Elucidating Potential Profibrotic Mechanisms of Emerging Biomarkers for Early Prognosis of Hepatic Fibrosis
Abstract
1. Introduction
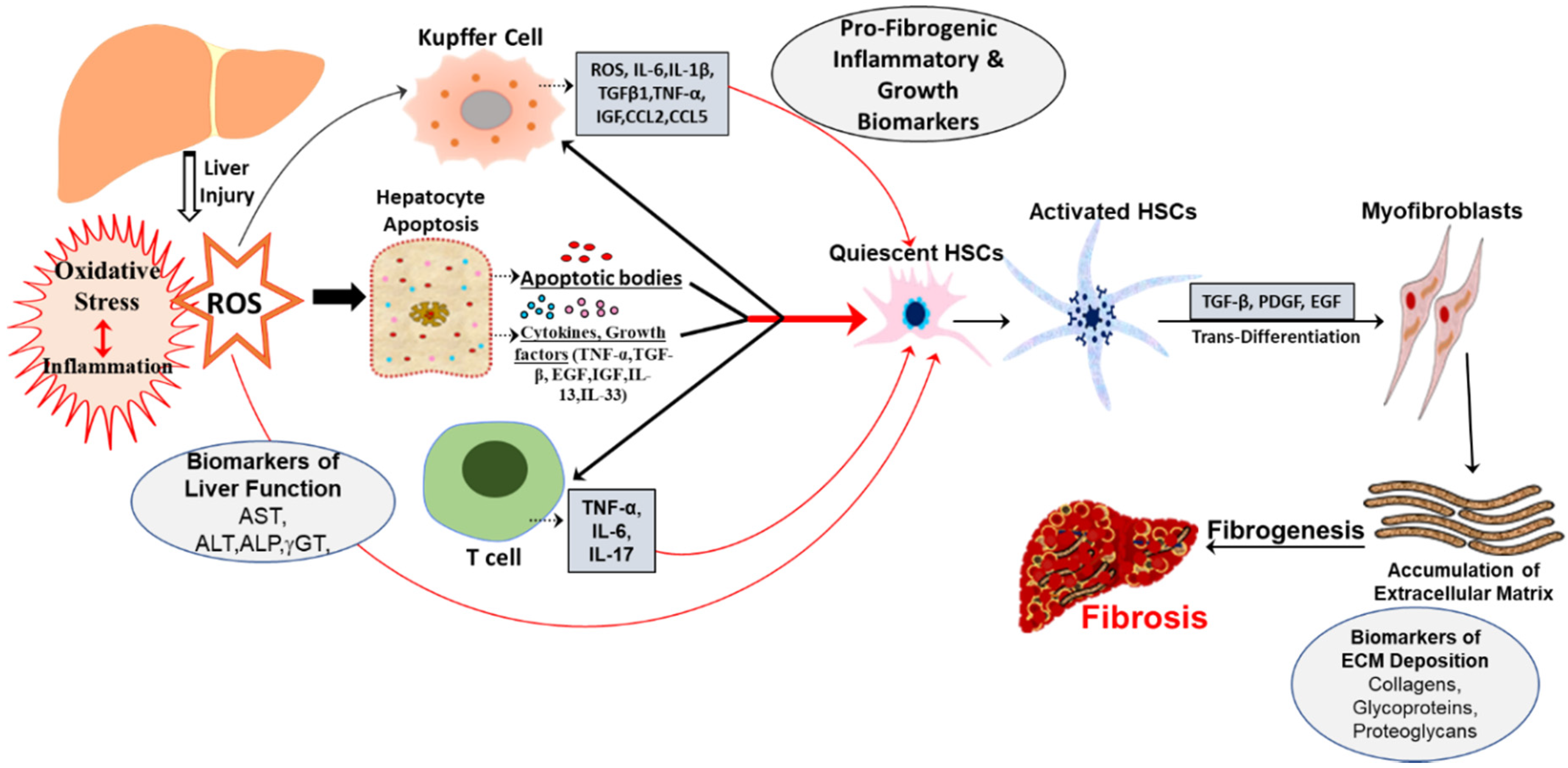
2. Oxidative Stress Mediated Biomarkers
3. Genetic and Epigenetic Markers
4. MicroRNAs
5. Exosomes
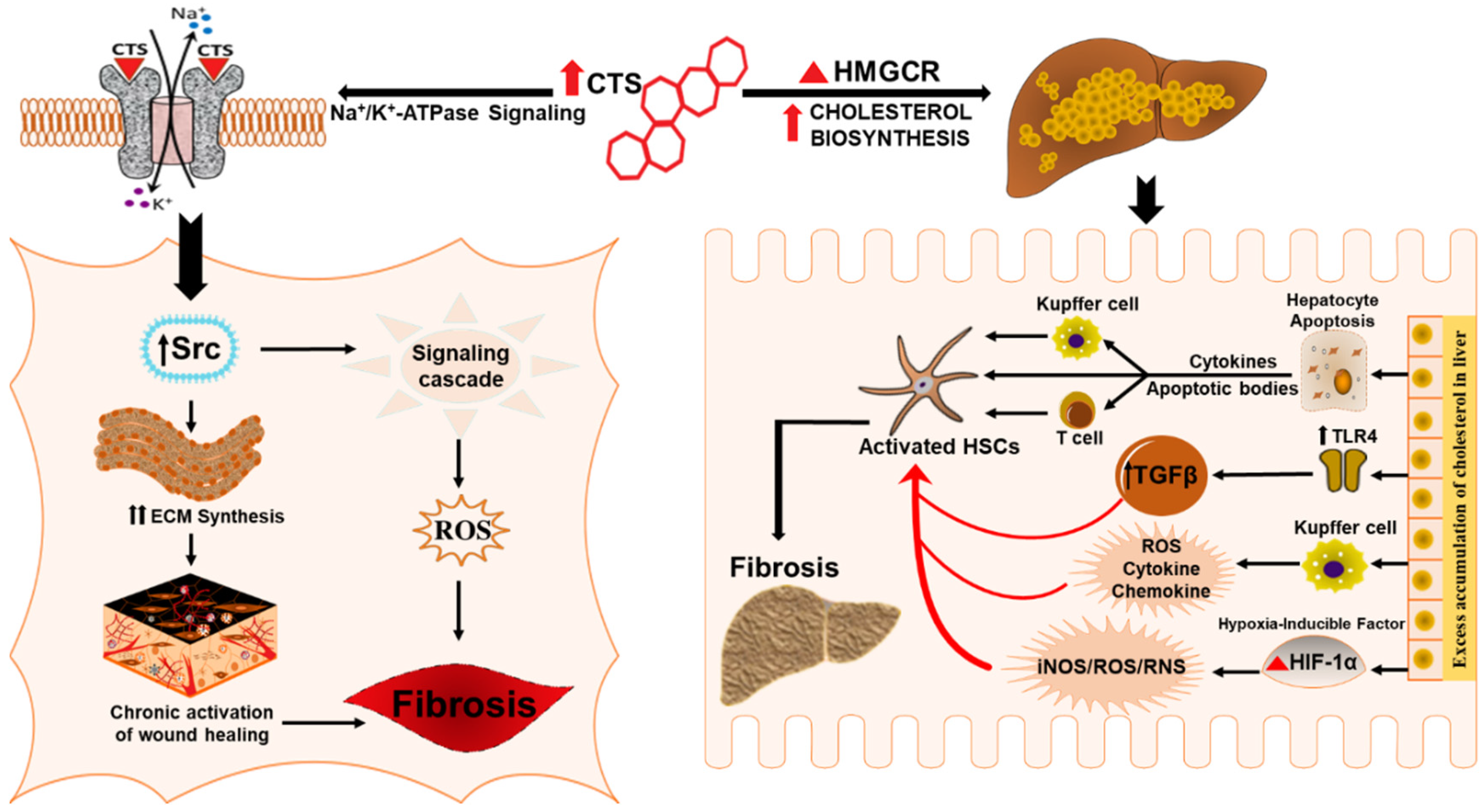
6. Potential Role of Cardiotonic Steroids in Mediating Hepatic Fibrosis
6.1. Cardiotonic Steroids in Wound Healing and Fibrosis
6.2. Cardiotonic Steroids Mediated Na/K-ATPase Signaling and Fibrosis
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Weiskirchen, R.; Weiskirchen, S.; Tacke, F. Recent advances in understanding liver fibrosis: Bridging basic science and individualized treatment concepts. F1000Res 2018, 7. [Google Scholar] [CrossRef]
- AbdulHameed, M.D.; Tawa, G.J.; Kumar, K.; Ippolito, D.L.; Lewis, J.A.; Stallings, J.D.; Wallqvist, A. Systems level analysis and identification of pathways and networks associated with liver fibrosis. PLoS ONE 2014, 9, e112193. [Google Scholar] [CrossRef]
- Sanchez-Valle, V.; Chavez-Tapia, N.C.; Uribe, M.; Mendez-Sanchez, N. Role of oxidative stress and molecular changes in liver fibrosis: A review. Curr. Med. Chem. 2012, 19, 4850–4860. [Google Scholar] [CrossRef]
- Urtasun, R.; Conde de la Rosa, L.; Nieto, N. Oxidative and nitrosative stress and fibrogenic response. Clin. Liver Dis. 2008, 12, 769–790, viii. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, R.; Zhao, S.; Liu, L.; Guo, S.; Wu, N.; Zhang, W.; Chen, P. Reactive oxygen species released from hypoxic hepatocytes regulates MMP-2 expression in hepatic stellate cells. Int. J. Mol. Sci. 2011, 12, 2434–2447. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, X.; Karsdal, M.A.; Leeming, D.J.; Genovese, F. Molecular serum markers of liver fibrosis. Biomark Insights 2012, 7, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Poli, G. Pathogenesis of liver fibrosis: Role of oxidative stress. Mol. Aspects Med. 2000, 21, 49–98. [Google Scholar] [CrossRef]
- Lee, U.E.; Friedman, S.L. Mechanisms of hepatic fibrogenesis. Best Pract. Res. Clin. Gastroenterol 2011, 25, 195–206. [Google Scholar] [CrossRef]
- Lee, Y.A.; Wallace, M.C.; Friedman, S.L. Pathobiology of liver fibrosis: A translational success story. Gut 2015, 64, 830–841. [Google Scholar] [CrossRef]
- Parola, M.; Robino, G. Oxidative stress-related molecules and liver fibrosis. J. Hepatol. 2001, 35, 297–306. [Google Scholar] [CrossRef]
- Hoffmann, C.; Djerir, N.E.H.; Danckaert, A.; Fernandes, J.; Roux, P.; Charrueau, C.; Lachages, A.M.; Charlotte, F.; Brocheriou, I.; Clement, K.; et al. Hepatic stellate cell hypertrophy is associated with metabolic liver fibrosis. Sci. Rep. 2020, 10, 3850. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Teo, E.K.; How, C.H.; Wong, T.Y.; Ang, T.L. A practical clinical approach to liver fibrosis. Singapore Med. J. 2018, 59, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.A. Reversibility of liver fibrosis. Gastroenterol. Hepatol. (N Y) 2013, 9, 737–739. [Google Scholar]
- Ramadori, G.; Veit, T.; Schwogler, S.; Dienes, H.P.; Knittel, T.; Rieder, H.; Meyer zum Buschenfelde, K.H. Expression of the gene of the alpha-smooth muscle-actin isoform in rat liver and in rat fat-storing (ITO) cells. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1990, 59, 349–357. [Google Scholar] [CrossRef]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Invest. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- Marra, F.; Svegliati-Baroni, G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J. Hepatol. 2018, 68, 280–295. [Google Scholar] [CrossRef]
- Udompap, P.; Kim, D.; Kim, W.R. Current and Future Burden of Chronic Nonmalignant Liver Disease. Clin. Gastroenterol. Hepatol. 2015, 13, 2031–2041. [Google Scholar] [CrossRef]
- Bertolani, C.; Marra, F. The role of adipokines in liver fibrosis. Pathophysiology 2008, 15, 91–101. [Google Scholar] [CrossRef]
- Trivedi, T.; Liu, J.; Probst, J.C.; Martin, A.B. The metabolic syndrome: Are rural residents at increased risk? J. Rural. Health 2013, 29, 188–197. [Google Scholar] [CrossRef]
- Sharma, D.L.; Lakhani, H.V.; Klug, R.L.; Snoad, B.; El-Hamdani, R.; Shapiro, J.I.; Sodhi, K. Investigating Molecular Connections of Non-alcoholic Fatty Liver Disease with Associated Pathological Conditions in West Virginia for Biomarker Analysis. J. Clin. Cell Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Pacheco, L.S.; Hernandez-Ontiveros, D.A.; Iniguez-Stevens, E.; Brodine, S.; Garfein, R.S.; Santibanez, M.; Fraga, M.A. Prevalence and correlates of diabetes and metabolic syndrome in a rural indigenous community in Baja California, Mexico. BMC Public Health 2018, 18, 1397. [Google Scholar] [CrossRef]
- Nallagangula, K.S.; Nagaraj, S.K.; Venkataswamy, L.; Chandrappa, M. Liver fibrosis: A compilation on the biomarkers status and their significance during disease progression. Future Sci. OA 2018, 4, FSO250. [Google Scholar] [CrossRef]
