Cytological and Gene Profile Expression Analysis Reveals Modification in Metabolic Pathways and Catalytic Activities Induce Resistance in Botrytis cinerea Against Iprodione Isolated From Tomato
Abstract
1. Introduction
2. Material and Methods
2.1. Collection of Samples and Chemicals
2.2. Sensitivity of B. cinerea to Iprodione
2.3. Evaluation of B. cinerea Mutants Resistant to Iprodione
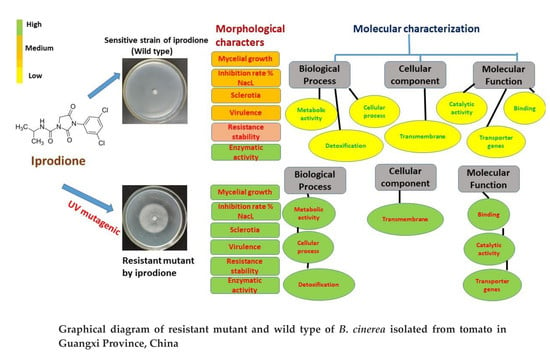
2.4. Characteristics of Iprodione Mutants and Sensitive Isolates
2.5. Enzymatic Activities of Iprodione Resistant Mutant and Its Corresponding Wild Type
2.6. Transcriptome Analysis of Wild and Mutant Strain
Differential Gene Expression
2.7. Quantitative Real Time PCR Analysis
2.8. DNA Extraction, Cloning and Sequence Analysis of the Tubulin Genes
2.9. Statistical Data Analysis
3. Results
3.1. Sensitivity of B. cinerea to Iprodione
3.2. In Vitro Iprodione-Induced B. cinerea Mutants
3.3. Morphology and Physiology of Mutants
3.3.1. Iprodione Resistant Mutant’s Mycelium Growth Rate and Sclerotia Formation
3.3.2. Cell Membrane Permeability Osmotic Sensitivity Pathogenicity and Enzymatic Activity of Mutants and its Wild Type
3.3.3. Cross Resistance
3.4. Transcriptomic Data Analysis of Iprodione-Resistant Mutant and Its Wild Type after Exposed to Iprodione In Vitro
3.4.1. Functional Annotation of Transcripts and Unigenes
3.4.2. Discovery of New Genes
3.4.3. Differential Genes Expression of Iprodione Resistant Mutant and its Wild Type After Exposure to Iprodione
3.5. qRT-PCR Amplification of Some Specific Genes
3.6. Detection of Mutations in Tubulin Genes in Iprodione Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; van Kan, J.A. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Yong, C.; Zhanquan, Z.; Boqiang, L.; Guozheng, Q.; Shiping, T. Pathogenic mechanisms and control strategies of Botrytis cinerea causing post-harvest decay in fruits and vegetables. Food Qual. Saf. 2018, 2, 111–119. [Google Scholar] [CrossRef]
- Amselem, J.; Cuomo, C.A.; Van Kan, J.A.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef] [PubMed]
- Tiedemann, B.; Have, A.; Hansen, M.E.; Tenberge, K. Infection Strategies of Botrytis cinerea and related necrotrophic pathogens. Fungal Pathol. 2013, 33–64. [Google Scholar]
- Maqsood, A.; Rehman, A.; Ahmad, I.; Nafees, M.; Ashraf, I.; Qureshi, R.; Jamil, M.; Rafay, M.; Hussain, T. Physiological attributes of fungi associated with stem end rot of mango (Mangifera indica L.) cultivars in postharvest fruit losses. Pak. J. Bot. 2014, 46, 1915–1920. [Google Scholar]
- Munhuweyi, K.; Lennox, C.L.; Meitz-Hopkins, J.C.; Caleb, O.J.; Opara, U.L. Major diseases of pomegranate (Punica granatum L.), their causes and management—A review. Sci. Horticult. 2016, 211, 126–139. [Google Scholar] [CrossRef]
- Tsitsigiannis, D.I.; Antoniou, P.P.; Tjamos, S.E.; Paplomatas, E.J. Major diseases of tomato, pepper and egg plant in green houses. Eur. J. Plant Sci. Biotechnol. 2008, 2, 106–124. [Google Scholar]
- Leroux, P.; Fritz, R.; Debieu, D.; Albertini, C.; Lanen, C.; Bach, J.; Gredt, M.; Chapeland, F. Mechanisms of resistance to fungicides in field strains of Botrytis cinerea. Pest Manag. Sci. 2002, 58, 876–888. [Google Scholar] [CrossRef]
- Xiang, F.; Han, Y.; Zeng, X.; Gu, Y.; Chen, F. The development situation and prospect of strawberry industry in western Hubei mountain districts. Hubei Agric. Sci. 2015, 54, 5930–5932. [Google Scholar]
