Abstract
TRANSPARENT TESTA GLABRA1 (TTG1) is a WD40 repeat protein. The phenotypes caused by loss-of-function of TTG1 were observed about half a century ago, but the TTG1 gene was identified only about twenty years ago. Since then, TTG1 has been found to be a plant-specific regulator with multiple roles and multiple functional mechanisms. TTG1 is involved in the regulation of cell fate determination, secondary metabolisms, accumulation of seed storage reserves, plant responses to biotic and abiotic stresses, and flowering time in plants. In some processes, TTG1 may directly or indirectly regulate the expression of downstream target genes via forming transcription activator complexes with R2R3 MYB and bHLH transcription factors. Whereas in other processes, TTG1 may function alone or interact with other proteins to regulate downstream target genes. On the other hand, the studies on the regulation of TTG1 are very limited. So far, only the B3-domain family transcription factor FUSCA3 (FUS3) has been found to regulate the expression of TTG1, phosphorylation of TTG1 affects its interaction with bHLH transcription factor TT2, and TTG1 proteins can be targeted for degradation by the 26S proteasome. Here, we provide an overview of TTG1, including the identification of TTG1, the functions of TTG1, the possible function mechanisms of TTG1, and the regulation of TTG1. We also proposed potential research directions that may shed new light on the regulation and functional mechanisms of TTG1 in plants.
1. Introduction
The phenotypes of Arabidopsis ttg (transparent testa, glabra) mutants were first reported about 50 years ago [1,2]. It was then proposed that the TRANSPARENT TESTA GLABRA (TTG) locus has pleiotropic roles in the regulation of trichome initiation, anthocyanin biosynthesis, and seed coat mucilage biosynthesis in Arabidopsis [3]. The TTG locus was mapped to chromosome 5 about 40 years ago [4]. About 30 years ago, it was proposed that TTG1 might encode an MYC transcription factor [5]. However, TTG1 gene was finally cloned about 20 years ago and was found to encode a WD40 repeat protein [6], a member of the WD40 repeat protein family, one of the largest protein families widely distributed in all eukaryotic cells. So far, more than 200 WD40 repeat proteins have been predicted in plants [7,8], and they are involved in the regulation of multiple processes in plants, such as signaling transduction, cell cycling, chromatin modification, transcriptional regulation, and RNA processing [7,9].
Accumulated evidence shows that, in addition to trichome initiation, anthocyanin biosynthesis, and seed coat mucilage production, TTG1 is also involved in the regulation of root hair formation [10], stomata development [11], seed development, post-embryonic development [3,6,12,13,14,15,16,17], proanthocyanidins (PAs) biosynthesis [3,13,17,18,19,20], accumulation of seed storage reserves such as fatty acids and proteins during the seed maturation process [15,16], as well as biotic and abiotic stress responses in plants [21,22,23].
In recent years, remarkable achievements have been made in understanding the roles and function mechanisms of TTG1 in regulating epidermal cell fate determination and secondary metabolism in Arabidopsis and other plants [10,11,12,13,14,15,16,17,18,19,20]. It has been proposed that TTG1 can interact with different R2R3 MYB and basic helix-loop-helix (bHLH) transcription factors to form multiple MYB-bHLH-WD40 (MBW) activator complexes to regulate the expression of downstream genes, thereby regulating cell fate determination including trichome initiation and root hair formation, and secondary metabolism including flavonoid biosynthesis and seed coat mucilage production [24,25,26,27,28,29]. For example, TTG1 regulates trichome initiation by forming MBW activator complexes with the R2R3 MYB transcription factor GLABRA1 (GL1), and the bHLH transcription factor GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3) to activate the expression of GLABRA2 (GL2) [25,30]. The same MBW complexes can also activate the expression of some R3 MYB genes. Whereas R3 MYBs can compete with GL1 for the binding of GL3 or EGL3, thus preventing the formation of the MBW activator complexes, resulting in inhibition of trichome initiation [25,26,27,31]. TTG1 regulates root hair formation by forming MBW complexes with the R2R3 MYB transcription factor WERWOLF (WER) and the bHLH transcription factor GL3 or EGL3 to activate the expression of GL2 [27,32,33]. TTG1 regulates flavonoid biosynthesis by forming MBW complexes with the R2R3 MYB transcription factors PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1), PAP2, MYB113, MYB114 or TRANSPARENT TESTA 2 (TT2) and the bHLH transcription factors TT8, GL3 or EGL3 to regulate the expression of the late biosynthesis genes in the flavonoid biosynthesis pathway [24,26,28,34,35,36,37,38].
Considering that some previous reviews have covered some aspects of the functions and function mechanisms of TTG1, for example, in the regulation of flavonoid biosynthesis [39,40] and evolution of cellular diversity as a component of MBW complexes [41]. We provide here an overview of TTG1, including the summary of its functions and possible functional mechanisms in regulating epidermal cell fate determination and secondary metabolism with an emphasis on recent progress, but with a specific focus on the identification history of TTG1, its functions that may not require the formation of the MBW complexes, the functions of TTG1 orthologs in other plants, and perspectives on potential future research directions.
2. Identification of TTG1
The Arabidopsis mutants showing a phenotype of yellow seeds and the absence of leaf trichomes were first reported in 1971 [1]. In 1978, the name ttg was assigned to these mutants [2]. In 1981, Koornneef found that the ttg mutants showed a pleiotropic phenotype including glabrous leaves, a transparent testa seed coat, reduced anthocyanin accumulation, and seed coat mucilage production [3]. In 1980, the gene TTG1 was found to be located on chromosome 5 and is closely linked to ms [42]. In 1983, Koornneef et al. mapped the TTG1 locus between the molecular marker MS1 and GA3 [4]. In 1999, Walkers et al. finally cloned TTG1 gene by using positional cloning [6]. They generated two sets of recombinants between ttg1-1 and the flanking genetic markers MS1 and GA3, and performed RFLP (restriction fragment length polymorphisms) analysis by using a variety of probes. A total of 439 recombinants were subjected to large-scale mapping, and EG20H2, a clone from an Arabidopsis genomic yeast artificial chromosome (YAC) library [43], was found to cover the TTG1 locus. Higher-resolution mapping of the region covered by EG20H2 showed that, when probed with g4556, a clone from an Arabidopsis genomic cosmid library [44], only a single recombinant could separate from ttg1-1, which suggested that the g4556 was very close to the TTG1 locus. The g4556 and YAC EG20H2 clones were then used to screen an Arabidopsis genomic lambda (λ) library [45], and several λ clones were obtained. DNA probing with the obtained λ clones showed that the deletion of the genomic fragment in λ8 could account for the phenotype of ttg1-13, a fast-neutron bombardment ttg mutant [46]. Complementation of the ttg1-1 mutant phenotype with entire and partial genomic fragments in λ8 confirmed that it contained the TTG1 locus, and sequence analysis showed that TTG1 encodes a WD40 repeat protein with high amino acid sequence identity and similarity to AN1 from petunia [6,47].