- Chin, J.L.; Pavlides, M.; Moolla, A.; Ryan, J.D. Non-invasive Markers of Liver Fibrosis: Adjuncts or Alternatives to Liver Biopsy? Front. Pharmacol. 2016, 7, 159. [Google Scholar] [CrossRef]
- Cequera, A.; Garcia de Leon Mendez, M.C. Biomarkers for liver fibrosis: Advances, advantages and disadvantages. Rev. Gastroenterol. Mex. 2014, 79, 187–199. [Google Scholar] [CrossRef]
- Grigorescu, M. Noninvasive biochemical markers of liver fibrosis. J. Gastrointestin. Liver Dis. 2006, 15, 149–159. [Google Scholar]
- Karsdal, M.A.; Daniels, S.J.; Holm Nielsen, S.; Bager, C.; Rasmussen, D.G.K.; Loomba, R.; Surabattula, R.; Villesen, I.F.; Luo, Y.; Shevell, D.; et al. Collagen biology and non-invasive biomarkers of liver fibrosis. Liver Int. 2020, 40, 736–750. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Houlihan, D.D.; Bentham, L.; Shaw, J.C.; Cramb, R.; Olliff, S.; Gill, P.S.; Neuberger, J.M.; Lilford, R.J.; Newsome, P.N. Presence and severity of non-alcoholic fatty liver disease in a large prospective primary care cohort. J. Hepatol. 2012, 56, 234–240. [Google Scholar] [CrossRef]
- Patel, K.; Sebastiani, G. Limitations of non-invasive tests for assessment of liver fibrosis. JHEP Rep. 2020, 2, 100067. [Google Scholar] [CrossRef]
- Li, S.; Tan, H.Y.; Wang, N.; Zhang, Z.J.; Lao, L.; Wong, C.W.; Feng, Y. The Role of Oxidative Stress and Antioxidants in Liver Diseases. Int. J. Mol. Sci. 2015, 16, 26087–26124. [Google Scholar] [CrossRef]
- Arauz, J.; Ramos-Tovar, E.; Muriel, P. Redox state and methods to evaluate oxidative stress in liver damage: From bench to bedside. Ann. Hepatol. 2016, 15, 160–173. [Google Scholar] [CrossRef]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Cichoz-Lach, H.; Michalak, A. Oxidative stress as a crucial factor in liver diseases. World J. Gastroenterol. 2014, 20, 8082–8091. [Google Scholar] [CrossRef]
- Crosas-Molist, E.; Fabregat, I. Role of NADPH oxidases in the redox biology of liver fibrosis. Redox Biol. 2015, 6, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hong, M.; Tan, H.Y.; Wang, N.; Feng, Y. Insights into the Role and Interdependence of Oxidative Stress and Inflammation in Liver Diseases. Oxid. Med. Cell. Longev. 2016, 2016, 4234061. [Google Scholar] [CrossRef]
- Ryan, K.A.; Smith, M.F., Jr.; Sanders, M.K.; Ernst, P.B. Reactive oxygen and nitrogen species differentially regulate Toll-like receptor 4-mediated activation of NF-kappa B and interleukin-8 expression. Infect. Immun. 2004, 72, 2123–2130. [Google Scholar] [CrossRef]
- Bubici, C.; Papa, S.; Pham, C.G.; Zazzeroni, F.; Franzoso, G. The NF-kappaB-mediated control of ROS and JNK signaling. Histol. Histopathol. 2006, 21, 69–80. [Google Scholar] [CrossRef]
- Novo, E.; Parola, M. The role of redox mechanisms in hepatic chronic wound healing and fibrogenesis. Fibrogenesis Tissue Repair 2012, 5, S4. [Google Scholar] [CrossRef]
- Morling, J.R.; Guha, I.N. Biomarkers of liver fibrosis. Clin. Liver Dis. (Hoboken) 2016, 7, 139–142. [Google Scholar] [CrossRef]
- Liang, S.; Kisseleva, T.; Brenner, D.A. The Role of NADPH Oxidases (NOXs) in Liver Fibrosis and the Activation of Myofibroblasts. Front. Physiol. 2016, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Kanno, K.; Ikuta, T.; Arihiro, K.; Sugiyama, A.; Kishikawa, N.; Tazuma, S. Enhancing hepatic fibrosis in spontaneously hypertensive rats fed a choline-deficient diet: A follow-up report on long-term effects of oxidative stress in non-alcoholic fatty liver disease. J. Hepatobiliary Pancreat. Sci. 2016, 23, 260–269. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Rippe, R.; Niemela, O.; Lin, M. Roles of oxidative stress in activation of Kupffer and Ito cells in liver fibrogenesis. J. Gastroenterol. Hepatol. 1995, 10 (Suppl. 1), S50–S53. [Google Scholar] [CrossRef]
- Zhangdi, H.J.; Su, S.B.; Wang, F.; Liang, Z.Y.; Yan, Y.D.; Qin, S.Y.; Jiang, H.X. Crosstalk network among multiple inflammatory mediators in liver fibrosis. World J. Gastroenterol. 2019, 25, 4835–4849. [Google Scholar] [CrossRef]
- Czaja, A.J. Hepatic inflammation and progressive liver fibrosis in chronic liver disease. World J. Gastroenterol. 2014, 20, 2515–2532. [Google Scholar] [CrossRef]
- Tacke, F. Functional role of intrahepatic monocyte subsets for the progression of liver inflammation and liver fibrosis in vivo. Fibrogenesis Tissue Repair 2012, 5, S27. [Google Scholar] [CrossRef]
- Meng, F.; Wang, K.; Aoyama, T.; Grivennikov, S.I.; Paik, Y.; Scholten, D.; Cong, M.; Iwaisako, K.; Liu, X.; Zhang, M.; et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 2012, 143, 765–776 e763. [Google Scholar] [CrossRef]
- Seki, E.; Schwabe, R.F. Hepatic inflammation and fibrosis: Functional links and key pathways. Hepatology 2015, 61, 1066–1079. [Google Scholar] [CrossRef]
- Brenner, D.A. Molecular pathogenesis of liver fibrosis. Trans. Am. Clin. Climatol. Assoc. 2009, 120, 361–368. [Google Scholar]