- Rosslenbroich, H.J.; Stuebler, D. Botrytis cinerea—History of chemical control and novel fungicides for its management. Crop Prot. 2000, 19, 557–561. [Google Scholar] [CrossRef]
- Leroux, P. Chemical control of Botrytis and its resistance to chemical fungicides. In Botrytis: Biology, Pathology and Control; Springer: Berlin, Germany, 2007; pp. 195–222. [Google Scholar]
- Kroes, R.; Galli, C.; Munro, I.; Schilter, B.; Tran, L.A.; Walker, R.; Würtzen, G. Threshold of toxicological concern for chemical substances present in the diet: A practical tool for assessing the need for toxicity testing. Food Chem. Toxicol. 2000, 38, 255–312. [Google Scholar] [CrossRef]
- Wang, X.; Gong, C.; Zhao, Y.; Shen, L. Transcriptome and resistance-related genes analysis of Botrytis cinerea B05. 10 strain to different selective pressures of cyprodinil and fenhexamid. Front. Microbiol. 2018, 9, 2591. [Google Scholar] [CrossRef] [PubMed]
- Hirooka, T.; Ishii, H. Chemical control of plant diseases. J. Gen. Plant Pathol. 2013, 79, 390–401. [Google Scholar] [CrossRef]
- Fernandez-Cornejo, J.; Nehring, R.F.; Osteen, C.; Wechsler, S.; Martin, A.; Vialou, A. Pesticide use in US agriculture: 21 selected crops, 1960–2008. USDA-ERS Econ. Inf. Bull. 2014, 124. [Google Scholar] [CrossRef]
- Grabke, A.; Fernández-Ortuño, D.; Amiri, A.; Li, X.; Peres, N.A.; Smith, P.; Schnabel, G. Characterization of iprodione resistance in Botrytis cinerea from strawberry and blackberry. Phytopathology 2014, 104, 396–402. [Google Scholar] [CrossRef]
- Hamada, M.S.; Yin, Y.; Ma, Z. Sensitivity to iprodione, difenoconazole and fludioxonil of Rhizoctonia cerealis isolates collected from wheat in China. Crop Prot. 2011, 30, 1028–1033. [Google Scholar] [CrossRef]
- Ma, Z.; Luo, Y.; Michailides, T.J. Resistance of Botryosphaeria dothidea from pistachio to iprodione. Plant Dis. 2001, 85, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Michailides, T.J. Characterization of iprodione-resistant Alternaria isolates from pistachio in California. Pestic. Biochem. Physiol. 2004, 80, 75–84. [Google Scholar] [CrossRef]
- Adnan, M.; Hamada, M.; Li, G.; Luo, C. Detection and molecular characterization of resistance to the dicarboximide and benzamide fungicides in Botrytis cinerea from tomato in Hubei Province, China. Plant Dis. 2018, 102, 1299–1306. [Google Scholar] [CrossRef]
- Adnan, M.; Hamada, M.S.; Hahn, M.; Li, G.Q.; Luo, C.X. Fungicide resistance of Botrytis cinerea from strawberry to procymidone and zoxamide in Hubei, China. Phytopathol. Res. 2019, 1, 17. [Google Scholar] [CrossRef]
- Sun, H.Y.; Wang, H.C.; Chen, Y.; Li, H.X.; Chen, C.J.; Zhou, M.G. Multiple resistance of Botrytis cinerea from vegetable crops to carbendazim, diethofencarb, procymidone, and pyrimethanil in China. Plant Dis. 2010, 94, 551–556. [Google Scholar] [CrossRef]
- Ma, Z.; Yan, L.; Luo, Y.; Michailides, T.J. Sequence variation in the two-component histidine kinase gene of Botrytis cinerea associated with resistance to dicarboximide fungicides. Pestic. Biochem. Physiol. 2007, 88, 300–306. [Google Scholar] [CrossRef]
- Marschall, R.; Schumacher, J.; Siegmund, U.; Tudzynski, P. Chasing stress signals–exposure to extracellular stimuli differentially affects the redox state of cell compartments in the wild type and signaling mutants of Botrytis cinerea. Fungal Genet. Biol. 2016, 90, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Avenot, H.F.; Quattrini, J.; Puckett, R.; Michailides, T.J. Different levels of resistance to cyprodinil and iprodione and lack of fludioxonil resistance in Botrytis cinerea isolates collected from pistachio, grape, and pomegranate fields in California. Crop Prot. 2018, 112, 274–281. [Google Scholar] [CrossRef]