TTG1 has four WD40 repeat motifs [4], therefore, it is a member of the WD40 protein family. WD40 proteins are one of the largest regulatory protein families conserved in all eukaryotic cells [9], and the only conserved feature of WD40 proteins is the presence of the WD40 motifs, which typically contain several copies of WD40 repeats, with each repeat containing 44–60 amino acid residues, with a glycine-histidine (GH) dipeptide at the N-terminal and Trp-Asp (WD) residues at the C-terminal [9]. WD40 proteins are able to provide a platform for interactions with other proteins and are involved in the regulation of growth and development, as well as some cellular functions such as signal transduction and transcriptional regulation [7,8,9].
After being summarized previously [39], a few more Arabidopsis ttg1 mutants were identified, and a complete list of Arabidopsis ttg1 mutants identified so far are presented in Table 1. By characterizing these mutants, it has been found that TTG1 regulates multiple aspects of plant growth and development, secondary metabolism, flowering time, and plant responses to environmental stresses.

Table 1.
The Arabidopsis ttg1 mutants identified so far.
3. Functions and Function Mechanisms of TTG1
TTG1 is a plant-specific WD40 protein widespread in angiosperms but not in gymnosperms and early evolved plants [58]. TTG1 has similar functions in different plant species. At least some of its functions including the regulation of trichome initiation, root hair formation, flavonoids biosynthesis, and seed coat mucilage production are achieved via interacting with specific R2R3 MYB and bHLH transcription factors to form different MBW complexes (Figure 1). However, it should be noted that there is a study showing that TTG1 and GL1 compete for binding of GL3 to form TTG1-GL3 or GL1-GL3 dimers, rather than simultaneously bind to GL3 to form TTG1-GL3-GL1 trimeric complex, the interaction of GL3 with GL1 and TTG1 can be suppressed by additional TTG1 and GL1 protein, and the competition/suppression occurs in a dosage-dependent manner [59].
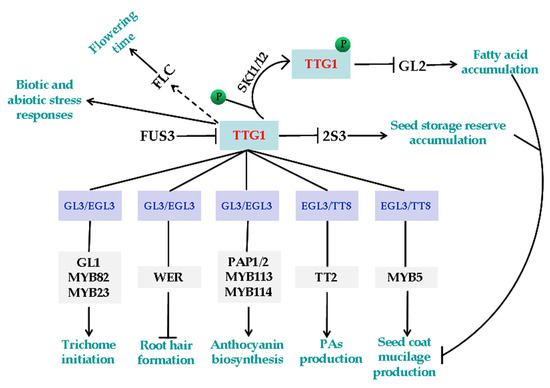
Figure 1.
The functions of TTG1. TTG1 regulates cell fate determination and secondary metabolism by forming MBW complexes with specific R2R3 MYB and bHLH transcription factors. The TTG1-GL3/EGL3-GL1/MYB23/MYB82 complexes regulate trichome initiation [25,30,60,61], the TTG1-GL3/EGL3-WER complexes regulate root hair formation [27,33], the TTG1-GL3/EGL3-PAP1/2/MYB113/114 complexes regulate anthocyanin biosynthesis [24,34,36], the TTG1-EGL3/TT8-TT2 complexes regulate proanthocyanidins (PAs) biosynthesis [24,28,34], and the TTG1-EGL3/TT8-MYB5 complexes regulate seed coat mucilage production [24]. TTG1 compromises the accumulation of seed storage reserves through inhibiting 2S3, and FUS3 can directly suppress the expression of TTG1. SK11/12 can phosphorylate TTG1, therefore inhibit GL2 expression, and affect fatty acid accumulation. TTG1 is also involved in regulating flowering, as well as biotic and abiotic stress responses.
3.1. Regulation of Epidermal Cell Fate Determination
In Arabidopsis, TTG1 regulates the specification of several different types of epidermal cells including trichomes, root hairs, and stomata. The ttg1-1 mutants do not produce trichomes [3,6], suggesting that TTG1 is involved in the regulation of trichome initiation. Further studies indicate that TTG1 regulates trichome initiation via associating with the R2R3 MYB transcription factor GL1 [62] and the bHLH transcription factors GL3 or EGL3 [34,63] to form MBW activator complexes (Figure 1), the complexes activate the expression of the downstream gene GL2 to promote trichome initiation and development [32]. The TTG1-GL3/EGL3-GL1 complexes can also regulate the expression of several single R3 MYB genes, whereas R3 MYBs can move from the trichome cells to the neighboring cells, where they compete with GL1 for binding of GL3 or EGL3 to prevent the formation of the MBW complexes, resulting in inhibition of trichome initiation [25,26,27,31,64]. TTG1 can also interact with the miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors SPL4 or SPL5, which affects the transcriptional activity of the MBW complexes and leads to inhibition of trichome initiation [65].
Different from trichome initiation, root hair formation follows a position-dependent manner. In wild-type Arabidopsis, hair cells (H cells; trichoblasts) can form root hairs, whereas hairless cells (N cells; atrichoblasts) cannot [66,67]. In the ttg1-1 mutants, the number of root hairs is increased due to ectopic root hair formation in N cells [10]. However, a decrease in root hair formation was observed in ttg1-23 and ttg1-24, two mutants with a single amino acid substitution in the TTG1 protein [51] (Table 1). Similar to the regulation of trichome initiation, TTG1 regulates root hair formation via forming MBW complexes, however, GL1 is replaced by another R2R3 MYB transcription factor, WER [68]. The TTG1-GL3/EGL3-WER complexes induce the expression of GL2 and some R3 MYBs. Different from their inhibitory function in regulating trichome initiation, R3 MYBs can promote root hair formation. R3 MYBs can move from N cell to H cell, where they compete with WER for binding of GL3 or EGL3, therefore, preventing the formation of the MBW complexes, resulting in the promotion of root hair formation [60,69,70,71]. In the ttg1-23 and ttg1-24 mutants, the single amino acid substitution in TTG1 abolished its interaction with the bHLH transcription factor GL3/EGL3, therefore, differentially affected the expression of the MBW complex target genes in the mutants. These results demonstrate that TTG1 may be able to balance the target genes expression to enable the plant to produce a proper amount of different types of epidermal cells during the root development [51].
