A Genomic Perspective on the Potential of Wild-Type Rumen Bacterium Enterobacter sp. LU1 as an Industrial Platform for Bio-Based Succinate Production
Abstract
:1. Introduction
2. Results and Discussion
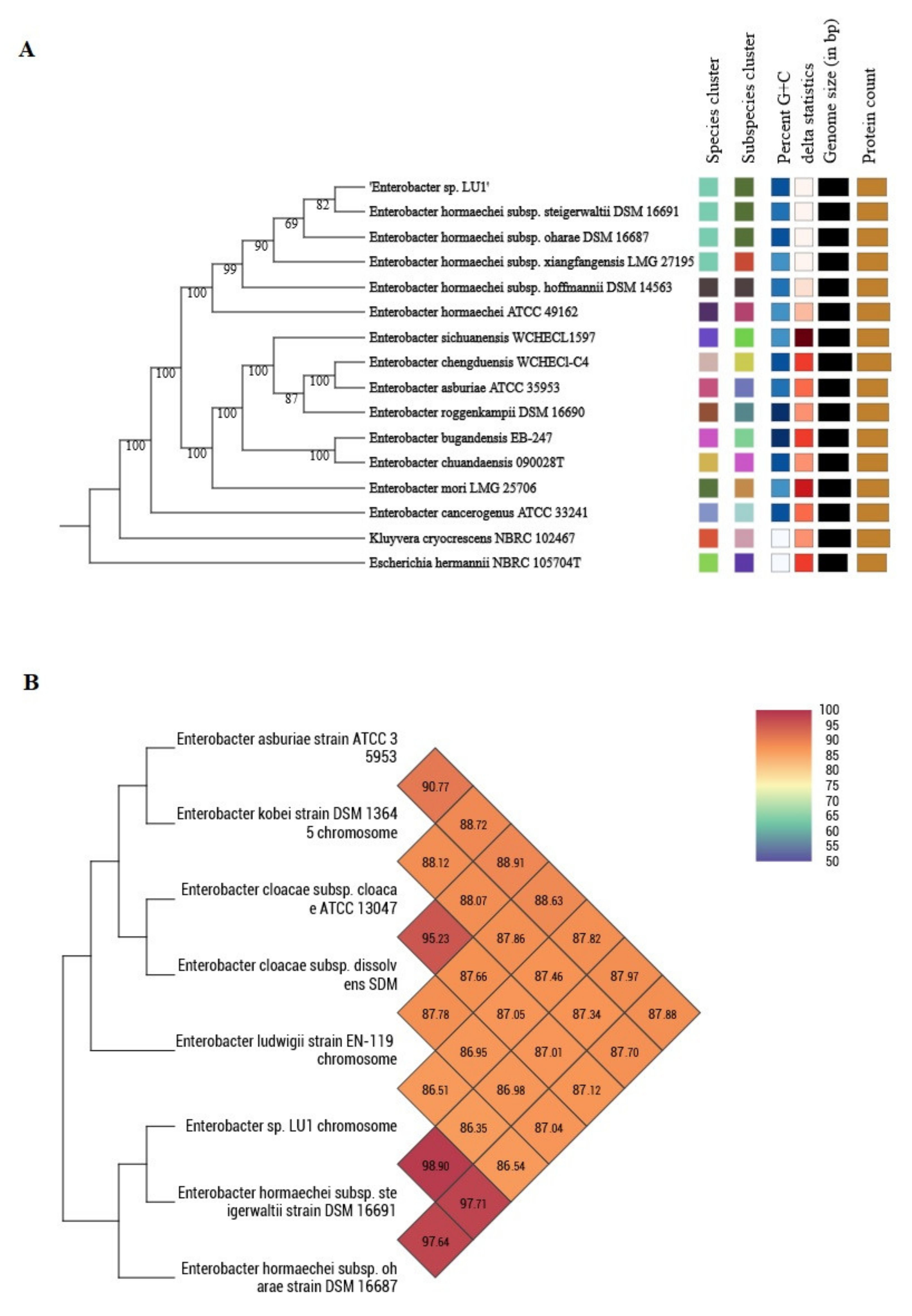
2.1. Phylogenomic Classification of Enterobacter sp. LU1
2.2. Assembly, Structure, and General Features of Enterobacter sp. LU1 Genome
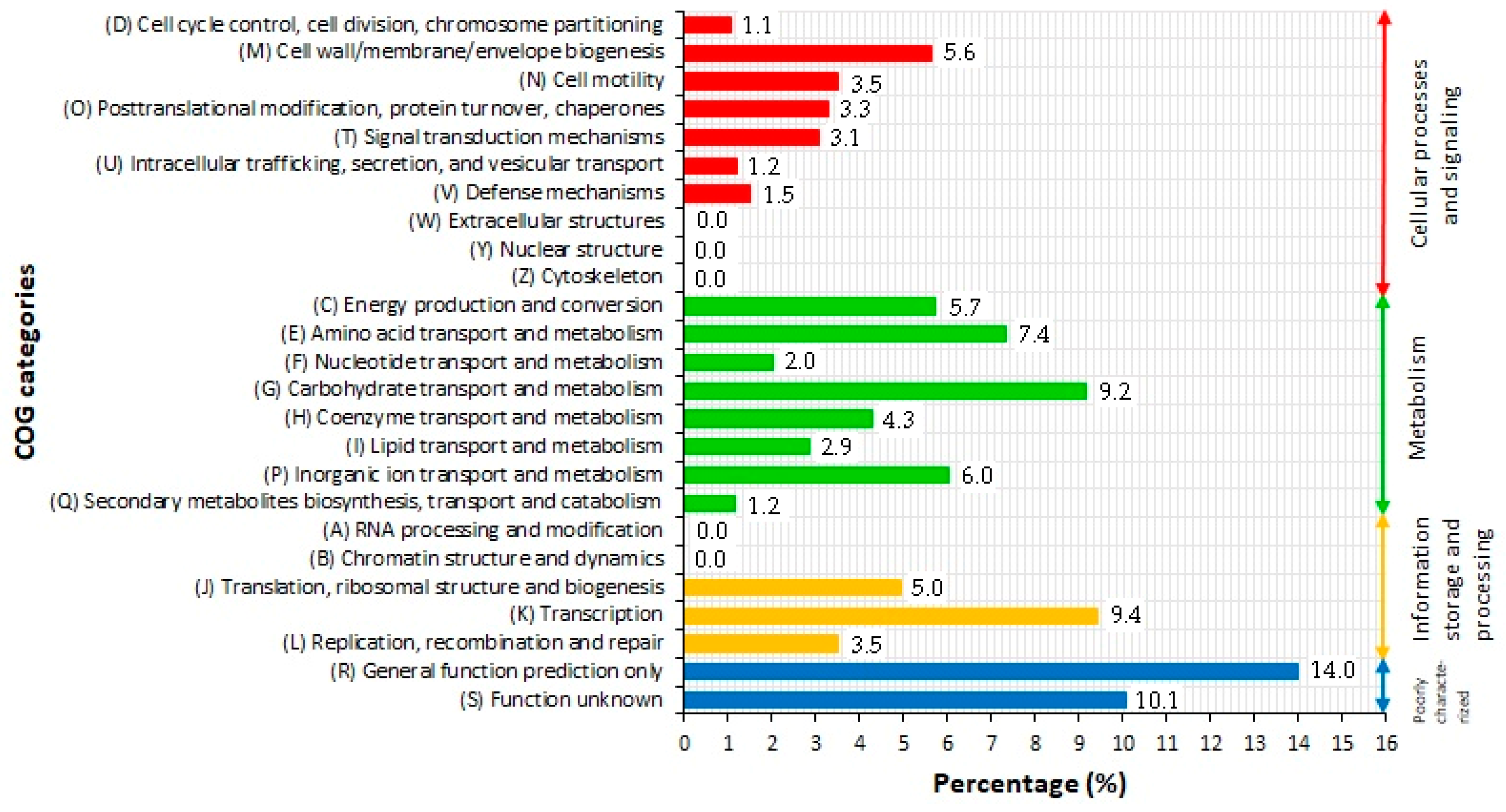
2.3. COG Classification of Predicted Genes
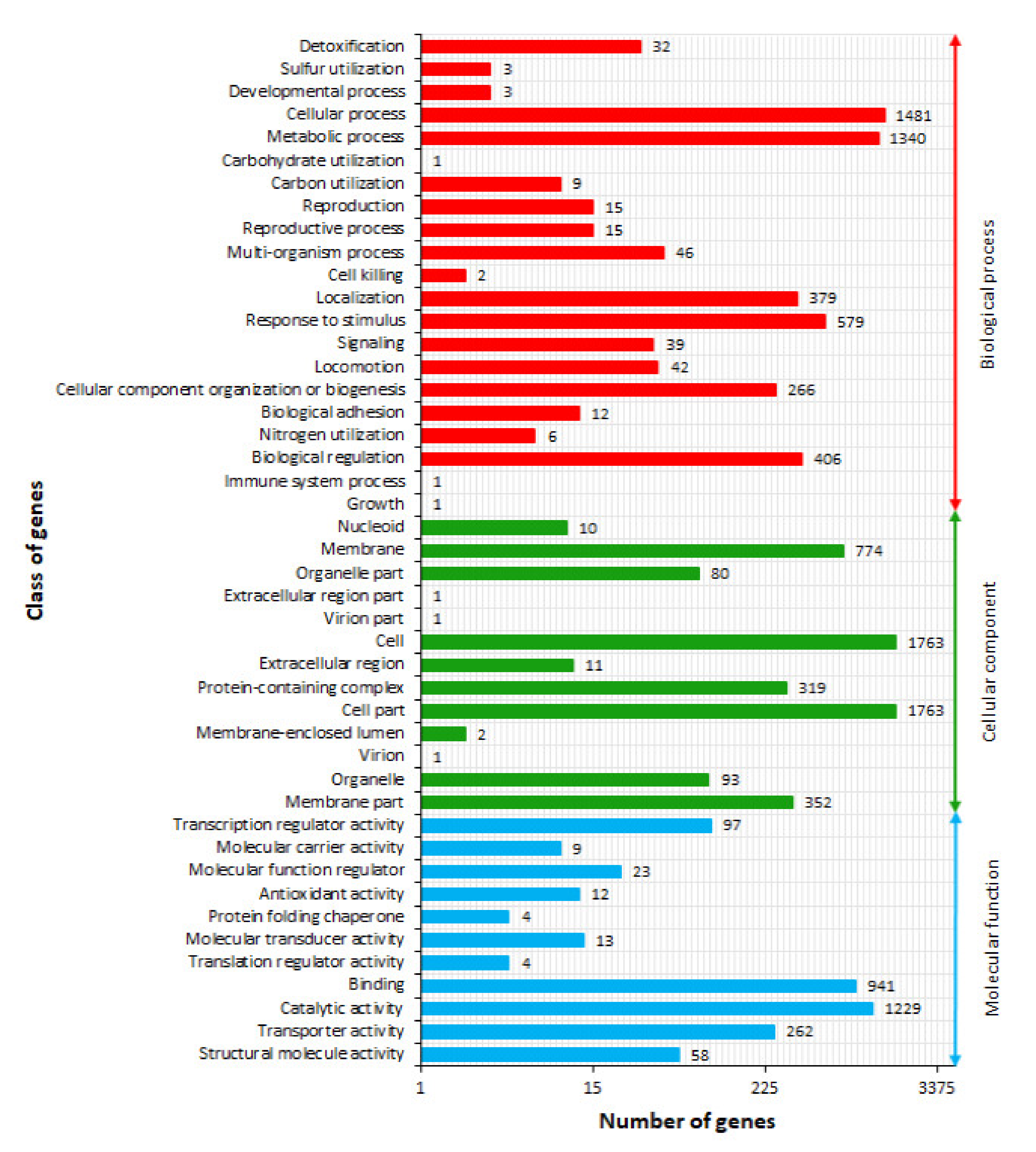
2.4. Gene Ontology Term Annotation
2.5. Transport and Metabolism of Carbon Sources
2.6. Glycerol Metabolism Pathways
2.7. Metabolic Routes for SA Biosynthesis
2.8. Dicarboxylic Acid Transporters
2.9. Osmotic and Oxidative Stress
2.10. Horizontal Gene Transfer and Acquired Antimicrobial Resistance Genes
2.11. Prophage
2.12. CRISPR
3. Conclusions
4. Materials and Methods
4.1. Strain
4.2. Growth Conditions
4.3. Genomic DNA Extraction
4.4. Re-Identification of Enterobacter sp. LU1
4.5. Hybrid Whole-Genome Sequencing
4.6. De novo Assembly and Complete Genome Annotation
4.7. Data Deposition
4.8. Phylogenomic Classification of Enterobacter sp. LU1
4.9. Functional Classification of Annotated Genes and Other Genome Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Podleśny, M.; Jarocki, P.; Wyrostek, J.; Czernecki, T.; Kucharska, J.; Nowak, A.; Targoński, Z. Enterobacter sp. LU1 as a novel succinic acid producer–co-utilization of glycerol and lactose. Microb. Biotechnol. 2017, 10, 492–501. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Wu, M.; Dai, Z.; Xin, F.; Zhou, J.; Dong, W.; Ma, J.; Jiang, M.; Zhang, W. Comprehensive investigation of succinic acid production by Actinobacillus succinogenes: A promising native succinic acid producer. Biofuels Bioprod. Biorefin. 2019. [Google Scholar] [CrossRef]