- Friedman, S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology 2008, 134, 1655–1669. [Google Scholar] [CrossRef]
- Friedman, S.L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 2000, 275, 2247–2250. [Google Scholar] [CrossRef]
- Duval, F.; Moreno-Cuevas, J.E.; Gonzalez-Garza, M.T.; Maldonado-Bernal, C.; Cruz-Vega, D.E. Liver fibrosis and mechanisms of the protective action of medicinal plants targeting inflammation and the immune response. Int. J. Inflam. 2015, 2015, 943497. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, Z.; Wang, F.S. Liver fibrosis: Mechanisms of immune-mediated liver injury. Cell Mol. Immunol. 2012, 9, 296–301. [Google Scholar] [CrossRef]
- Del Campo, J.A.; Gallego, P.; Grande, L. Role of inflammatory response in liver diseases: Therapeutic strategies. World J. Hepatol. 2018, 10, 1–7. [Google Scholar] [CrossRef]
- Weber, S.; Gressner, O.A.; Hall, R.; Grunhage, F.; Lammert, F. Genetic determinants in hepatic fibrosis: From experimental models to fibrogenic gene signatures in humans. Clin. Liver Dis. 2008, 12, 747–757, vii. [Google Scholar] [CrossRef]
- Krawczyk, M.; Mullenbach, R.; Weber, S.N.; Zimmer, V.; Lammert, F. Genome-wide association studies and genetic risk assessment of liver diseases. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 669–681. [Google Scholar] [CrossRef]
- Hillebrandt, S.; Wasmuth, H.E.; Weiskirchen, R.; Hellerbrand, C.; Keppeler, H.; Werth, A.; Schirin-Sokhan, R.; Wilkens, G.; Geier, A.; Lorenzen, J.; et al. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat. Genet. 2005, 37, 835–843. [Google Scholar] [CrossRef]
- Wasmuth, H.E.; Lammert, F.; Zaldivar, M.M.; Weiskirchen, R.; Hellerbrand, C.; Scholten, D.; Berres, M.L.; Zimmermann, H.; Streetz, K.L.; Tacke, F.; et al. Antifibrotic effects of CXCL9 and its receptor CXCR3 in livers of mice and humans. Gastroenterology 2009, 137, 309–319, 319 e301–e303. [Google Scholar] [CrossRef] [PubMed]
- Wasmuth, H.E.; Zaldivar, M.M.; Berres, M.L.; Werth, A.; Scholten, D.; Hillebrandt, S.; Tacke, F.; Schmitz, P.; Dahl, E.; Wiederholt, T.; et al. The fractalkine receptor CX3CR1 is involved in liver fibrosis due to chronic hepatitis C infection. J. Hepatol. 2008, 48, 208–215. [Google Scholar] [CrossRef]
- Krawczyk, M.; Grunhage, F.; Zimmer, V.; Lammert, F. Variant adiponutrin (PNPLA3) represents a common fibrosis risk gene: Non-invasive elastography-based study in chronic liver disease. J. Hepatol. 2011, 55, 299–306. [Google Scholar] [CrossRef]
- Hall, R.A.; Liebe, R.; Hochrath, K.; Kazakov, A.; Alberts, R.; Laufs, U.; Bohm, M.; Fischer, H.P.; Williams, R.W.; Schughart, K.; et al. Systems genetics of liver fibrosis: Identification of fibrogenic and expression quantitative trait loci in the BXD murine reference population. PLoS ONE 2014, 9, e89279. [Google Scholar] [CrossRef]
- Hillebrandt, S.; Goos, C.; Matern, S.; Lammert, F. Genome-wide analysis of hepatic fibrosis in inbred mice identifies the susceptibility locus Hfib1 on chromosome 15. Gastroenterology 2002, 123, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Zeremski, M.; Dimova, R.; Astemborski, J.; Thomas, D.L.; Talal, A.H. CXCL9 and CXCL10 chemokines as predictors of liver fibrosis in a cohort of primarily African-American injection drug users with chronic hepatitis C. J. Infect. Dis. 2011, 204, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Sousa, M.A.; Gomez-Moreno, A.Z.; Pineda-Tenor, D.; Medrano, L.M.; Sanchez-Ruano, J.J.; Fernandez-Rodriguez, A.; Artaza-Varasa, T.; Saura-Montalban, J.; Vazquez-Moron, S.; Ryan, P.; et al. CXCL9-11 polymorphisms are associated with liver fibrosis in patients with chronic hepatitis C: A cross-sectional study. Clin. Transl. Med. 2017, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Moran-Salvador, E.; Mann, J. Epigenetics and Liver Fibrosis. Cell Mol. Gastroenterol. Hepatol. 2017, 4, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Qin, C.Y.; Zhao, Z.H.; Fan, Y.C.; Wang, K. Epigenetic modifications in hepatic stellate cells contribute to liver fibrosis. Tohoku J. Exp. Med. 2013, 229, 35–43. [Google Scholar] [CrossRef]
- Etheridge, A.; Lee, I.; Hood, L.; Galas, D.; Wang, K. Extracellular microRNA: A new source of biomarkers. Mutat. Res. 2011, 717, 85–90. [Google Scholar] [CrossRef]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef]
- Jiang, X.P.; Ai, W.B.; Wan, L.Y.; Zhang, Y.Q.; Wu, J.F. The roles of microRNA families in hepatic fibrosis. Cell Biosci. 2017, 7, 34. [Google Scholar] [CrossRef]
- Yan, G.; Li, B.; Xin, X.; Xu, M.; Ji, G.; Yu, H. MicroRNA-34a Promotes Hepatic Stellate Cell Activation via Targeting ACSL1. Med. Sci. Monit. 2015, 21, 3008–3015. [Google Scholar] [CrossRef]
- Meng, F.; Glaser, S.S.; Francis, H.; Yang, F.; Han, Y.; Stokes, A.; Staloch, D.; McCarra, J.; Liu, J.; Venter, J.; et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am. J. Pathol. 2012, 181, 804–817. [Google Scholar] [CrossRef]
