Synechococcus elongatus PCC 7942. Biosci. Biotechnol. Biochem. 2008, 72, 1394–1397. [Google Scholar] [CrossRef]
- Lv, F.; Zhou, J.; Zeng, L.; Xing, D. β-cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis. J. Exp. Bot. 2015, 66, 4719–47132. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef]
- Wang, L.; Tsuda, K.; Truman, W.; Sato, M.; Nguyen, L.V.; Katagiri, F.; Glazebrook, J. CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant. J. 2011, 67, 1029–1041. [Google Scholar] [CrossRef]
- Sun, T.; Li, Y.; Zhang, Q.; Ding, Y.; Zhang, Y.; Zhang, Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 2015, 6, 10159. [Google Scholar] [CrossRef]
- van Verk, M.C.; Bol, J.F.; Linthorst, H.J.M. WRKY Transcription Factors Involved in Activation of SA Biosynthesis Genes. BMC Plant. Biol. 2011, 11, 89. [Google Scholar] [CrossRef]
- Nomura, H.; Komori, T.; Uemura, S.; Kanda, Y.; Shimotani, K.; Nakai, K.; Furuichi, T.; Takebayashi, K.; Sugimoto, T.; Sano, S.; et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 2012, 3, 926. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant. Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, B.W.; Du, L.; Wang, H.; Yang, T. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant. Physiol. 2013, 163, 531–542. [Google Scholar] [CrossRef]
- Nawrath, C.; Métraux, J.P. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant. Cell 1999, 11, 1393–1404. [Google Scholar] [PubMed]
- Gross, J.; Won, K.C.; Lezhneva, L.; Falk, J.; Krupinska, K.; Shinozaki, K.; Seki, M.; Herrmann, R.G.; Meurer, J. A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J. Biol. Chem. 2006, 281, 17189–17196. [Google Scholar] [CrossRef]
- Strawn, M.A.; Marr, S.K.; Inoue, K.; Inada, N.; Zubieta, C.; Wildermuth, M.C. Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 2007, 282, 5919–5933. [Google Scholar] [CrossRef] [PubMed]
- Garcion, C.; Lohmann, A.; Lamodière, E.; Catinot, J.; Buchala, A.; Doermann, P.; Métraux, J.P. Characterization and biological function of the Isochorismate Synthase2 gene of Arabidopsis. Plant. Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant. Signal. Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef]
- Brodersen, P.; Malinovsky, F.G.; Hématy, K.; Newman, M.A.; Mundy, J. The role of salicylic acid in the induction of cell death in Arabidopsis acd11. Plant. Physiol. 2005, 138, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Radojičić, A.; Li, X.; Zhang, Y. Salicylic acid: A double-edged sword for programed cell death in plants. Front. Plant. Sci. 2018, 9, 1133. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Vásquez, A.; Salinas, P.; Holuigue, L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant. Sci. 2015, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant. J. 2007, 51, 941–954. [Google Scholar] [CrossRef]
- Hamel, L.P.; Sekine, K.T.; Wallon, T.; Sugiwaka, Y.; Kobayashi, K.; Moffett, P. The chloroplastic protein THF1 interacts with the Coiled-Coil domain of the disease resistance protein Nˊ and regulates light-dependent cell death. Plant. Physiol. 2016, 171, 658–674. [Google Scholar] [CrossRef]
- Chandra-Shekara, A.C.; Gupte, M.; Navarre, D.; Raina, S.; Raina, R.; Klessig, D.; Kachroo, P. Light-dependent hypersensitive response and resistance signaling against Turnip Crinkle Virus in Arabidopsis. Plant. J. 2006, 45, 320–334. [Google Scholar] [CrossRef]
- Chen, L.J.; Ren, H.; Deng, X.G.; Li, Y.N.; Cha, W.Q.; Lin, H.H.; Xi, D.H. Effects of light intensity on the susceptibility of Nicotiana tabacum to cucumber mosaic virus. J. Gen. Plant. Pathol. 2015, 81, 399–408. [Google Scholar] [CrossRef]