Unlike trichomes and root hairs, stomata are developed from precursor cells. The ttg1-1 mutants produced more ectopic stomata on the hypocotyls, but stomata distribution in cotyledons and leaves remained unchanged [11]. As the gl2 mutants also produced more ectopic stomata on the hypocotyls [11], TTG1 might regulate stomata development via regulating the expression of GL2. Previous studies showed that the TTG1 may function epistatic to GL2 in regulating root hair formation [10,32]. This could also be the case for TTG1 in regulating stomata development [11].
Available evidence shows that TTG1 orthologs in several other plants are also able to regulate epidermal cell fate determination. For example, transgenic Arabidopsis plants ectopically expressing the Limonium bicolor TTG1 (LbTTG1) produced more trichomes and fewer root hairs [21]; expression of TTG1 ortholog genes from other plant species including the Gossypium hirsutum TTG1 (GhTTG1) and GhTTG3, the Malus domestica TTG1 (MdTTG1), and the maize PAC restored trichome defect phenotype in the ttg1 mutants [72,73,74]; the Arabis alpina ttg1 (aattg1) mutants showed a glabrous phenotype and produced more root hairs [75]; expression of the Cucumis sativus TTG1 (CsTTG1) in cucumber promoted fruit trichome and spine formation, whereas silencing of CsTTG1 inhibited fruit spine initiation. However, TTG1 in other plants may use different mechanisms to regulate epidermal cell fate determination, as molecular and genetic analysis show that even though CsTTG1 acts in parallel to CsGL1 in regulating fruit trichome initiation, CsTTG1 can directly interact with CsGL1, whereas in Arabidopsis, TTG1 directly interacts GL3 but not GL1 [76].
3.2. Regulation of Flavonoid Biosynthesis
Flavonoids including proanthocyanidins (PAs) and anthocyanins are secondary metabolites in higher plants [19,34,35]. The ttg1-1 mutants do not accumulate anthocyanins and can produce yellow seeds [3,6,18,24], suggesting that TTG1 is involved in the regulation of PA and anthocyanin biosynthesis. Similar to the regulation of cell fate determination, TTG1 in Arabidopsis regulates flavonoid biosynthesis via forming MBW complexes. TTG1 can form MBW complexes with the R2R3 MYB transcription factors PAP1, PAP2, MYB113, MYB114 or TT2, and the bHLH transcription factors GL3, EGL3 or TT8 [24,26,28,34,35,36,37,38,77,78]. It has been shown that different MBW complexes regulate the biosynthesis of different flavonoids. The TTG1-TT8/GL3-PAP1/PAP2/MYB113/MYB114 complexes regulate anthocyanin biosynthesis by activating the expression of late biosynthetic genes including DIHYDROFLAVONOL 4-REDUCTASE (DFR), ANTHOCYANIDIN SYNTHASE (ANS) and UDP-GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE (UF3GT) in the anthocyanin biosynthetic pathway [36,78,79]. Whereas the TTG1-GL3/TT8–TT2 complexes regulate PA accumulation by activating the expression of DFR, ANS, BANYULS (BAN), TT19, and TT12 [28,38,79]. Whole genome-wide identification studies of the TTG1-dependent MBW complex target genes found that the TTG1-dependent MBW complexes are able to bind directly to the promoters of TTG2, TT8, FLAVANONE 3-HYDROXYLASE (F3′H), DFR, and ANS genes to regulate their expression [80].
The TTG1 orthologs in other plant species have also been found to regulate flavonoid biosynthesis. For example, ectopic expression of the Freesia hybrida TTG1 (FhTTG1) in the ttg1-1 mutants partially restored the anthocyanin biosynthesis deficient phenotype [81]. Ectopic expression of CsWD40, a TTG1 homolog gene in Camellia sinensis and tobacco increased anthocyanin accumulation [82]. Salvia miltiorrhiza TTG1 (SmTTG1) is also involved in the regulation of anthocyanin accumulation [83]. It has been shown that FhTTG1 is able to interact with FhTT8L and FhGL3L [81], and CsWD40 is able to interact with CsGL3/CsTT8 and CsAN2/CsMYB5e [82], suggesting that TTG1 in these plants may use similar mechanisms as in Arabidopsis to regulate flavonoid biosynthesis. On the other hand, the bHLH transcription factor in Arabidopsis often functions as a bridge between MYB transcription factors and TTG1 [28,84,85]. However, in both bimolecular fluorescence complementation (BiFC) and yeast two-hybrid (Y2H) experiments, SmMYB111 interacted with SmTTG1 and SmbHLH51, but SmTTG1 did not interact with SmbHLH51 [82]. This suggests that SmMYB111 may function as a bridge between SmTTG1 and SmbHLH51 to form a SmTTG1-SmMYB111-SmbHLH51 complex in S. miltiorrhiza.
3.3. Regulation of Seed Coat Mucilage Production
In addition to the defects on cell fate determination and flavonoid biosynthesis, the ttg1-1 mutants also have defects in seed coat mucilage production [3], suggesting that TTG1 is involved in the regulation of seed coat mucilage production. Similar to that in the regulation of cell fate determination and flavonoid biosynthesis, TTG1 regulates seed coat mucilage production via forming MBW complexes. TTG1 can form MBW complexes with the bHLH transcription factors TT8 or EGL3 and the R2R3 MYB transcription factors MYB5 or TT2 [24,86,87,88]. The MBW complexes can activate the expression of TTG2 and GL2 [89,90], GL2, in turn, activates the expression of MUCILAGE MODIFIED 4 (MUM4), a mucilage biosynthesis gene, leading to the production of seed coat mucilage [90]. It should be noted that not all the MBW complex components play an equal role in regulating seed coat mucilage production, as an example, MYB5 plays a key role in the regulation of seed coat mucilage production, whereas TT2 has only a minor effect [38,91,92]. It should be also noted that some regulators such as APETALA2 (AP2), NAC-REGULATED SEED MORPHOLOGY1(NARS1), NARS2, MYB52, MYB61, and LUENIG HOMOLOG (LUH) can regulate seed coat mucilage production independent of TTG1 [17,29,93,94,95].