- Boutz, D.R.; Collins, P.J.; Suresh, U.; Lu, M.; Ramirez, C.M.; Fernandez-Hernando, C.; Huang, Y.; Abreu Rde, S.; Le, S.Y.; Shapiro, B.A.; et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J. Biol. Chem. 2011, 286, 18066–18078. [Google Scholar] [CrossRef]
- Trebicka, J.; Anadol, E.; Elfimova, N.; Strack, I.; Roggendorf, M.; Viazov, S.; Wedemeyer, I.; Drebber, U.; Rockstroh, J.; Sauerbruch, T.; et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 2013, 58, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Tijsen, A.J.; van der Made, I.; van den Hoogenhof, M.M.; Wijnen, W.J.; van Deel, E.D.; de Groot, N.E.; Alekseev, S.; Fluiter, K.; Schroen, B.; Goumans, M.J.; et al. The microRNA-15 family inhibits the TGFbeta-pathway in the heart. Cardiovasc. Res. 2014, 104, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Pan, Q.; Li, D.G.; Sun, H.; Liu, B.W. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J. Hepatol. 2009, 50, 766–778. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wei, X.X.; Wang, T.B.; Zhou, Y.C.; Liu, A.M.; Zhang, G.W. Increased miR-16 expression induced by hepatitis C virus infection promotes liver fibrosis through downregulation of hepatocyte growth factor and Smad7. Arch. Virol. 2015, 160, 2043–2050. [Google Scholar] [CrossRef]
- Sekiya, Y.; Ogawa, T.; Iizuka, M.; Yoshizato, K.; Ikeda, K.; Kawada, N. Down-regulation of cyclin E1 expression by microRNA-195 accounts for interferon-beta-induced inhibition of hepatic stellate cell proliferation. J. Cell Physiol. 2011, 226, 2535–2542. [Google Scholar] [CrossRef]
- Sun, X.; He, Y.; Ma, T.T.; Huang, C.; Zhang, L.; Li, J. Participation of miR-200a in TGF-beta1-mediated hepatic stellate cell activation. Mol. Cell Biochem. 2014, 388, 11–23. [Google Scholar] [CrossRef]
- Murakami, Y.; Toyoda, H.; Tanaka, M.; Kuroda, M.; Harada, Y.; Matsuda, F.; Tajima, A.; Kosaka, N.; Ochiya, T.; Shimotohno, K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE 2011, 6, e16081. [Google Scholar] [CrossRef]
- Hyun, J.; Wang, S.; Kim, J.; Rao, K.M.; Park, S.Y.; Chung, I.; Ha, C.S.; Kim, S.W.; Yun, Y.H.; Jung, Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat. Commun. 2016, 7, 10993. [Google Scholar] [CrossRef]
- Roderburg, C.; Mollnow, T.; Bongaerts, B.; Elfimova, N.; Vargas Cardenas, D.; Berger, K.; Zimmermann, H.; Koch, A.; Vucur, M.; Luedde, M.; et al. Micro-RNA profiling in human serum reveals compartment-specific roles of miR-571 and miR-652 in liver cirrhosis. PLoS ONE 2012, 7, e32999. [Google Scholar] [CrossRef]
- Chen, L.; Brenner, D.A.; Kisseleva, T. Combatting Fibrosis: Exosome-Based Therapies in the Regression of Liver Fibrosis. Hepatol. Commun. 2019, 3, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Wetmore, B.A.; Brees, D.J.; Singh, R.; Watkins, P.B.; Andersen, M.E.; Loy, J.; Thomas, R.S. Quantitative analyses and transcriptomic profiling of circulating messenger RNAs as biomarkers of rat liver injury. Hepatology 2010, 51, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Gonzalez, E.; Berisa, A.; Gil, D.; Embade, N.; Valle, M.; Luka, Z.; Elortza, F.; Wagner, C.; et al. Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Proteomics Clin. Appl. 2010, 4, 416–425. [Google Scholar] [CrossRef]
- Alhomrani, M.; Correia, J.; Zavou, M.; Leaw, B.; Kuk, N.; Xu, R.; Saad, M.I.; Hodge, A.; Greening, D.W.; Lim, R.; et al. The Human Amnion Epithelial Cell Secretome Decreases Hepatic Fibrosis in Mice with Chronic Liver Fibrosis. Front. Pharmacol. 2017, 8, 748. [Google Scholar] [CrossRef]
- Seo, W.; Eun, H.S.; Kim, S.Y.; Yi, H.S.; Lee, Y.S.; Park, S.H.; Jang, M.J.; Jo, E.; Kim, S.C.; Han, Y.M.; et al. Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by gammadelta T cells in liver fibrosis. Hepatology 2016, 64, 616–631. [Google Scholar] [CrossRef]
- Chen, L.; Charrier, A.; Zhou, Y.; Chen, R.; Yu, B.; Agarwal, K.; Tsukamoto, H.; Lee, L.J.; Paulaitis, M.E.; Brigstock, D.R. Epigenetic regulation of connective tissue growth factor by MicroRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology 2014, 59, 1118–1129. [Google Scholar] [CrossRef]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef]
- Charrier, A.; Chen, R.; Chen, L.; Kemper, S.; Hattori, T.; Takigawa, M.; Brigstock, D.R. Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor (CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery 2014, 156, 548–555. [Google Scholar] [CrossRef]
- Welker, M.W.; Reichert, D.; Susser, S.; Sarrazin, C.; Martinez, Y.; Herrmann, E.; Zeuzem, S.; Piiper, A.; Kronenberger, B. Soluble serum CD81 is elevated in patients with chronic hepatitis C and correlates with alanine aminotransferase serum activity. PLoS ONE 2012, 7, e30796. [Google Scholar] [CrossRef]