- Liu, L.; Li, J. Communications between the endoplasmic reticulum and other organelles during abiotic stress response in plants. Front. Plant. Sci. 2019, 10, 749. [Google Scholar] [CrossRef]
- Barton, K.A.; Wozny, M.R.; Mathur, N.; Jaipargas, E.A.; Mathur, J. Chloroplast behaviour and interactions with other organelles in Arabidopsis thaliana pavement cells. J. Cell Sci. 2018, 131, jcs202275. [Google Scholar] [CrossRef] [PubMed]
- Schattat, M.H.; Griffiths, S.; Mathur, N.; Barton, K.; Wozny, M.R.; Dunn, N.; Greenwood, J.S.; Mathur, J. Differential coloring reveals that plastids do not form networks for exchanging macromolecules. Plant. Cell 2012, 24, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.; Xiao, Y.; Wang, J.Z.; Baidoo, E.E.; Keasling, J.D.; Shen, Z.; Briggs, S.P.; Dehesh, K. Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2015, 112, 6212–6217. [Google Scholar] [CrossRef] [PubMed]
- Benn, G.; Bjornson, M.; Ke, H.; De Souza, A.; Balmond, E.I.; Shaw, J.T.; Dehesh, K. Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc. Natl. Acad. Sci. USA 2016, 113, 8855–8860. [Google Scholar] [CrossRef]
- Park, C.J.; Park, J.M. Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front. Plant. Sci. 2019, 10, 399. [Google Scholar] [CrossRef]
- Chiumenti, M.; Catacchio, C.R.; Miozzi, L.; Pirovano, W.; Ventura, M.; Pantaleo, V. A Short Indel-Lacking-Resistance Gene Triggers Silencing of the Photosynthetic Machinery Components Through TYLCSV-Associated Endogenous siRNAs in Tomato. Front. Plant. Sci. 2018, 9, 1470. [Google Scholar] [CrossRef]
- Boekel, J.; Chilton, J.M.; Cooke, I.R.; Horvatovich, P.L.; Jagtap, P.D.; Käll, L.; Lehtiö, J.; Lukasse, P.; Moerland, P.D.; Griffin, T.J. Multi-omic data analysis using Galaxy. Nat. Biotechnol. 2015, 33, 137–139. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.B.; Dangl, J.L. Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics 1999, 151, 305–319. [Google Scholar] [PubMed]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant. J. 2003, 33, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, H.B.; Scholthof, K.B.; Jackson, A.O. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant. Cell 1995, 7, 1157–1172. [Google Scholar]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Daudi, A.; O’Brien, J. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. BIO-PROTOCOL 2012, 2, e263. [Google Scholar] [CrossRef]




| GO Terms | Total | DEGs | Fold Enrich. | FDR |
|---|---|---|---|---|
| Immune system process | 372 | 101 | 2.73 | <0.01 |
| Innate immune response | 300 | 83 | 2.78 | <0.01 |
| Response to stimulus | 5327 | 880 | 1.66 | <0.01 |
| Response to stress | 3079 | 547 | 1.79 | <0.01 |
| Response to osmotic stress | 545 | 105 | 1.94 | <0.01 |
| Response to salt stress | 469 | 90 | 1.93 | <0.01 |
| Defense response | 1005 | 231 | 2.31 | <0.01 |
| Response to wounding | 207 | 59 | 2.86 | <0.01 |
| Response to oxidative stress | 392 | 83 | 2.13 | <0.01 |
| Response to hormone | 1360 | 235 | 1.74 | <0.01 |
| Response to SA | 206 | 41 | 2 | <0.01 |
| Response to JA | 217 | 46 | 2.13 | <0.01 |
| JA-mediated signaling pathway | 67 | 23 | 3.45 | <0.01 |
| Response to ABA | 524 | 103 | 1.98 | <0.01 |
| Response to ER stress | 97 | 37 | 3.83 | <0.01 |
| ERAD pathway | 60 | 17 | 2.85 | 0.01 |
| ER unfolded protein response | 36 | 13 | 3.63 | 0.01 |
| Metabolic process | 7731 | 992 | 1.29 | <0.01 |