In addition to directly regulate seed coat mucilage production as described above, TTG1 can also indirectly regulate seed coat mucilage production via regulating fatty acid accumulation in seeds [15,96]. As the content of total proteins and fatty acids is increased in the ttg1 mutant embryos, the dry weight of the ttg1 mutant embryos is significantly increased compared to the wild-type plants. Molecular and genetic analysis indicated that TTG1 suppresses the accumulation of seed storage proteins partially through inhibiting the expression of the 2S albumin precursor gene 2S3. On the other hand, TTG1 is a direct target of the seed maturation master regulator FUSCA3 (FUS3), which can inhibit the expression of TTG1 in developing seeds [15]. The SHAGGY-like kinase 11 (SK11) and SK12 are able to phosphorylate TTG1 at the serine 215, therefore preventing the interaction of TTG1 with TT2, leads to an increase in fatty acid biosynthesis in the embryo, but a decrease in seed coat mucilage production in the sk11 sk12 mutants [16].
Ectopic expression of Setaria italica TTG1 (SiTTG1) in the ttg1-13 mutant successfully restored its phenotypes of a high content of fatty acids, transparent seed coat, storage protein contents, as well as seed coat mucilage defects, indicating that SiTTG1 has similar functions to TTG1 in regulating seed coat mucilage production and fatty acid accumulation [97].
3.4. Regulation of Flowering Time
Plant flowering is an important trait for the transition from the vegetative phase to the reproductive phase. In long-day conditions, the ttg1 mutants flowered earlier and the TTG1 over-expression plants flowered later than the wild-type plants [98], suggesting that TTG1 is involved in the regulation of flowering time. It is known that FLOWERING LOCUS C (FLC) can repress the expression of FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1) by binding to their promoter regions [99]. Paffendorf et al. found that TTG1 can activate the expression of FLC [98]. They also found that TTG1 may also regulate the circadian clock component genes. By screening yeast two-hybrid libraries, the author identified PSEUDO RESPONSE REGULATOR 5 (PRR5) and bHLH92 as TTG1 interactors, and they found that TTG1 is able to modulate bHLH92 localization [94].
It has previously been reported that the expression of PRR5 can be directly regulated by LIGHT-REGULATED WD 1 (LWD1) [100]. Whereas, LWD1 and LWD2 are two closely related WD40 repeat proteins to TTG1, and they function redundantly in the photoperiodic pathway to regulate flowering time in Arabidopsis, as the lwd1 lwd2 double mutants flowered early in long-day condition and the expression level of FT was increased in the mutants [101]. These results indicate that TTG1, LWD1, and LWD2 may function in the same pathway to regulate flowering time in plants. On the other hand, the lwd1 lwd2 ttg1 triple mutants lack detectable circadian rhythms, indicating that TTG1 is a regulator of the circadian system [102]. However, TTG1 could rescue the phenotypes of ttg1 and lwd1 lwd2 mutants, but LWD1 and LWD2 could not rescue the epidermal defects of ttg1 mutants, suggesting that subfunctionalization happened following the divergence of the TTG1 and LWD proteins in angiosperms [102].
3.5. Regulation of Biotic and Abiotic Stress Responses
Some available experimental evidence suggests that TTG1 is involved in the regulation of plant response to abiotic stresses. First, the seed germination rate of the ttg1 mutants is lower than Col in response to NaCl treatment, and NaCl treatment also affects leaf and root development in the ttg1 mutant seedlings, indicating that TTG1 is involved in the regulation of plant response to salt stress [21,97]. Second, the ttg1 mutants are also sensitive to sucrose stresses [96]. Third, ectopic expression of SiTTG1 completely restores the salt and sucrose sensitivity phenotypes observed in the ttg1 mutants, indicating that SiTTG1 and TTG1 have a similar function in regulating plant response to salt and sucrose stresses [97]. Forth, ectopic expression of LbTTG1 in Arabidopsis enhanced salt stress tolerance [21]. Under NaCl treatment, the transgenic plants accumulated less Na+ and malondialdehyde (MDA), but more K+, proline, and soluble sugar when compared with that in the wild-type plants, and an elevated expression level of salt-tolerance marker genes including SALT OVERLY SENSITIVE 1 (SOS1), SOS2, SOS3, and PYRPOLINE-5-CARBOXYLATE SYNTHASE 1 (P5CS1) was observed in the LbTTG1 transgenic plants [21]. Fifth, ectopic expression of TaWD40D, the Triticum aestivum TTG1 also enhanced salt and osmotic stress tolerance in Arabidopsis [22]. In addition, the expression of CsWD40 is induced by ABA and sucrose treatments, indicating that CsWD40 may be involved in the regulation of abiotic stress responses in tea plants [82].
Recently, it has been shown that SAR DEFICIENT 4 (SARD4), RECEPTOR LIKE PROTEIN (RLP3), RLP27, RLP26, and PATHOGENESIS-RELATED GENE 6 (PR6) regulate plant immunity in a TTG1-dependent manner [103]. It has also been reported that the TTG1 ortholog Tannin1 (Tan1) is responsible for pathogen-induced color variation in Sorghum bicolor [104], and tobacco TTG1 (NtTTG1) physically interacts with the oomycete-specific effector ParA1 to regulate plant immune responses, including reactive oxygen species (ROS) production and programmed cell death [23]. These experiments support that TTG1 plays a role in regulating plant response to biotic stresses.
3.6. Other Functions
In addition to the functions mentioned above, TTG1 may also have a few other functions. TTG1 can regulate wax ester biosynthesis by suppressing the expression of LONG-CHAIN ACYL-COA SYNTHETASE 3 (LACS3), FATTY ACID REDUCTASE 6 (FAR6), WSD1, LIPID TRANSFER PROTEIN 5 (LTP5), LTP10 and ATP-BINDING CASSETTE G23 (ABCG23) [103]. TTG1 can also regulate cutin biosynthesis by regulating the expression of CYTOCHROME P450, FAMILY 96, SUBFAMILY A, POLYPEPTIDE 11 (CYP96A11), and CUS4 [103]. The expression of several hormone modification genes including UDT-DEPENDENT GLYCOSYLTRANSFERASE 75 D (UGT75D), BETA GLUCOSIDASE 18 (BGLU18), and BRASSINOSTEROID INACTIVATOR 1 (BIA1) is regulated by TTG1, indicating that TTG1 may regulate hormone metabolism [103]. It has been reported that the Salvia miltiorrhiza MYB111 (SmMYB111) positively regulates the biosynthesis of phenolic acids, Sal B, and rosmarinic acid (RA) by forming the SmTTG1−SmbHLH51−SmMYB111 transcription complex, suggesting that TTG1 may play a role in regulating the biosynthesis of phenolic acids, Sal B, and RA [83].