- Khalaf, F.K.; Dube, P.; Mohamed, A.; Tian, J.; Malhotra, D.; Haller, S.T.; Kennedy, D.J. Cardiotonic Steroids and the Sodium Trade Balance: New Insights into Trade-Off Mechanisms Mediated by the Na(+)/K(+)-ATPase. Int. J. Mol. Sci. 2018, 19, 2576. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.J.; Shrestha, K.; Sheehey, B.; Li, X.S.; Guggilam, A.; Wu, Y.; Finucan, M.; Gabi, A.; Medert, C.M.; Westfall, K.; et al. Elevated Plasma Marinobufagenin, An Endogenous Cardiotonic Steroid, Is Associated With Right Ventricular Dysfunction and Nitrative Stress in Heart Failure. Circ. Heart Fail. 2015, 8, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, D. The role of cardiotonic steroids in the pathogenesis of cardiomyopathy in chronic kidney disease. Nephron. Clin. Pract. 2014, 128, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xie, Z.J. The sodium pump and cardiotonic steroids-induced signal transduction protein kinases and calcium-signaling microdomain in regulation of transporter trafficking. Biochim. Biophys. Acta 2010, 1802, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Schoner, W.; Scheiner-Bobis, G. Role of endogenous cardiotonic steroids in sodium homeostasis. Nephrol. Dial. Transplant. 2008, 23, 2723–2729. [Google Scholar] [CrossRef]
- Schoner, W.; Scheiner-Bobis, G. Endogenous and exogenous cardiac glycosides: Their roles in hypertension, salt metabolism, and cell growth. Am. J. Physiol. Cell Physiol. 2007, 293, C509–C536. [Google Scholar] [CrossRef]
- Elkareh, J.; Periyasamy, S.M.; Shidyak, A.; Vetteth, S.; Schroeder, J.; Raju, V.; Hariri, I.M.; El-Okdi, N.; Gupta, S.; Fedorova, L.; et al. Marinobufagenin induces increases in procollagen expression in a process involving protein kinase C and Fli-1: Implications for uremic cardiomyopathy. Am. J. Physiol. Renal Physiol. 2009, 296, F1219–F1226. [Google Scholar] [CrossRef]
- Fedorova, L.V.; Raju, V.; El-Okdi, N.; Shidyak, A.; Kennedy, D.J.; Vetteth, S.; Giovannucci, D.R.; Bagrov, A.Y.; Fedorova, O.V.; Shapiro, J.I.; et al. The cardiotonic steroid hormone marinobufagenin induces renal fibrosis: Implication of epithelial-to-mesenchymal transition. Am. J. Physiol. Renal Physiol. 2009, 296, F922–F934. [Google Scholar] [CrossRef]
- Bagrov, A.Y.; Shapiro, J.I.; Fedorova, O.V. Endogenous cardiotonic steroids: Physiology, pharmacology, and novel therapeutic targets. Pharmacol. Rev. 2009, 61, 9–38. [Google Scholar] [CrossRef]
- Manunta, P.; Ferrandi, M.; Bianchi, G.; Hamlyn, J.M. Endogenous ouabain in cardiovascular function and disease. J. Hypertens. 2009, 27, 9–18. [Google Scholar] [CrossRef]
- Kennedy, D.J.; Khalaf, F.K.; Sheehy, B.; Weber, M.E.; Agatisa-Boyle, B.; Conic, J.; Hauser, K.; Medert, C.M.; Westfall, K.; Bucur, P.; et al. Telocinobufagin, a Novel Cardiotonic Steroid, Promotes Renal Fibrosis via Na(+)/K(+)-ATPase Profibrotic Signaling Pathways. Int. J. Mol. Sci. 2018, 19, 2566. [Google Scholar] [CrossRef] [PubMed]
- Elkareh, J.; Kennedy, D.J.; Yashaswi, B.; Vetteth, S.; Shidyak, A.; Kim, E.G.; Smaili, S.; Periyasamy, S.M.; Hariri, I.M.; Fedorova, L.; et al. Marinobufagenin stimulates fibroblast collagen production and causes fibrosis in experimental uremic cardiomyopathy. Hypertension 2007, 49, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Kolmakova, E.V.; Haller, S.T.; Kennedy, D.J.; Isachkina, A.N.; Budny, G.V.; Frolova, E.V.; Piecha, G.; Nikitina, E.R.; Malhotra, D.; Fedorova, O.V.; et al. Endogenous cardiotonic steroids in chronic renal failure. Nephrol. Dial. Transplant. 2011, 26, 2912–2919. [Google Scholar] [CrossRef]
- El-Okdi, N.; Smaili, S.; Raju, V.; Shidyak, A.; Gupta, S.; Fedorova, L.; Elkareh, J.; Periyasamy, S.; Shapiro, A.P.; Kahaleh, M.B.; et al. Effects of cardiotonic steroids on dermal collagen synthesis and wound healing. J. Appl. Physiol. (1985) 2008, 105, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, O.V.; Fadeev, A.V.; Grigorova, Y.N.; Marshall, C.A.; Zernetkina, V.; Kolodkin, N.I.; Agalakova, N.I.; Konradi, A.O.; Lakatta, E.G.; Bagrov, A.Y. Cardiotonic Steroids Induce Vascular Fibrosis Via Pressure-Independent Mechanism in NaCl-Loaded Diabetic Rats. J. Cardiovasc. Pharmacol. 2019, 74, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tian, J.; Haas, M.; Shapiro, J.I.; Askari, A.; Xie, Z. Ouabain interaction with cardiac Na+/K+-ATPase initiates signal cascades independent of changes in intracellular Na+ and Ca2+ concentrations. J. Biol. Chem. 2000, 275, 27838–27844. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.T.; Yan, Y.; Drummond, C.A.; Xie, J.; Tian, J.; Kennedy, D.J.; Shilova, V.Y.; Xie, Z.; Liu, J.; Cooper, C.J.; et al. Rapamycin Attenuates Cardiac Fibrosis in Experimental Uremic Cardiomyopathy by Reducing Marinobufagenin Levels and Inhibiting Downstream Pro-Fibrotic Signaling. J. Am. Heart Assoc. 2016, 5. [Google Scholar] [CrossRef]
- Campia, I.; Gazzano, E.; Pescarmona, G.; Ghigo, D.; Bosia, A.; Riganti, C. Digoxin and ouabain increase the synthesis of cholesterol in human liver cells. Cell Mol. Life Sci. 2009, 66, 1580–1594. [Google Scholar] [CrossRef]