- Zeikus, J.G.; Jain, M.K.; Elankovan, P. Biotechnology of succinic acid production and markets for derived industrial products. Appl. Microbiol. Biotechnol. 1999, 51, 545–552. [Google Scholar] [CrossRef]
- Bozell, J.J.; Petersen, G.R. Technology development for the production of biobased products from biorefinery carbohydrates—The US Department of Energy’s “top 10” revisited. Green Chem. 2010, 12, 539–554. [Google Scholar] [CrossRef]
- Cok, B.; Tsiropoulos, I.; Roes, A.L.; Patel, M.K. Succinic acid production derived from carbohydrates: An energy and greenhouse gas assessment of a platform chemical toward a bio-based economy. Biofuels Bioprod. Biorefin. 2014, 8, 16–29. [Google Scholar] [CrossRef]
- Ahn, J.H.; Jang, Y.S.; Lee, S.Y. Production of succinic acid by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 2016, 42, 54–66. [Google Scholar] [CrossRef]
- Pateraki, C.; Patsalou, M.; Vlysidis, A.; Kopsahelis, N.; Webb, C.; Koutinas, A.A.; Koutinas, M. Actinobacillus succinogenes: Advances on succinic acid production and prospects for development of integrated biorefineries. Biochem. Eng. J. 2016, 112, 285–303. [Google Scholar] [CrossRef]
- Thuy, N.T.H.; Kongkaew, A.; Flood, A.; Boontawan, A. Fermentation and crystallization of succinic acid from Actinobacillus succinogenes ATCC55618 using fresh cassava root as the main substrate. Bioresour. Technol. 2017, 233, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, J.; Idris, A.; Aziz, R.A. Recent advances in production of succinic acid from lignocellulosic biomass. Appl. Microbiol. Biotechnol. 