- Panebianco, A.; Castello, I.; Cirvilleri, G.; Perrone, G.; Epifani, F.; Ferrara, M.; Polizzi, G.; Walters, D.R.; Vitale, A. Detection of Botrytis cinerea field isolates with multiple fungicide resistance from table grape in Sicily. Crop Prot. 2015, 77, 65–73. [Google Scholar] [CrossRef]
- Marie, C.; White, T.C. Genetic basis of antifungal drug resistance. Curr. Fungal Infect. Rep. 2009, 3, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, A.; Wu, H.; Kamran, M.; Altaf, H.; Mustafa, A.; Ahmar, S.; Hong, N.T.T.; Tariq, K.; He, Q.; Chen, J.T. Variations in growth, physiology, and antioxidative defense responses of two tomato (Solanum lycopersicum L.) cultivars after co-infection of Fusarium oxysporum and Meloidogyne incognita. Agronomy 2020, 10, 159. [Google Scholar] [CrossRef]
- Fernández-Ortuño, D.; Pérez-García, A.; Chamorro, M.; de la Peña, E.; de Vicente, A.; Torés, J.A. Resistance to the SDHI fungicides boscalid, fluopyram, fluxapyroxad, and penthiopyrad in Botrytis cinerea from commercial strawberry fields in Spain. Plant Dis. 2017, 101, 1306–1313. [Google Scholar] [CrossRef]
- Luo, B.; Young, V.G., Jr.; Gladfelter, W.L. Si-C bond cleavage in the reaction of gallium chloride with lithium bis (trimethylsilyl) amide and thermolysis of base adducts of dichloro (trimethylsilyl) amido gallium compounds. J. Organomet. Chem. 2002, 649, 268–275. [Google Scholar] [CrossRef]
- Fan, H.; Yu, G.; Liu, Y.; Zhang, X.; Liu, J.; Zhang, Y.; Rollins, J.A.; Sun, F.; Pan, H. An atypical forkhead-containing transcription factor SsFKH1 is involved in sclerotial formation and is essential for pathogenicity in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2017, 18, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Cao, Y.; Guo, Z.F.; Chen, M.; Chen, X.; Guo, Z. Menaquinone biosynthesis in Escherichia coli: Identification of 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate as a novel intermediate and re-evaluation of MenD activity. Biochemistry 2007, 46, 10979–10989. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, C.; Grintzalis, K.; Zervoudakis, G.; Papapostolou, I. Mechanism of coomassie brilliant blue G-250 binding to proteins: A hydrophobic assay for nanogram quantities of proteins. Anal. Bioanal. Chem. 2008, 391, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xu, Y.; Zhang, R.; Wang, X.; Yang, C. Comprehensive transcriptome profiling of soybean leaves in response to simulated acid rain. Ecotoxicol. Environ. Saf. 2018, 158, 18–27. [Google Scholar] [CrossRef]
- Feng, J.; Meyer, C.A.; Wang, Q.; Liu, J.S.; Shirley Liu, X.; Zhang, Y. GFOLD: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012, 28, 2782–2788. [Google Scholar] [CrossRef] [PubMed]
- Cai, N.; Liu, C.; Feng, Z.; Li, X.; Qi, Z.; Ji, M.; Qin, P.; Ahmed, W.; Cui, Z. Design, synthesis, and SAR of novel 2-glycinamide cyclohexyl sulfonamide derivatives against Botrytis cinerea. Molecules 2018, 23, 740. [Google Scholar] [CrossRef]
- Leroch, M.; Kretschmer, M.; Hahn, M. Fungicide resistance phenotypes of Botrytis cinerea isolates from commercial vineyards in South West Germany. J. Phytopathol. 2011, 159, 63–65. [Google Scholar] [CrossRef]
- Beckerman, J.L.; Sundin, G.W.; Rosenberger, D.A. Do some IPM concepts contribute to the development of fungicide resistance? Lessons learned from the apple scab pathosystem in the United States. Pest Manag. Sci. 2015, 71, 331–342. [Google Scholar] [CrossRef]