4. Regulation of TTG1
4.1. Transcriptional Regulation
Studies on the transcriptional regulation of TTG1 are very limited. So far, only the B3-domain family transcription factor FUS3 has been identified as a regulator of TTG1. Tsuchiya et al. found that the expression level of TTG1 is increased in the embryos of the fus3 mutants, and the fus3 mutant phenotypes including defects in anthocyanin biosynthesis and seed storage proteins accumulation are recovered in the fus3 ttg1 double mutant, suggesting that expression of TTG1 is down-regulated by FUS3 [14]. By using a dexamethasone-inducible transgenic plant system, Chen et al. found that TTG1 may be an immediate target of FUS3, by using ChIP-PCR and an electrophoretic mobility shift assay (EMSA), they found that the FUS3 can bind directly to the consensus binding site of B3-domain family transcription factors in the promoter region of TTG1, indicating that TTG1 is a direct target of FUS3 [15].
4.2. Posttranscriptional Regulation
Protein degradation by the ubiquitin/26S proteasome system (UPS) pathway affects protein functions. Patra et al. demonstrated that the TTG1 proteins are short-lived and can be targeted by UPS for degradation [105]. They generated transgenic plants overexpressing FLAG targeted TTG1 in the ttg1 (Salk_104152) mutants and used anti-FLAG antibodies to examine TTG1 fusion protein in the transgenic plants. They found that the amount of the TTG1 fusion protein was depleted significantly when the seedlings were treated with cycloheximide (CHX), a protein synthesis inhibitor. Whereas depletion of TTG1 fusion protein was inhibited by combined treatment of CHX and MG132, a peptide aldehyde that can selectively inhibit the proteolytic activity of the 26S proteasome, suggesting that TTG1 can be targeted by UPS for degradation [105].
Protein phosphorylation can also affect protein functions. Both SK11 and SK12 and the GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2) have been shown to be able to phosphorylate TTG1. SK11 and SK12 can interact with TTG1 and phosphorylate TTG1 at serine 215, thus preventing the interaction of TTG1 with TT2, resulting in inhibition of the MBW complex formation [16]. Previous studies have shown that the GSK3-like kinases may be involved in BR-mediated root epidermal cell fate determination by acting upstream of the TTG1–GL3/EGL3–WER MBW complexes [106]. By using GST pull-down and BiFC to test the interaction between the MBW complex component proteins and the GSK3-like kinase BIN2, Cheng et al. found that BIN2 can interact with TTG1, and in vitro kinase assays confirmed that BIN2 could also phosphorylate TTG1 [107].
5. Challenges and Future Perspectives
As mentioned above, TTG1 has been found to regulate multiple aspects of plant growth and development, secondary metabolism, as well as plant response to environmental stimuli, and the functional mechanisms of TTG1 have also been extensively studied. Yet, more efforts are required to further elucidate the regulation and functional mechanisms of TTG1 in plants.
It has been shown that in some processes, such as trichome initiation, root-hairs formation, flavonoid biosynthesis, and seed coat mucilage production, TTG1 needs to interact with specific R2R3 MYB and bHLH transcription factors to form different MBW complexes to regulate the expression of downstream genes [24,25,26,27,28]. However, it has also been shown that GL1 and TTG1 may compete for binding with GL3 to form GL1-GL3 and GL3-TTG1 dimers [58], and dimers formed by an R2R3 MYB and a bHLH transcription factor are sufficient to activate the expression of GL2 and some R3 MYB genes [108], ectopic expression of maize R, a bHLH transcription factor gene complemented all the phenotypes of ttg1 mutants [5], and TT2 binds to GL2 promoter independent of TTG1, but in the absence of TT2, TTG1 can still bind to GL2 promoter [16]. This evidence suggests that formation of the MBW complexes may not be necessary for TTG1 to regulate downstream gene expression. Considering that regulators other than bHLH transcription factors, such as PRR5 and ParA1, which are involved in the regulation of flowering time and plant immune responses, respectively [23,98], have been shown to be able to interact with TTG1, it will be of interest to examine if TTG1 may interact with other regulators to regulate the cell fate determination, flavonoid biosynthesis, and seed coat mucilage production. On the other hand, CsTTG1 interacts directly with CsGL1 to regulate fruit trichome initiation in C. sativa [76], and SmTTG1 interacts with SmMYB111 to regulate phenolic acid biosynthesis complex in S. miltiorrhiza [83], it will be of interest to examine why different plants may use the different bridge proteins to form MBW complexes.
In addition, TTG1 interacts with different bHLH transcription factors to regulate specification of different cell types and different secondary metabolisms [24,25,26,27,28,29]. It has been shown that phosphorylation of TTG1 by SK11 and SK12 at the serine 215 is able to prevent the interaction of TTG1 with TT2, therefore, affecting fatty acids biosynthesis in embryo, mucilage, and flavonoid production in the seed coat [16], so it will be meaningful to detect whether the phosphorylation of TTG1 by SK11 and SK12 may also affect its interaction with other interactors involved in the regulation of cell fate determination, therefore, affecting its functions in regulating cell fate determination, or if different kinases may be involved in these processes.
Some experiments suggest the TTG1 may be required for the stabilization of the bHLH-MYB dimers, as the absence of TTG1 weakened the interaction between MYB and bHLH transcription factor in transfected protoplasts [28,63]. Consistent with this, it has been shown that the ttg1-10, a mutant with a point mutation in the 5′-UTR region showed a transparent testa seed coat phenotype, but anthocyanin synthesis and trichome initiation was largely unaffected, suggesting that reduced TTG1 protein level may have a big effect on the stabilization of the TT8-TT2 dimer, but not the PAP1/PAP2/GL1-GL3 dimers [46]. TTG1 proteins are short-lived and can be targeted for UPS degradation [105], but the ligases involved remain unknown. Identification of the ligases and the examination of how the degradation of TTG1 may affect the stability of the bHLH-MYB dimers will also help to understand the functional mechanisms of TTG1.
The regulation of TTG1 expression also needs to be further studied. FUS3 has been identified as a negative regulator of TTG1 [15]. Identification and characterization of positive regulators of TTG1 may help further explore the functional mechanisms of TTG1. It should be noted that similar to the fus3 mutant, the leafy cotyledon 1 (lec1) and lec2 mutants can produce cotyledon trichomes [109], LEC2 is also a B3-domain family transcription factor [110], and is able to interact with FUS3 [111], it is worthwhile to examine if LEC1 and LEC2 may also regulate the expression of TTG1.
Funding
This research was supported by the National Key R&D Program of China (2016YFD0101900) and a startup funding from Linyi University (LYDX2019BS039).