- Campia, I.; Sala, V.; Kopecka, J.; Leo, C.; Mitro, N.; Costamagna, C.; Caruso, D.; Pescarmona, G.; Crepaldi, T.; Ghigo, D.; et al. Digoxin and ouabain induce the efflux of cholesterol via liver X receptor signalling and the synthesis of ATP in cardiomyocytes. Biochem. J. 2012, 447, 301–311. [Google Scholar] [CrossRef]
- Pasqualini, J.R. Enzymes involved in the formation and transformation of steroid hormones in the fetal and placental compartments. J. Steroid Biochem. Mol. Biol. 2005, 97, 401–415. [Google Scholar] [CrossRef]
- Tirosh, O. Hypoxic Signaling and Cholesterol Lipotoxicity in Fatty Liver Disease Progression. Oxid. Med. Cell. Longev. 2018, 2018, 2548154. [Google Scholar] [CrossRef]
- Pavlovic, D. Endogenous cardiotonic steroids and cardiovascular disease, where to next? Cell Calcium 2020, 86, 102156. [Google Scholar] [CrossRef] [PubMed]
- Laredo, J.; Hamilton, B.P.; Hamlyn, J.M. Secretion of endogenous ouabain from bovine adrenocortical cells: Role of the zona glomerulosa and zona fasciculata. Biochem. Biophys. Res. Commun. 1995, 212, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Trapani, L.; Segatto, M.; Pallottini, V. Regulation and deregulation of cholesterol homeostasis: The liver as a metabolic “power station”. World J. Hepatol. 2012, 4, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Yang, H.; Song, B.L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Hou, W.; Syn, W.K. Role of Metabolism in Hepatic Stellate Cell Activation and Fibrogenesis. Front. Cell Dev. Biol. 2018, 6, 150. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Maher, J.J. Lipids in liver disease: Looking beyond steatosis. Gastroenterology 2012, 142, 8–11. [Google Scholar] [CrossRef]
- Teratani, T.; Tomita, K.; Suzuki, T.; Oshikawa, T.; Yokoyama, H.; Shimamura, K.; Tominaga, S.; Hiroi, S.; Irie, R.; Okada, Y.; et al. A high-cholesterol diet exacerbates liver fibrosis in mice via accumulation of free cholesterol in hepatic stellate cells. Gastroenterology 2012, 142, 152–164 e110. [Google Scholar] [CrossRef]
- Tomita, K.; Teratani, T.; Suzuki, T.; Shimizu, M.; Sato, H.; Narimatsu, K.; Usui, S.; Furuhashi, H.; Kimura, A.; Nishiyama, K.; et al. Acyl-CoA:cholesterol acyltransferase 1 mediates liver fibrosis by regulating free cholesterol accumulation in hepatic stellate cells. J. Hepatol. 2014, 61, 98–106. [Google Scholar] [CrossRef]
- Furuhashi, H.; Tomita, K.; Teratani, T.; Shimizu, M.; Nishikawa, M.; Higashiyama, M.; Takajo, T.; Shirakabe, K.; Maruta, K.; Okada, Y.; et al. Vitamin A-coupled liposome system targeting free cholesterol accumulation in hepatic stellate cells offers a beneficial therapeutic strategy for liver fibrosis. Hepatol. Res. 2018, 48, 397–407. [Google Scholar] [CrossRef]
- Arguello, G.; Balboa, E.; Arrese, M.; Zanlungo, S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim. Biophys. Acta 2015, 1852, 1765–1778. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.; Ho, S.L.; Jeng, Y.M.; Lai, Y.S.; Chen, Y.H.; Lu, S.C.; Chen, H.L.; Chang, P.Y.; Hu, R.H.; Lee, P.H. Accumulation of free cholesterol and oxidized low-density lipoprotein is associated with portal inflammation and fibrosis in nonalcoholic fatty liver disease. J. Inflamm. (Lond) 2019, 16, 7. [Google Scholar] [CrossRef] [PubMed]
- Anavi, S.; Eisenberg-Bord, M.; Hahn-Obercyger, M.; Genin, O.; Pines, M.; Tirosh, O. The role of iNOS in cholesterol-induced liver fibrosis. Lab. Invest. 2015, 95, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Pratt, R.D.; Brickman, C.R.; Cottrill, C.L.; Shapiro, J.I.; Liu, J. The Na/K-ATPase Signaling: From Specific Ligands to General Reactive Oxygen Species. Int. J. Mol. Sci. 2018, 19, 2600. [Google Scholar] [CrossRef]
- Fuerstenwerth, H. On the differences between ouabain and digitalis glycosides. Am. J. Ther. 2014, 21, 35–42. [Google Scholar] [CrossRef]
- Laursen, M.; Gregersen, J.L.; Yatime, L.; Nissen, P.; Fedosova, N.U. Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex. Proc. Natl. Acad. Sci. USA 2015, 112, 1755–1760. [Google Scholar] [CrossRef]
- Takara, K.; Takagi, K.; Tsujimoto, M.; Ohnishi, N.; Yokoyama, T. Digoxin up-regulates multidrug resistance transporter (MDR1) mRNA and simultaneously down-regulates steroid xenobiotic receptor mRNA. Biochem. Biophys. Res. Commun. 2003, 306, 116–120. [Google Scholar] [CrossRef]
- Mijatovic, T.; Kiss, R. Cardiotonic steroids-mediated Na+/K+-ATPase targeting could circumvent various chemoresistance pathways. Planta. Med. 2013, 79, 189–198. [Google Scholar] [CrossRef]
- Kau, M.M.; Kan, S.F.; Wang, J.R.; Wang, P.S. Inhibitory effects of digoxin and ouabain on aldosterone synthesis in human adrenocortical NCI-H295 cells. J. Cell Physiol. 2005, 205, 393–401. [Google Scholar] [CrossRef]
- Neri, G.; De Toni, R.; Tortorella, C.; Rebuffat, P.; Bova, S.; Cargnelli, G.; Petrelli, L.; Spinazzi, R.; Nussdorfer, G.G. Ouabain chronic infusion enhances the growth and steroidogenic capacity of rat adrenal zona glomerulosa: The possible involvement of the endothelin system. Int. J. Mol. Med. 2006, 18, 315–319. [Google Scholar]