- Xu, S.; Wang, J.; Wang, H.; Bao, Y.; Li, Y.; Govindaraju, M.; Yao, W.; Chen, B.; Zhang, M. Molecular characterization of carbendazim resistance of Fusarium species complex that causes sugarcane pokkah boeng disease. BMC Genom. 2019, 20, 115. [Google Scholar] [CrossRef]
- Cai, M.; Lin, D.; Chen, L.; Bi, Y.; Xiao, L.; Liu, X.L. M233I mutation in the β-tubulin of Botrytis cinerea confers resistance to zoxamide. Sci. Rep. 2015, 5, 16881. [Google Scholar] [CrossRef]
- Veloukas, T.; Kalogeropoulou, P.; Markoglou, A.; Karaoglanidis, G. Fitness and competitive ability of Botrytis cinerea field isolates with dual resistance to SDHI and QoI fungicides, associated with several sdh B and the cyt b G143A mutations. Phytopathology 2014, 104, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.H.; Jiao, X.L.; Hao, J.J.; Chen, A.J.; Gao, W.W. Characterization of resistance to multiple fungicides in Botrytis cinerea populations from Asian ginseng in northeastern China. Eur. J. Plant Pathol. 2016, 144, 467–476. [Google Scholar] [CrossRef]
- Guo, L.N.; Xiao, M.; Cao, B.; Qu, F.; Zhan, Y.L.; Hu, Y.J.; Wang, X.R.; Liang, G.W.; Gu, H.T.; Qi, J. Epidemiology and antifungal susceptibilities of yeast isolates causing invasive infections across urban Beijing, China. Future Microbiol. 2017, 12, 1075–1086. [Google Scholar] [CrossRef]
- Chen, J.Y.; Xiao, H.L.; Gui, Y.J.; Zhang, D.D.; Li, L.; Bao, Y.M.; Dai, X.F. Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front. Microbiol. 2016, 7, 1709. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, S.; Qin, T.; Li, N.; Niu, Y.; Li, D.; Yuan, Y.; Geng, H.; Xiong, L.; Liu, D. Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms. BMC Genom. 2015, 16, 855. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, J. Role of phospholipases in fungal fitness, pathogenicity, and drug development–lessons from Cryptococcus neoformans. Front. Microbiol. 2010, 1, 125. [Google Scholar] [CrossRef]
- Gilbert, N.M.; Donlin, M.J.; Gerik, K.J.; Specht, C.A.; Djordjevic, J.T.; Wilson, C.F.; Sorrell, T.C.; Lodge, J.K. KRE genes are required for β-1, 6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol. Microbiol. 2010, 76, 517–534. [Google Scholar] [CrossRef]
- Luczaj, J.; Leavitt, S.; Csank, A.; Panyushkina, I.; Wright, W. Comment on “Non-Mineralized Fossil Wood” by George E. Mustoe (Geosciences, 2018). Geosciences 2018, 8, 462. [Google Scholar] [CrossRef]
- Zerillo, M.M.; Adhikari, B.N.; Hamilton, J.P.; Buell, C.R.; Lévesque, C.A.; Tisserat, N. Carbohydrate-active enzymes in Pythium and their role in plant cell wall and storage polysaccharide degradation. PLoS ONE 2013, 8, e72572. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, X.; Dong, Y.; Qiu, D. The Botrytis cinerea xylanase BcXyl1 modulates plant immunity. Front. Microbiol. 2018, 9, 2535. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Han, C.; Chen, J.; Li, H.; He, K.; Liu, A.; Li, D. Fungal cellulase is an elicitor but its enzymatic activity is not required for its elicitor activity. Mol. Plant Pathol. 2015, 16, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Noda, J.; Brito, N.; González, C. The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol. 2010, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Brito, N.; Espino, J.J.; González, C. The endo-β-1, 4-xylanase Xyn11A is required for virulence in Botrytis cinerea. Mol. Plant-Microbe Interact. 2006, 19, 25–32. [Google Scholar] [CrossRef]