Acknowledgments
We thank all the researchers in this field and apologize to the colleagues whose work could not be included due to the space constraints and specific focus of this review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bürger, D. Die morphologischen Mutanten des Göttinger Arabidopsis-Sortiments, einschlieβlich der Mutanten mit abweichender Samenfarbe. Arab. Inf. Serv. 1971, 8, 36–42. [Google Scholar]
- Koornneef, M.; Van der Veen, J.H. Gene localization with trisomics in Arabidopsis thaliana. Arab. Inf. Serv. 1978, 15, 38–43. [Google Scholar]
- Koornneef, M. The complex syndrome of ttg mutants. Arab. Inf. Serv. 1981, 18, 45–51. [Google Scholar]
- Koornneef, M.; van Eden, J.; Hanhart, C.J.; Stam, P.; Braaksma, F.J.; Feenstra, W.J. Linkage map of Arabidopsis thaliana. J. Hered. 1983, 74, 265–272. [Google Scholar] [CrossRef]
- Lloyd, A.M.; Walbot, V.; Davis, R.W. Arabidopsis and Nicotiana anthocyanin production activated by maize anthocyanin-specific regulators R and C1. Science 1992, 258, 1773–1775. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Davison, P.A.; Bolognesiwinfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef]
- Van Nocker, S.; Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genom. 2003, 4, 50. [Google Scholar] [CrossRef]
- Ouyang, Y.; Huang, X.; Lu, Z.; Yao, J. Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genom. 2012, 13, 100. [Google Scholar] [CrossRef]
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The ancient regulatory protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef]
- Galway, M.E.; Masucci, J.D.; Lloyd, A.M.; Walbot, V.; Davis, R.W.; Schiefelbein, J.W. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 1994, 166, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Berger, F.; Linstead, P.; Dolan, L.; Haseloff, J. Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol. 1998, 194, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Debeaujon, I.; Leon-Kloosterziel, K.M.; Koornneef, M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000, 122, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Western, T.L.; Burn, J.; Tan, W.L.; Skinner, D.J.; Martin-McCaffffrey, L.; Moffffatt, B.A.; Haughn, G.W. Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 2001, 127, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, Y.; Nambara, E.; Naito, S.; McCourt, P. The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 2004, 37, 73–81. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, B.; Li, C.; Kulaveerasingam, H.; Chew, F.T.; Yu, H. TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol. 2015, 169, 391–402. [Google Scholar] [CrossRef]
- Li, C.; Zhang, B.; Chen, B.; Ji, L.; Hao, Y. Site-specifific phosphorylation of TRANSPARENT TESTA GLABRA1 mediates carbon partitioning in Arabidopsis seeds. Nat. Commun. 2018, 9, 571. [Google Scholar] [CrossRef]
- Penfield, S.; Meissner, R.C.; Shoue, D.A.; Carpita, N.C.; Bevan, M.W. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 2001, 13, 2777–2791. [Google Scholar] [CrossRef]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Ausubel, F.M.; Goodman, H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995, 8, 659–671. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Ann. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Windsor, J.B.; Symonds, V.V.; Mendenhall, J.; Lloyd, A.M. Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 2000, 22, 483–493. [Google Scholar] [CrossRef]
- Yuan, F.; Leng, B.; Zhang, H.; Wang, X.; Han, G.; Wang, B. A WD40-repeat protein from the Recretohalophyte Limonium bicolor enhances trichome formation and salt tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 1456. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Li, M.; Dong, Z.; Ji, H.; Li, X. Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 2015, 34, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, R.; Chen, L.; Wang, Y.; Liang, Y.; Wu, X.; Li, B.; Wu, J.; Liang, Y.; Wang, X.; et al. Nicotiana tabacum TTG1 contributes to ParA1-induced signalling and cell death in leaf trichomes. J. Cell Sci. 2009, 122, 2673–2685. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Schiefelbein, J. Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 2003, 6, 74–78. [Google Scholar] [CrossRef]
- Lin, Q.; Aoyama, T. Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: Update on the roles of the classic regulator. J. Integr. Plant Biol. 2012, 54, 729–737. [Google Scholar]
- Wang, S.; Chen, J.G. Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 2014, 5, 133. [Google Scholar] [CrossRef]
- Baudry, A.; Heim, M.A.; Debreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Golz, J.F.; Allen, P.J.; Li, S.F.; Parish, R.W.; Jayawardana, N.; Bacic, A.; Doblin, M.S. Layers of regulation–insights into the role of transcription factors controlling mucilage production in the Arabidopsis thaliana seed coat. Plant Sci. 2018, 272, 179–192. [Google Scholar] [CrossRef]
- Pesch, M.; Hulskamp, M. Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 2004, 14, 422–427. [Google Scholar] [CrossRef]
- Schellmann, S.; Schnittger, A.; Kirik, V.; Wada, T.; Okada, K.; Beermann, A.; Thumfahrt, J.; Jürgens, G.; Hulskamp, M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002, 21, 5036–5046. [Google Scholar] [CrossRef] [PubMed]
- Masucci, J.D.; Rerie, W.G.; Foreman, D.R.; Zhang, M.; Galway, M.E.; Marks, M.D.; Schiefelbein, J. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 1996, 122, 1253–1260. [Google Scholar]
- Schiefelbein, J.; Huang, L.; Zheng, X. Regulation of epidermal cell fate in Arabidopsis roots: The importance of multiple feedback loops. Front. Plant Sci. 2014, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [PubMed]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive Organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Zhang, B.; Schrader, A. TRANSPARENT TESTA GLABRA 1-dependent regulation of flavonoid biosynthesis. Plants 2017, 6, 65. [Google Scholar] [CrossRef]
- Broun, P. Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005, 8, 272–279. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Braaksma, F.J.; Feenstra, W.J. The localization of the suppressor gene su and the nitrate reductase deficient mutant B73. Arab. Inf. Serv. 1980, 17, 96–98. [Google Scholar]