- Pu, H.F.; Wang, S.W.; Tseng, C.I.; Huang, H.L.; Lin, C.W.; Hsu, J.M.; Chen, M.J.; Chow, Y.C.; Wang, P.S. Mechanisms of digoxin and digitoxin on the production of corticosterone in zona fasciculata-reticularis cells of ovariectomized rats. J. Cell Biochem. 2006, 97, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, G.N. The Role of Cholesterol in the Pathogenesis of NASH. Trends Endocrinol. Metab. 2016, 27, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Bellanti, F.; Mitarotonda, D.; Tamborra, R.; Blonda, M.; Iannelli, G.; Petrella, A.; Sanginario, V.; Iuliano, L.; Vendemiale, G.; Serviddio, G. Oxysterols induce mitochondrial impairment and hepatocellular toxicity in non-alcoholic fatty liver disease. Free Radic. Biol. Med. 2014, 75 (Suppl. 1), S16–S17. [Google Scholar] [CrossRef] [PubMed]
- Caballero, F.; Fernandez, A.; De Lacy, A.M.; Fernandez-Checa, J.C.; Caballeria, J.; Garcia-Ruiz, C. Enhanced free cholesterol, SREBP-2 and StAR expression in human NASH. J. Hepatol. 2009, 50, 789–796. [Google Scholar] [CrossRef]
- Subramanian, S.; Goodspeed, L.; Wang, S.; Kim, J.; Zeng, L.; Ioannou, G.N.; Haigh, W.G.; Yeh, M.M.; Kowdley, K.V.; O’Brien, K.D.; et al. Dietary cholesterol exacerbates hepatic steatosis and inflammation in obese LDL receptor-deficient mice. J. Lipid Res. 2011, 52, 1626–1635. [Google Scholar] [CrossRef]
- Malhotra, P.; Aloman, C.; Ankireddy, A.; Khadra, H.; Ooka, K.; Gill, R.K.; Saksena, S.; Dudeja, P.K.; Alrefai, W.A. Overactivation of intestinal sterol response element-binding protein 2 promotes diet-induced nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 313, G376–G385. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Subramanian, S.; Chait, A.; Haigh, W.G.; Yeh, M.M.; Farrell, G.C.; Lee, S.P.; Savard, C. Cholesterol crystallization within hepatocyte lipid droplets and its role in murine NASH. J. Lipid Res. 2017, 58, 1067–1079. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Haigh, W.G.; Thorning, D.; Savard, C. Hepatic cholesterol crystals and crown-like structures distinguish NASH from simple steatosis. J. Lipid Res. 2013, 54, 1326–1334. [Google Scholar] [CrossRef]
- Baffy, G. Kupffer cells in non-alcoholic fatty liver disease: The emerging view. J. Hepatol. 2009, 51, 212–223. [Google Scholar] [CrossRef]
- Mesarwi, O.A.; Shin, M.K.; Bevans-Fonti, S.; Schlesinger, C.; Shaw, J.; Polotsky, V.Y. Hepatocyte Hypoxia Inducible Factor-1 Mediates the Development of Liver Fibrosis in a Mouse Model of Nonalcoholic Fatty Liver Disease. PLoS ONE 2016, 11, e0168572. [Google Scholar] [CrossRef]
- Zhang, C.; Bian, M.; Chen, X.; Jin, H.; Zhao, S.; Yang, X.; Shao, J.; Chen, A.; Guo, Q.; Zhang, F.; et al. Oroxylin A prevents angiogenesis of LSECs in liver fibrosis via inhibition of YAP/HIF-1alpha signaling. J. Cell Biochem. 2018, 119, 2258–2268. [Google Scholar] [CrossRef] [PubMed]
- Moczydlowska, J.; Miltyk, W.; Hermanowicz, A.; Lebensztejn, D.M.; Palka, J.A.; Debek, W. HIF-1 alpha as a Key Factor in Bile Duct Ligation-Induced Liver Fibrosis in Rats. J. Invest. Surg. 2017, 30, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Anavi, S.; Madar, Z.; Tirosh, O. Non-alcoholic fatty liver disease, to struggle with the strangle: Oxygen availability in fatty livers. Redox Biol. 2017, 13, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Mederacke, I.; Hsu, C.C.; Troeger, J.S.; Huebener, P.; Mu, X.; Dapito, D.H.; Pradere, J.P.; Schwabe, R.F. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 2013, 4, 2823. [Google Scholar] [CrossRef] [PubMed]
- Puche, J.E.; Saiman, Y.; Friedman, S.L. Hepatic stellate cells and liver fibrosis. Compr. Physiol. 2013, 3, 1473–1492. [Google Scholar] [CrossRef]
- Gandhi, C.R. Hepatic stellate cell activation and pro-fibrogenic signals. J. Hepatol. 2017, 67, 1104–1105. [Google Scholar] [CrossRef]
- Friedman, S.L. Mechanisms of disease: Mechanisms of hepatic fibrosis and therapeutic implications. Nat. Clin. Pract. Gastroenterol. Hepatol. 2004, 1, 98–105. [Google Scholar] [CrossRef]
- Pinzani, M.; Rombouts, K. Liver fibrosis: From the bench to clinical targets. Dig. Liver Dis. 2004, 36, 231–242. [Google Scholar] [CrossRef]
- Parola, M.; Marra, F.; Pinzani, M. Myofibroblast - like cells and liver fibrogenesis: Emerging concepts in a rapidly moving scenario. Mol. Aspects Med. 2008, 29, 58–66. [Google Scholar] [CrossRef]
- Parola, M.; Pinzani, M. Hepatic wound repair. Fibrogenesis Tissue Repair 2009, 2, 4. [Google Scholar] [CrossRef]
- Hinz, B.; Phan, S.H.; Thannickal, V.J.; Galli, A.; Bochaton-Piallat, M.L.; Gabbiani, G. The myofibroblast: One function, multiple origins. Am. J. Pathol. 2007, 170, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Darby, I.A.; Laverdet, B.; Bonte, F.; Desmouliere, A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 2014, 7, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Orlov, S.N.; La, J.; Smolyaninova, L.V.; Dulin, N.O. Na+,K+-ATPase as a Target for Treatment of Tissue Fibrosis. Curr. Med. Chem. 2019, 26, 564–575. [Google Scholar] [CrossRef]