2014, 98, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Argenzio, O.; D’Ambrosio, S.; Lama, L.; Finore, I.; Finamore, R.; Pepe, O.; Faraco, C.; Schiraldi, C. Production of succinic acid from Basfia succiniciproducens up to the pilot scale from Arundo donax hydrolysate. Bioresour. Technol. 2016, 222, 355–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimini, D.; Zaccariello, L.; D’Ambrosio, S.; Lama, L.; Ruoppolo, G.; Pepe, O.; Faraco, V.; Schiraldi, C. Improved production of succinic acid from Basfia succiniciproducens growing on A. donax and process evaluation through material flow analysis. Biotechnol. Biofuels 2019, 12, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szczerba, H.; Komoń-Janczara, E.; Dudziak, K.; Waśko, A.; Targoński, Z. A novel biocatalyst, Enterobacter aerogenes LU2, for efficient production of succinic acid using whey permeate as a cost-effective carbon source. Biotechnol. Biofuels 2020, 13, 1–12. [Google Scholar] [CrossRef]
- Ventorino, V.; Robertiello, A.; Cimini, D.; Argenzio, O.; Schiraldi, C.; Montella, S.; Faraco, V.; Ambrosanio, A.; Viscardi, S.; Pepe, O. Bio-based succinate production from Arundo donax hydrolysate with the new natural succinic acid-producing strain Basfia succiniciproducens BPP7. Bioenergy Res. 2017, 10, 488–498. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.; Hong, H.; Chang, P.; Lee, S.Y.; Hong, S.H.; Chang, H.N. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl. Microbiol. Biotechnol. 