- Zhang, L.; Van Kan, J.A. Botrytis cinerea mutants deficient in D-galacturonic acid catabolism have a perturbed virulence on Nicotiana benthamiana and Arabidopsis, but not on tomato. Mol. Plant Pathol. 2013, 14, 19–29. [Google Scholar] [CrossRef]
- Črešnar, B.; Petrič, Š. Cytochrome P450 enzymes in the fungal kingdom. BBA Proteins Proteom. 2011, 1814, 29–35. [Google Scholar] [CrossRef]
- Durairaj, P.; Hur, J.S.; Yun, H. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb. Cell Fact. 2016, 15, 125. [Google Scholar] [CrossRef]
- Muñoz, C.; Talquenca, S.G.; Volpe, M.L. Tetra primer ARMS-PCR for identification of SNP in β-tubulin of Botrytis cinerea, responsible of resistance to benzimidazole. J. Microbiol. Methods 2009, 78, 245–246. [Google Scholar] [CrossRef]
- Sameer, W.M. Control of the root rot diseases of wheat using fungicides alone and in combinations with selenium. Al-Azhar Bull. Sci. 2018, 29, 27–37. [Google Scholar]
- Lata Panwar, S.; Krishnamurthy, S.; Gupta, V.; Alarco, A.M.; Raymond, M.; Sanglard, D.; Prasad, R. CaALK8, an alkane assimilating cytochrome P450, confers multidrug resistance when expressed in a hypersensitive strain of Candida albicans. Yeast 2001, 18, 1117–1129. [Google Scholar] [CrossRef]
- Alias, Z. The role of glutathione transferases in the development of insecticide resistance. Insectic. Resist. 2016, 315. [Google Scholar] [CrossRef][Green Version]
- Kolawole, A.O.; Ajele, J.O.; Sirdeshmuhk, R. Studies on glutathione transferase of cowpea storage bruchid, Callosobrochus maculatus F. Pestic. Biochem. Physiol. 2011, 100, 212–220. [Google Scholar] [CrossRef]
- Holmes, A.R.; Keniya, M.V.; Ivnitski-Steele, I.; Monk, B.C.; Lamping, E.; Sklar, L.A.; Cannon, R.D. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrob. Agents Chemother. 2012, 56, 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; Zwiers, L.H.; De Waard, M.A. Secretion of natural and synthetic toxic compounds from filamentous fungi by membrane transporters of the ATP-binding cassette and major facilitator superfamily. Eur. J. Plant Pathol. 2002, 108, 719–734. [Google Scholar] [CrossRef]
- Mernke, D.; Dahm, S.; Walker, A.S.; Lalève, A.; Fillinger, S.; Leroch, M.; Hahn, M. Two promoter rearrangements in a drug efflux transporter gene are responsible for the appearance and spread of multidrug resistance phenotype MDR2 in Botrytis cinerea isolates in French and German vineyards. Phytopathology 2011, 101, 1176–1183. [Google Scholar] [CrossRef]
- De Waard, M.A.; Andrade, A.C.; Hayashi, K.; Schoonbeek, H.j.; Stergiopoulos, I.; Zwiers, L.H. Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 2006, 62, 195–207. [Google Scholar] [CrossRef]








| Strain | Raw Reads | Clean Reads | Clean Bases | Mapped Reads | Q 20 Avg (%) | GC Avg (%) |
|---|---|---|---|---|---|---|
| M0 | 47,684,718 | 47,218,908 | 7,057,947,521 | 44,704,018 (95.8%) | ||
| M-ipro | 45,648,745 | 46,083,059 | 6,885,980,894 | 44,251,477 (96%) | ||
| W0 | 47,109,858 | 46,628,979 | 6,970,197,563 | 44,597,601 (95.62%) | 97.76 | 47.23 |
| W-ipro | 46,618,817 | 46,096,018 | 6,885,797,287 | 44,052,162 (96.22%) | ||
| Total | 187,062,138 | 186,026,964 |
| Strain | All DEG | DEG Upregulated | DEG Downregulated |
|---|---|---|---|
| M-ipro vs. W-ipro | 1897 | 886 | 1011 |
| M0 vs. W0 | 1707 | 890 | 817 |
| M-ipro vs. M0 | 281 | 166 | 115 |
| W-ipro vs. W0 | 99 | 85 | 14 |
| Gene ID | Log2 Fold Change Value | Function Annotation | |||
|---|---|---|---|---|---|
| M-ipro vs. W-ipro | M0 vs. W0 | M-ipro vs. M0 | W-ipro vs. W0 | ||