- Wynn, T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef]
- Parsons, C.J.; Takashima, M.; Rippe, R.A. Molecular mechanisms of hepatic fibrogenesis. J. Gastroenterol. Hepatol. 2007, 22 (Suppl. 1), S79–S84. [Google Scholar] [CrossRef]
- Artlett, C.M. Inflammasomes in wound healing and fibrosis. J. Pathol. 2013, 229, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Elpek, G.O. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J. Gastroenterol. 2014, 20, 7260–7276. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- George, J.; Tsuchishima, M.; Tsutsumi, M. Molecular mechanisms in the pathogenesis of N-nitrosodimethylamine induced hepatic fibrosis. Cell Death Dis. 2019, 10, 18. [Google Scholar] [CrossRef]
- Sodhi, K.; Srikanthan, K.; Goguet-Rubio, P.; Nichols, A.; Mallick, A.; Nawab, A.; Martin, R.; Shah, P.T.; Chaudhry, M.; Sigdel, S.; et al. pNaKtide Attenuates Steatohepatitis and Atherosclerosis by Blocking Na/K-ATPase/ROS Amplification in C57Bl6 and ApoE Knockout Mice Fed a Western Diet. Sci. Rep. 2017, 7, 193. [Google Scholar] [CrossRef]
- Kennedy, D.J.; Vetteth, S.; Periyasamy, S.M.; Kanj, M.; Fedorova, L.; Khouri, S.; Kahaleh, M.B.; Xie, Z.; Malhotra, D.; Kolodkin, N.I.; et al. Central role for the cardiotonic steroid marinobufagenin in the pathogenesis of experimental uremic cardiomyopathy. Hypertension 2006, 47, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.T.; Drummond, C.A.; Yan, Y.; Liu, J.; Tian, J.; Malhotra, D.; Shapiro, J.I. Passive immunization against marinobufagenin attenuates renal fibrosis and improves renal function in experimental renal disease. Am. J. Hypertens. 2014, 27, 603–609. [Google Scholar] [CrossRef]
- Paczula, A.; Wiecek, A.; Piecha, G. Cardiotonic Steroids-A Possible Link Between High-Salt Diet and Organ Damage. Int. J. Mol. Sci. 2019, 20, 590. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante-Silva, L.H.A.; Lima, E.A.; Carvalho, D.C.M.; de Sales-Neto, J.M.; Alves, A.K.A.; Galvao, J.; da Silva, J.S.F.; Rodrigues-Mascarenhas, S. Corrigendum: Much More than a Cardiotonic Steroid: Modulation of Inflammation by Ouabain. Front. Physiol. 2018, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.X.; Shapiro, A.P.; Shapiro, J.I. The Trade-Off between Dietary Salt and Cardiovascular Disease; A Role for Na/K-ATPase Signaling? Front. Endocrinol. (Lausanne) 2014, 5, 97. [Google Scholar] [CrossRef]
- Lingrel, J.B. The physiological significance of the cardiotonic steroid/ouabain-binding site of the Na,K-ATPase. Annu. Rev. Physiol. 2010, 72, 395–412. [Google Scholar] [CrossRef]
- Yan, Y.; Shapiro, A.P.; Mopidevi, B.R.; Chaudhry, M.A.; Maxwell, K.; Haller, S.T.; Drummond, C.A.; Kennedy, D.J.; Tian, J.; Malhotra, D.; et al. Protein Carbonylation of an Amino Acid Residue of the Na/K-ATPase alpha1 Subunit Determines Na/K-ATPase Signaling and Sodium Transport in Renal Proximal Tubular Cells. J. Am. Heart Assoc. 2016, 5. [Google Scholar] [CrossRef]
- Cui, X.; Xie, Z. Protein Interaction and Na/K-ATPase-Mediated Signal Transduction. Molecules 2017, 22, 990. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, T.; Wang, H.; Li, Z.; Loreaux, E.; Lingrel, J.B.; Xie, Z. Regulation of intracellular cholesterol distribution by Na/K-ATPase. J. Biol. Chem. 2009, 284, 14881–14890. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Ye, Q.; Tian, J.; Jing, R.; Xie, Z. Regulation of alpha1 Na/K-ATPase expression by cholesterol. J. Biol. Chem. 2011, 286, 15517–15524. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zehra, M.; Curry, J.C.; Pillai, S.S.; Lakhani, H.V.; Edwards, C.E.; Sodhi, K. Elucidating Potential Profibrotic Mechanisms of Emerging Biomarkers for Early Prognosis of Hepatic Fibrosis. Int. J. Mol. Sci. 2020, 21, 4737. https://doi.org/10.3390/ijms21134737
Zehra M, Curry JC, Pillai SS, Lakhani HV, Edwards CE, Sodhi K. Elucidating Potential Profibrotic Mechanisms of Emerging Biomarkers for Early Prognosis of Hepatic Fibrosis. International Journal of Molecular Sciences. 2020; 21(13):4737. https://doi.org/10.3390/ijms21134737
Chicago/Turabian StyleZehra, Mishghan, James C. Curry, Sneha S. Pillai, Hari Vishal Lakhani, Cory E. Edwards, and Komal Sodhi. 2020. "Elucidating Potential Profibrotic Mechanisms of Emerging Biomarkers for Early Prognosis of Hepatic Fibrosis" International Journal of Molecular Sciences 21, no. 13: 4737. https://doi.org/10.3390/ijms21134737
APA StyleZehra, M., Curry, J. C., Pillai, S. S., Lakhani, H. V., Edwards, C. E., & Sodhi, K. (2020). Elucidating Potential Profibrotic Mechanisms of Emerging Biomarkers for Early Prognosis of Hepatic Fibrosis. International Journal of Molecular Sciences, 21(13), 4737. https://doi.org/10.3390/ijms21134737