- Grill, E.; Somerville, C. Construction and characterization of a yeast artificial chromosome library of Arabidopsis which is suitable for chromosome walking. Mol. Gen. Genet. 1991, 226, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.G.; Giraudat, J.; Den Boer, B.; Moonan, F.; Loos, W.; Hauge, B.M.; Goodman, H.M. Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell 1989, 1, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.M.; Coates, D.; Fricker, M.D.; Evans, D.E. Genomic sequence of a calnexin homolog from Arabidopsis thaliana. Plant Physiol. 1994, 106, 1691. [Google Scholar] [CrossRef]
- Larkin, J.C.; Walker, J.D.; Bolognesi-Winfifield, A.C.; Gray, J.C.; Walker, A.R. Allele-specifific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 1999, 151, 1591–1604. [Google Scholar]
- de Vetten, N.; Quattrocchio, F.; Mol, J.; Koes, R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997, 11, 1422–1434. [Google Scholar] [CrossRef]
- Larkin, J.C.; Oppenheimer, D.G.; Lloyd, A.M.; Paparozzi, E.T.; Marks, M.D. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell 1994, 6, 1065–1076. [Google Scholar] [CrossRef]
- Appelhagen, I.; Thiedig, K.; Nordholt, N.; Schmidt, N.; Huep, G.; Sagasser, M.; Weisshaar, B. Update on transparent testa mutants from Arabidopsis thaliana: Characterization of new alleles from an isogenic collection. Planta 2014, 240, 955–970. [Google Scholar] [CrossRef]
- Rosso, M.G.; Li, Y.; Strizhov, N.; Reiss, B.; Dekker, K.; Weisshaar, B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003, 53, 247–259. [Google Scholar] [CrossRef]
- Long, Y.; Schiefelbein, J. Novel TTG1 mutants modify root-hair pattern formation in Arabidopsis. Front. Plant Sci. 2020, 11, 383. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Lee, Y.S.; Mukundi, E.; Gomez-Cano, F.; Rivero, L.; Grotewold, E. Diversity of genetic lesions characterizes new Arabidopsis flavonoid pigment mutant alleles from T-DNA collections. Plant Sci. 2020, 291, 110335. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Sano, R.; Wada, T.; Takabayashi, J.; Okada, K. Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 2009, 136, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.K.; Khurana, J.P. Molecular characterization of transparent testa (tt) mutants of Arabidopsis thaliana (ecotype Estland) impaired in flavonoid biosynthetic pathway. Plant Sci. 2003, 165, 1321–1332. [Google Scholar] [CrossRef]
- Taheri, A.; Gao, P.; Yu, M.; Cui, D.; Regan, S.; Parkin, I.; Gruber, M. A landscape of hairy and twisted: Hunting for new trichome mutants in the Saskatoon Arabidopsis T-DNA population. Plant Biol. 2015, 17, 384–394. [Google Scholar] [CrossRef]
- Robinson, S.J.; Tang, L.H.; Mooney, B.A.; McKay, S.J.; Clarke, W.E.; Links, M.G.; Karcz, S.; Regan, S.; Wu, Y.Y.; Gruber, M.Y.; et al. An archived activation tagged population of Arabidopsis thaliana to facilitate forward genetics approaches. BMC Plant Biol. 2009, 9, 101. [Google Scholar] [CrossRef]
- Shikazono, N.; Tanaka, A.; Watanabe, H.; Tano, S. Rearrangements of the DNA in carbon ion-induced mutants of Arabidopsis thaliana. Genetics 2001, 157, 379–387. [Google Scholar]
- Miller, J.C.; Chezem, W.R.; Clay, N.K. Ternary WD40 repeat-containing protein complexes: Evolution, composition and roles in plant immunity. Front. Plant Sci. 2015, 6, 1108. [Google Scholar] [CrossRef]
- Pesch, M.; Schultheis, I.; Klopffleisch, K.; Uhrig, J.F.; Koegl, M.; Clemen, C.S.; Simon, R.; Weidtkamp-Peters, S.; Hulskamp, M. TRANSPARENT TESTA GLABRA1 and GLABRA1 compete for binding to GLABRA3 in Arabidopsis. Plant Physiol. 2015, 168, 584–597. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Li, Y.; Ai, Q.; Yu, D. MYB82 functions in regulation of trichome development in Arabidopsis. J. Exp. Bot. 2014, 65, 3215–3223. [Google Scholar] [CrossRef]
- Kirik, V.; Schnittger, A.; Radchuk, V.; Adler, K.; Hulskamp, M.; Baumlein, H. Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev. Biol. 2001, 235, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar]
- Ishida, T.; Kurata, T.; Okada, K.; Wada, T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 2008, 59, 364–386. [Google Scholar] [CrossRef] [PubMed]
- Ioannidi, E.; Rigas, S.; Tsitsekian, D.; Daras, G.; Alatzas, A.; Makris, A.; Tanou, G.; Argiriou, A.; Alexandrou, D.; Poethig, S.; et al. Trichome patterning control involves TTG1 interaction with SPL transcription factors. Plant Mol. Biol. 2016, 92, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Schiefelbein, J. Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 2000, 124, 1525–1531. [Google Scholar] [CrossRef]
- Dolan, L.; Costa, S. Evolution and genetics of root hair stripes in the root epidermis. J. Exp. Bot. 2001, 52, 413–417. [Google Scholar] [CrossRef]
- Lee, M.M.; Schiefelbein, J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 1999, 99, 473–483. [Google Scholar] [CrossRef]
- Tominaga-Wada, R.; Ishida, T.; Wada, T. New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Bio. 2011, 286, 67–106. [Google Scholar]
- Schiefelbein, J.; Kwak, S.H.; Wieckowski, Y.; Barron, C.; Bruex, A. The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 2009, 60, 1515–1521. [Google Scholar] [CrossRef]
- Grebe, M. The patterning of epidermal hairs in Arabidopsis updated. Curr. Opin. Plant Biol. 2012, 15, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.A.; Walker, A.R.; Timmis, J.N.; Orford, S.J. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 2005, 57, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Brueggemann, J.; Weisshaar, B.; Sagasser, M. A WD40-repeat gene from Malus x domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1. Plant Cell Rep. 2010, 29, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.C.; Strahle, J.T.; Selinger, D.A.; Chandler, V.L. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 2004, 16, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Chopra, D.; Wolff, H.; Span, J.; Schellmann, S.; Coupland, G.; Albani, M.C.; Schrader, A.; Hülskamp, M. Analysis of TTG1 function in Arabis alpina. BMC Plant Biol. 2014, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yin, S.; Liu, X.; Liu, B.; Yang, S.; Xue, S.; Cai, Y.; Black, K.; Liu, H.; Dong, M.; et al. CsTTG1 encodes a WD-repeat protein that regulates fruit wart formation in Cucumis sativus through interaction with the homeodomain-leucine Zipper I protein Mict. Plant Physiol. 2016, 171, 1156–1168. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.L.; Shi, M.Z.; Xie, D.Y. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/ TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta 2012, 236, 825–837. [Google Scholar] [CrossRef]