| Cellular respiration | 80 | 21 | 2.64 | 0.01 |
| Protein glycosylation | 104 | 24 | 2.32 | 0.01 |
| Protein autophosphorylation | 183 | 46 | 2.53 | <0.01 |
| JA metabolic process | 50 | 14 | 2.81 | 0.03 |
| JA biosynthetic process | 26 | 10 | 3.87 | 0.02 |
| Regulation of SA metabolic process | 20 | 10 | 5.02 | 0.01 |
| Regulation of SA biosynthetic process | 12 | 8 | 6.7 | 0.01 |
| Cellular process | 10,043 | 1299 | 1.3 | <0.01 |
| Cell death | 112 | 32 | 2.87 | <0.01 |
| Programmed cell death | 91 | 27 | 2.98 | <0.01 |
| Plant-type hypersensitive response | 57 | 25 | 4.41 | <0.01 |
| Regulation of cell death | 79 | 22 | 2.8 | <0.01 |
| Negative regulation of cell death | 33 | 13 | 3.96 | <0.01 |
| Plant organ development | 946 | 141 | 1.5 | <0.01 |
| Root morphogenesis | 253 | 43 | 1.71 | 0.04 |
| Leaf senescence | 103 | 29 | 2.83 | <0.01 |
| GO Terms | Total | DEGs | Fold Enrich. | FDR |
|---|---|---|---|---|
| Metabolic process | 7731 | 1278 | 1.3 | <0.01 |
| Carbohydrate metabolic process | 668 | 146 | 1.72 | <0.01 |
| Lipid metabolic process | 728 | 175 | 1.89 | <0.01 |
| Cellular AA* metabolic process | 333 | 85 | 2.01 | <0.01 |
| Photosynthesis | 177 | 102 | 4.54 | <0.01 |
| Cofactor metabolic process | 416 | 129 | 2.44 | <0.01 |
| Pigment metabolic process | 123 | 48 | 3.07 | <0.01 |
| Vitamin metabolic process | 77 | 31 | 3.17 | <0.01 |
| Cellular process | 10,043 | 1592 | 1.25 | <0.01 |
| Cell wall organization | 279 | 60 | 1.69 | 0.01 |
| Cell cycle | 486 | 106 | 1.72 | <0.01 |
| Cellular homeostasis | 311 | 62 | 1.57 | 0.04 |
| Cellular component organization | 2503 | 454 | 1.43 | <0.01 |
| Plastid organization | 298 | 136 | 3.6 | <0.01 |
| Response to stimulus | 5327 | 903 | 1.34 | <0.01 |
| Response to osmotic stress | 545 | 103 | 1.49 | 0.01 |
| Response to oxidative stress | 392 | 87 | 1.75 | <0.01 |
| Response to salt stress | 469 | 91 | 1.53 | 0.01 |
| Response to auxin | 310 | 72 | 1.83 | <0.01 |
| Response to light stimulus | 680 | 198 | 2.29 | <0.01 |
| Rhythmic process | 122 | 43 | 2.78 | <0.01 |
| Circadian rhythm | 109 | 42 | 3.04 | <0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, S.; Bhor, S.A.; Tanaka, K.; Sakamoto, H.; Yaeno, T.; Kaya, H.; Kobayashi, K. Impaired Expression of Chloroplast HSP90C Chaperone Activates Plant Defense Responses with a Possible Link to a Disease-Symptom-Like Phenotype. Int. J. Mol. Sci. 2020, 21, 4202. https://doi.org/10.3390/ijms21124202
Islam S, Bhor SA, Tanaka K, Sakamoto H, Yaeno T, Kaya H, Kobayashi K. Impaired Expression of Chloroplast HSP90C Chaperone Activates Plant Defense Responses with a Possible Link to a Disease-Symptom-Like Phenotype. International Journal of Molecular Sciences. 2020; 21(12):4202. https://doi.org/10.3390/ijms21124202
Chicago/Turabian StyleIslam, Shaikhul, Sachin Ashok Bhor, Keisuke Tanaka, Hikaru Sakamoto, Takashi Yaeno, Hidetaka Kaya, and Kappei Kobayashi. 2020. "Impaired Expression of Chloroplast HSP90C Chaperone Activates Plant Defense Responses with a Possible Link to a Disease-Symptom-Like Phenotype" International Journal of Molecular Sciences 21, no. 12: 4202. https://doi.org/10.3390/ijms21124202
APA StyleIslam, S., Bhor, S. A., Tanaka, K., Sakamoto, H., Yaeno, T., Kaya, H., & Kobayashi, K. (2020). Impaired Expression of Chloroplast HSP90C Chaperone Activates Plant Defense Responses with a Possible Link to a Disease-Symptom-Like Phenotype. International Journal of Molecular Sciences, 21(12), 4202. https://doi.org/10.3390/ijms21124202