2002, 58, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Podleśny, M.; Wyrostek, J.; Kucharska, J.; Jarocki, P.; Komon-Janczara, E.; Targonski, Z. A new strategy for effective succinic acid production by Enterobacter sp. LU1 using a medium based on crude glycerol and whey permeate. Molecules 2019, 24, 4543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Lei, L.; Zheng, L.; Xiao, X.; Tang, H.; Luo, C. Genome sequencing of gut symbiotic Bacillus velezensis LC1 for bioethanol production from bamboo shoots. Biotechnol. Biofuels 2020, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Szczerba, H.; Komoń-Janczara, E.; Krawczyk, M.; Dudziak, K.; Nowak, A.; Kuzdraliński, A.; Waśko, A.; Targoński, Z. Genome analysis of a wild rumen bacterium Enterobacter aerogenes LU2—A novel bio-based succinic acid producer. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Rappert, S.; Song, L.; Sabra, W.; Wang, W.; Zeng, A.P. Draft genome sequence of type strain Clostridium pasteurianum DSM 525 (ATCC 6013), a promising producer of chemicals and fuels. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.F.; Wang, W.J.; Ji, X.J. Draft genome sequence of tetracycline-resistant Klebsiella oxytoca CCTCC M207023 producing 2,3-butanediol isolated from China. J. Glob. Antimicrob. Resist. 2020, 20, 160–162. [Google Scholar] [CrossRef]
- Fan, L.; Li, M.; Li, Y.; Fan, X.; Liu, Y.; Lv, Y. Draft genome sequence of thermophilic Bacillus sp. TYF-LIM-B05 directly producing ethanol from various carbon sources including lignocellulose. Curr. Microbiol. 2020, 77, 491. [Google Scholar] [CrossRef]
- Todd, S.M.; Settlage, R.E.; Lahmers, K.K.; Slade, D.J. Fusobacterium genomics using minion and illumina sequencing enables genome completion and correction. mSphere 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Monahan, L.G.; Demaere, M.Z.; Cummins, M.L.; Djordjevic, S.P.; Roy Chowdhury, P.; Darling, A.E. High contiguity genome sequence of a multidrug-resistant hospital isolate of Enterobacter hormaechei. Gut Pathog. 