| BCIN_02g08890 | 9.104 | 8.746 | 0.565 | 0 | Integral cellular component domain |
| BCIN_13g03170 | 8.222 | 5.529 | 0.378 | −0.231 | Glycerophospholipid metabolism |
| BCIN_06g07150 | 6.213 | 5.503 | 0.12 | −0.599 | Energy production and conservation |
| BCIN_09g01190 | 4.878470612 | 6.006 | −0.511 | 1.2044 | Zinc finger proteins |
| BCIN_13g04220 | 8.618 | −7.08119 | 8.576 | −21.308 | Chaperones proteins |
| BCIN_08g06520 | 7.317 | 4.154955 | 1.07079 | −4.216 | Oxygenase super family |
| Bcpg2 | 4.978 | −0.967 | 2.896255 | −2.978 | Glycosyl hydrolase family |
| Bcoah | 4.878 | 6.600 | −0.511 | 1.204 | Isocitrtae lyase family |
| BcatrD | 2.529 | 1.66 | 0.266 | −0.249 | ABC transporter proteins |
| Bclga1 | 2.266 | 0.745 | 0.865 | −0.705 | Glutathione S transferase proteins |
| BCIN_12g0243 | 2.597 | 3.064 | −0.407 | −0.046 | Catalyze aminotransferase acid |
| Bcrds2 | 1.229 | 1.092 | −0.062 | −0.192 | Drug sensitivity proteins |
| Bcltf1 | 0.531 | 1.292 | −1.108 | −0.3241 | Sexual development transcription factor NsdD |
| BCIN_06g02680 | 2.00 | 0.835 | 1.292 | −1.108 | Phenylalanine aminomutase enzyme |
| BCIN_08g02390 | 2.837039 | 2.628415 | 0.436 | 0.223 | Heat shock binding proteins |
| BCIN_02g01500 | 1.929 | 0.199 | 0.318 | 1.779 | Carbohydrate transport and metabolism |
| BCIN_04g01200 | 3.721 | 2.744 | 1.035 | 0.061 | Steroid biosynthesis proteins |
| BCIN_08g01540 | 2.119 | 1.637 | 0.0715 | −0.411 | Steroid biosynthesis proteins |
| BCIN_03g03480 | 4.555 | 4.296 | 0.285 | 0.019 | Xylanase proteins |
| Bcpgx1 | 2.536 | 1.212 | 0.496 | −0.823 | Polygalacturose proteins |
| BCIN_02g04800 | 4.861 | 4.796 | 0.611 | 0.536 | Amino acid metabolism proteins |
| BCIN_01g01440 | 2.811 | −1.449 | 2.354 | −1.871 | Dioxygenase TDA family |
| BCIN_12g04510 | 1.087 | 0.736 | 0.193 | −0.165 | Histidine kinase activity proteins |
| BCIN_01g03510 | 4.531 | 3.321 | −2.06396 | 0.7321 | Cytochrome P450 |
| BCIN_02g08880 | 9.495 | 8.718 | 0.024 | −0.752 | Unknown function hypothetical proteins |
| BCIN_02g08890 | 9.104 | 8.745 | 0.565 | 0 | Unknown function hypothetical proteins |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maqsood, A.; Wu, C.; Ahmar, S.; Wu, H. Cytological and Gene Profile Expression Analysis Reveals Modification in Metabolic Pathways and Catalytic Activities Induce Resistance in Botrytis cinerea Against Iprodione Isolated From Tomato. Int. J. Mol. Sci. 2020, 21, 4865. https://doi.org/10.3390/ijms21144865
Maqsood A, Wu C, Ahmar S, Wu H. Cytological and Gene Profile Expression Analysis Reveals Modification in Metabolic Pathways and Catalytic Activities Induce Resistance in Botrytis cinerea Against Iprodione Isolated From Tomato. International Journal of Molecular Sciences. 2020; 21(14):4865. https://doi.org/10.3390/ijms21144865
Chicago/Turabian StyleMaqsood, Ambreen, Chaorong Wu, Sunny Ahmar, and Haiyan Wu. 2020. "Cytological and Gene Profile Expression Analysis Reveals Modification in Metabolic Pathways and Catalytic Activities Induce Resistance in Botrytis cinerea Against Iprodione Isolated From Tomato" International Journal of Molecular Sciences 21, no. 14: 4865. https://doi.org/10.3390/ijms21144865
APA StyleMaqsood, A., Wu, C., Ahmar, S., & Wu, H. (2020). Cytological and Gene Profile Expression Analysis Reveals Modification in Metabolic Pathways and Catalytic Activities Induce Resistance in Botrytis cinerea Against Iprodione Isolated From Tomato. International Journal of Molecular Sciences, 21(14), 4865. https://doi.org/10.3390/ijms21144865