- Shi, M.Z.; Xie, D.Y. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017, 36, 1–34. [Google Scholar] [CrossRef]
- Wei, Z.; Cheng, Y.; Zhou, C.; Li, D.; Gao, X.; Zhang, S.; Chen, M. Genome-wide identification of direct targets of the TTG1-bHLH-MYB complex in regulating trichome formation and flavonoid accumulation in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, e5014. [Google Scholar] [CrossRef]
- Shan, X.; Li, Y.; Yang, S.; Gao, R.; Zhou, L.; Bao, T.; Han, T.; Wang, S.; Gao, X.; Wang, L. A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol. Biochem. 2019, 141, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hou, H.; Jiang, X.; Wang, P.; Dai, X.; Chen, W.; Gao, L.; Xia, T. A WD40 repeat protein from Camellia sinensis regulates anthocyanin and proanthocyanidin accumulation through the formation of MYB–bHLH–WD40 ternary complexes. Int. J. Mol. Sci. 2018, 19, e1686. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, Y.; Kuang, J.; Wang, H.; Du, T.; Huang, Y.; Zhang, Y.; Cao, X.; Wang, Z. SmMYB111 Is a key factor to phenolic acid biosynthesis and interacts with both SmTTG1 and SmbHLH51 in Salvia miltiorrhiza. J. Agric. Food Chem. 2018, 66, 8069–8078. [Google Scholar] [CrossRef]
- Wan, S.; Li, C.; Ma, X.; Luo, K. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017, 36, 1263–1276. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.; de Vos, R.C.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Francoz, E.; Ranocha, P.; Burlat, V.; Dunand, C. Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci. 2015, 20, 515–524. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Yang, B.; Schmidt, M.H.; Günl, M.; Usadel, B. Starting to gel: How Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int. J. Mol. Sci. 2015, 16, 3452–3473. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Western, T.L.; Young, D.S.; Dean, G.H.; Tan, W.L.; Samuels, A.L.; Haughn, G.W. MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 2004, 134, 296–306. [Google Scholar] [CrossRef]
- Li, S.F.; Milliken, O.N.; Pham, H.; Seyit, R.; Napoli, R.; Preston, J.; Koltunow, A.M.; Parish, R.W. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 2009, 21, 72–89. [Google Scholar] [CrossRef]
- Gonzalez, A.; Mendenhall, J.; Huo, Y.; Lloyd, A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009, 325, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Den Boer, B.G.; Van Montagu, M.; Okamuro, J.k. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed]
- Kunieda, T.; Mitsuda, N.; Ohme-takagi, M.; Takeda, S.; Aida, M.; Tasaka, M.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. NAC Family Proteins NARS1/NAC2 and NARS2/NAM in the Outer Integument Regulate Embryogenesis in Arabidopsis. Plant Cell 2008, 20, 2631–2642. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Ren, A.; Tang, X.; Qi, G.; Xu, Z.; Chai, G.; Hu, R.; Zhou, G.; Kong, Y. MYB52 negatively regulates pectin demethylesterification in seed coat mucilage. Plant Physiol. 2018, 176, 2737–2749. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Katavic, V.; Yu, Y.; Kunst, L.; Haughn, G. Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J. 2012, 69, 37–46. [Google Scholar] [CrossRef]
- Liu, K.; Qi, S.; Li, D.; Jin, C.; Gao, C.; Duan, S.; Feng, B.; Chen, M. TRANSPARENT TESTA GLABRA 1 ubiquitously regulates plant growth and development from Arabidopsis to foxtail millet (Setaria italica). Plant Sci. 2017, 254, 60–69. [Google Scholar] [CrossRef]
- Paffendorf, B.A.; Qassrawi, R.; Meys, A.M.; Trimborn, L.; Schrader, A. TRANSPARENT TESTA GLABRA 1 participates in flowering time regulation in Arabidopsis thaliana. Peer J. 2020, 8, e8303. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Wood, C.C.; Robertson, M.; James-Peacock, W.; Dennis, E.S. The Arabidopsis FLC protein interacts directly in vivowith SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006, 46, 183–192. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.F.; Nakamichi, N.; Sakakibara, H.; Nam, H.G.; Wu, S.H. LIGHTREGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 2011, 23, 486–498. [Google Scholar] [CrossRef]
- Wu, J.F.; Wang, Y.; Wu, S.H. Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 2008, 148, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Airoldi, C.A.; Hearn, T.J.; Brockington, S.F.; Webb, A.A.; Glover, B.J. TTG1 proteins regulate circadian activity as well as epidermal cell fate and pigmentation. Nat. Plants 2019, 5, 1145–1153. [Google Scholar] [CrossRef]
- Li, S.F.; Allen, P.J.; Napoli, R.S.; Browne, R.G.; Pham, H.; Parish, R.W. MYB-bHLH-TTG1 regulates Arabidopsis seed coat biosynthesis pathways directly and indirectly via multiple tiers of transcription factors. Plant Cell Physiol. 2020, 61, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Yazawa, T.; Kasuga, S.; Sawada, Y.; Ogata, J.; Ando, T.; Kanamori, H.; Yonemaru, J.; Wu, J.; Hirai, Y.M.; et al. Expression level of a flavonoid 3′-hydroxylase gene determines pathogen induced color variation in Sorghum. BMC Res. Notes 2014, 7, 761. [Google Scholar] [CrossRef] [PubMed]
- Patra, B.; Pattanaik, S.; Yuan, L. Proteolytic degradation of the flavonoid regulators, TRANSPARENT TESTA8 and TRANSPARENT TESTA GLABRA1, in Arabidopsis is mediated by the ubiquitin/26Sproteasome system. Plant Signal. Behav. 2013, 8, e25901. [Google Scholar] [CrossRef]
- Kuppusamy, K.T.; Chen, A.Y.; Nemhauser, J.L. Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2009, 106, 8073–8076. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhu, W.; Chen, Y.; Ito, S.; Asami, T.; Wang, X. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 2014, 3, e02525. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hubbard, L.; Chang, Y.; Guo, J.; Schiefelbein, J.; Chen, J.G. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 2008, 8, 81. [Google Scholar] [CrossRef]
- Meinke, D.W.; Franzmann, L.H.; Nickle, T.C.; Yeung, E.C. Leafy Cotyledon Mutants of Arabidopsis. Plant Cell 1994, 6, 1049–1064. [Google Scholar] [CrossRef]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 1180–11811. [Google Scholar] [CrossRef]
- Tang, L.P.; Zhou, C.; Wang, S.S.; Yuan, J.; Zhang, X.S.; Su, Y.H. FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 2017, 213, 1740–1754. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).