2019, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Type Strain Genome Server (TYGS) Platform. Available online: https://tygs.dsmz.de (accessed on 27 June 2020).
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Makuwa, S.C.; Serepa-Dlamini, M.H. Data on draft genome sequence of Bacillus sp. strain MHSD28, a bacterial endophyte isolated from Dicoma anomala. Data Brief 2019, 26, 104524. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in bacterial taxonomy: Impact on the genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Hoffmann, H.; Stindl, S.; Ludwig, W.; Stumpf, A.; Mehlen, A.; Monget, D.; Pierard, D.; Ziesing, S.; Heesemann, J.; Roggenkamp, A.; et al. Enterobacter hormaechei subsp. oharae subsp. nov., E. hormaechei subsp. hormaechei comb. nov., and E. hormaechei subsp. steigerwaltii subsp. nov., three new subspecies of clinical importance. J. Clin. Microbiol. 2005, 43, 3297–3303. [Google Scholar] [CrossRef] [Green Version]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [Green Version]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Trujillo, M.E.; Bacigalupe, R.; Pujic, P.; Igarashi, Y.; Benito, P.; Riesco, R.; Médigue, C.; Normand, P. Genome features of the endophytic Actinobacterium Micromonospora lupini strain Lupac 08: On the process of adaptation to an endophytic life style? PLoS ONE 2014, 9, e108522. [Google Scholar] [CrossRef] [Green Version]
- CGView Software. Available online: http://wishart.biology.ualberta.ca/cgview/ (accessed on 27 June 2020).
- Tatusov, R.L. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrés-Barrao, C.; Lafi, F.F.; Alam, I.; de Zélicourt, A.; Eida, A.A.; Bokhari, A.; Alzubaidy, H.; Bajic, V.B.; Hirt, H.; Saad, M.M. Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 2017, 8, 2023. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Gao, F.; Lin, Y. Evolutionary conservation analysis between the essential and nonessential genes in bacterial genomes. Sci. Rep. 2015, 5, 13210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef]
- Yu, Y.; Zhu, X.; Xu, H.; Zhang, X. Construction of an energy-conserving glycerol utilization pathways for improving anaerobic succinate production in Escherichia coli. Metab. Eng. 2019, 56, 181–189. [Google Scholar] [CrossRef]
- Murarka, A.; Dharmadi, Y.; Yazdani, S.S.; Gonzalez, R. Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl. Environ. Microbiol. 2008, 74, 1124–1135. [Google Scholar] [CrossRef] [Green Version]
- Thapa, L.P.; Lee, S.J.; Yang, X.; Lee, J.H.; Choi, H.S.; Park, C.; Kim, S.W. Improved bioethanol production from metabolic engineering of Enterobacter aerogenes ATCC 29007. Process Biochem. 2015, 50, 2051–2060. [Google Scholar] [CrossRef]
- Li, C.; Lesnik, K.L.; Liu, H. Microbial conversion of waste glycerol from biodiesel production into value-added products. Energies 2013, 6, 4739–4768. [Google Scholar] [CrossRef]
- Cheng, K.-K.; Wang, G.-Y.; Zeng, J.; Zhang, J.-A. Improved succinate production by metabolic engineering. Biomed. Res. Int. 2013, 2013, 538790. [Google Scholar] [CrossRef] [Green Version]
- Tajima, Y.; Kaida, K.; Hayakawa, A.; Fukui, K.; Nishio, Y.; Hashiguchi, K.; Fudou, R.; Matsui, K.; Usuda, Y.; Sode, K. Study of the role of anaerobic metabolism in succinate production by Enterobacter aerogenes. Appl. Microbiol. Biotechnol. 2014, 98, 7803–7813. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Laivenieks, M.; Schindler, B.D.; McKinlay, A.A.; Siddaramappa, S.; Challacombe, J.F.; Lowry, S.R.; Clum, A.; Lapidus, A.L.; Burkhart, K.B.; et al. A genomic perspective on the potential of Actinobacillus succinogenes for industrial succinate production. BMC Genomics 2010, 11, 680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valentini, M.; Storelli, N.; Lapouge, K. Identification of C 4-dicarboxylate transport systems in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2011, 193, 4307–4316. [Google Scholar] [CrossRef] [Green Version]
- Karinou, E.; Compton, E.L.R.; Morel, M.; Javelle, A. The Escherichia coli SLC26 homologue YchM (DauA) is a C4-dicarboxylic acid transporter. Mol. Microbiol. 2013, 87, 623–640. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, R.; Gross, R.; Simon, J.; Unden, G.; Kroger, A. Transport of C4-dicarboxylates Wolinella succinogenes. J. Bacteriol. 2000, 182, 5757–5764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janausch, I.G.; Zientz, E.; Tran, Q.H.; Kröger, A.; Unden, G. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta Bioenerg. 2002, 1553, 39–56. [Google Scholar] [CrossRef] [Green Version]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Liu, W.-Y.; Wong, C.-F.; Chung, K.M.-K.; Jiang, J.-W.; Leung, F.C.-C. Comparative genome analysis of Enterobacter cloacae. PLoS ONE 2013, 8, e74487. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, L.; Pei, J.; Pang, H.; Guo, Y.; Lin, L.; Huang, R. Whole genome sequencing reveals a novel CRISPR system in industrial Clostridium acetobutylicum. J. Ind. Microbiol. Biotechnol. 2014, 41, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, T.; Zhu, L.; Huang, H.; Jiang, L. Insights from the complete genome sequence of Clostridium tyrobutyricum provide a platform for biotechnological and industrial applications. J. Ind. Microbiol. Biotechnol. 2017, 44, 1245–1260. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [Green Version]
- Medina-Aparicio, L.; Dávila, S.; Rebollar-Flores, J.E.; Calva, E.; Hernández-Lucas, I. The CRISPR-Cas system in Enterobacteriaceae. Pathog. Dis. 2018, 76, fty002. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [Green Version]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K.; Van Bel, M. PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 2017, 33, 2946–2947. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Nakao, M.; Takagi, T. TogoWS: Integrated SOAP and REST APIs for interoperable bioinformatics Web services. Nucleic Acids Res. 2010, 38, W706–W711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapid Annotations Using Subsystems Technology (RAST). Available online: http://rast.nmpdr.org/ (accessed on 27 June 2020).
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef] [Green Version]


| Properties | |
|---|---|
| Strain | Enterobacter sp. LU1 |
| Sample source | Goat rumen |
| Sequencing type | Hybrid (short/long read sequencing) |
| Sequencing platforms | Illumina MiSeq/ONT MinION |
| Library type | 1 library with 400 bp insert/1D long read library |
| Fold average coverage | 98 ×/150 × |
| Assembly method | Unicycler v. 0.4.7 |
| Assembly level | Complete genome |
| Annotation method | Best-placed reference protein set; GeneMarkS-2+ |
| Annotation pipeline | NCBI Prokaryotic Genome Annotation Pipeline |
| BioProject | PRJNA562627 |
| BioSample | SAMN1264612 |
| Accession number | CP043438 |
| Genome Features | Enterobacter sp. LU1 | K. aerogenes LU2 | Enterobacteriaceae Average b | Enterobacteriaceae Range b |
|---|---|---|---|---|
| Genome size (bp) | 4,636,526 | 5,0626,51 | 5,133,067 | 4,641,652–5,452,368 |
| G+C (%) | 55.6 | 55 | 53.3 | 50.6–57.5 |
| No. of contigs | 1 | 1 | 2.5 | 1–10 |
| No. of plasmids | 0 | 0 | 1.5 | 0–9 |
| Total genes | 4425 | 4986 | 5045 | 4532–5523 |
| Total CDSs | 4311 | 4868 | – | – |
| Protein-coding genes | 4283 | 4741 | 4804 | 4242–5300 |
| Gene density (gene/kb) | 0.954 | 0.985 | 0.983 | 0.951–1.082 |
| 5S rRNAs | 9 | 8 | 24 | 19–25 |
| 16S rRNAs | 8 | 7 | ||
| 23S rRNAs | 8 | 7 | ||
| tRNAs | 84 | 86 | 84 | 76–88 |
| ncRNAs | 5 | 10 | – | – |
| Pseudogenes | 49 | 127 | 115 | 1–205 |
| CRISPR arrays | 2 | 0 | – | – |
| Prophages | 3 | 1 | – | – |
| Region | Region Length (kb) | Completeness | Total Proteins | Region Position | GC (%) |
|---|---|---|---|---|---|
| 1 | 70.3 | Intact | 67 | 1,116,715–1,187,045 | 52.96 |
| 2 | 20.9 | Intact | 29 | 2,196,837–2,217,799 | 52.75 |
| 3 | 49.8 | Intact | 50 | 3,424,122–3,473,981 | 50.02 |
| Region | Start | End | Spacers | Repeat Consensus/Cas Genes | Evidence Level |
|---|---|---|---|---|---|
| 1 | 1,143,757 | 1,143,850 | 1 | TGTCCAGCCGATGTCCAGCCAGTGTCCA | 1 |
| 2 | 1,566,815 | 1,567,745 | 15 | GTTCACTGCCGTGCAGGCAGCTTAGAAA | 4 |
| 3 | 1,571,432 | 1,572,180 | 12 | GTGCACTGCCGTACAGGCAGCTTAGAAA | 4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szczerba, H.; Dudziak, K.; Krawczyk, M.; Targoński, Z. A Genomic Perspective on the Potential of Wild-Type Rumen Bacterium Enterobacter sp. LU1 as an Industrial Platform for Bio-Based Succinate Production. Int. J. Mol. Sci. 2020, 21, 4835. https://doi.org/10.3390/ijms21144835
Szczerba H, Dudziak K, Krawczyk M, Targoński Z. A Genomic Perspective on the Potential of Wild-Type Rumen Bacterium Enterobacter sp. LU1 as an Industrial Platform for Bio-Based Succinate Production. International Journal of Molecular Sciences. 2020; 21(14):4835. https://doi.org/10.3390/ijms21144835
Chicago/Turabian StyleSzczerba, Hubert, Karolina Dudziak, Mariusz Krawczyk, and Zdzisław Targoński. 2020. "A Genomic Perspective on the Potential of Wild-Type Rumen Bacterium Enterobacter sp. LU1 as an Industrial Platform for Bio-Based Succinate Production" International Journal of Molecular Sciences 21, no. 14: 4835. https://doi.org/10.3390/ijms21144835
APA StyleSzczerba, H., Dudziak, K., Krawczyk, M., & Targoński, Z. (2020). A Genomic Perspective on the Potential of Wild-Type Rumen Bacterium Enterobacter sp. LU1 as an Industrial Platform for Bio-Based Succinate Production. International Journal of Molecular Sciences, 21(14), 4835. https://doi.org/10.3390/ijms21144835





