Development of a Single Construct System for Site-Directed RNA Editing Using MS2-ADAR
Abstract
1. Introduction
2. Results
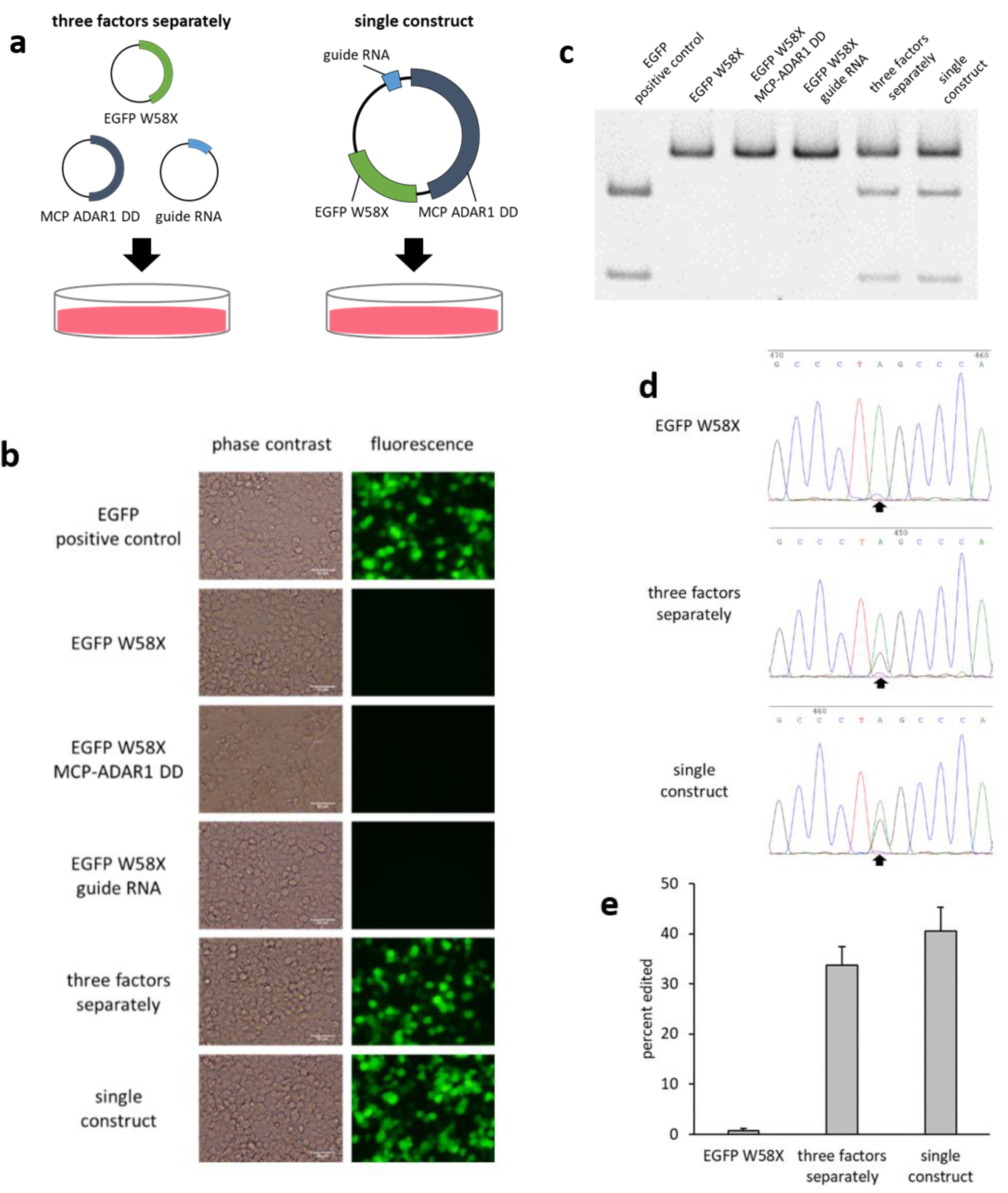
2.1. System for Introducing an Equal Number of Copies of the Three Factors
2.2. Comparison of the Three Types of MCPs
2.3. Nuclear Localization of MCP-ADAR1 DD
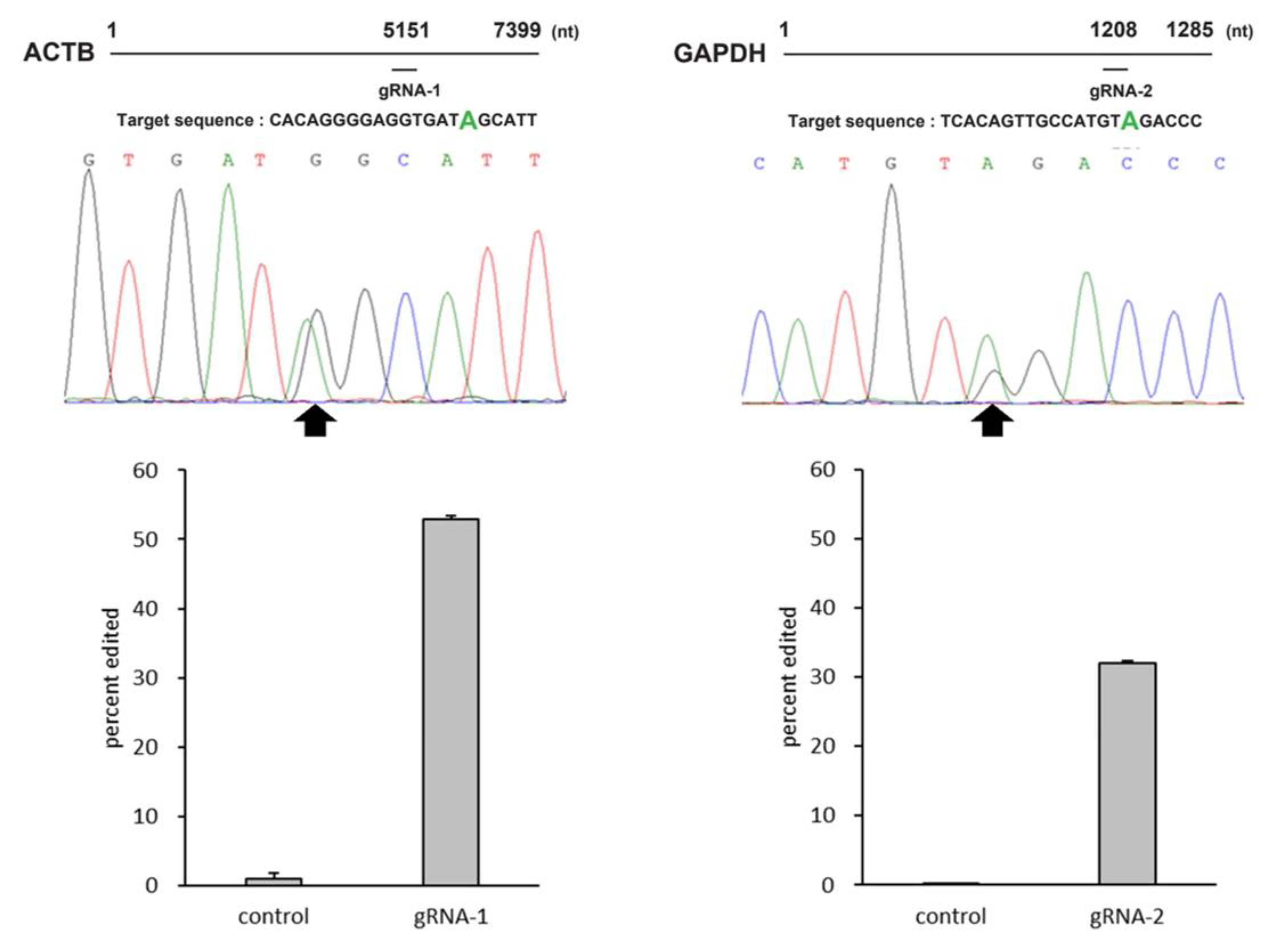
2.4. Applicability of MS2-ADAR to Endogenous Gene Editing
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Cell Culture
4.3. Transfection
4.4. Determination of Editing Efficiency
4.5. RFLP Analysis
4.6. Endogenous Gene Editing Assay
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gori, J.L.; Hsu, P.D.; Maeder, M.L.; Shen, S.; Welstead, G.G.; Bumcrot, D. Delivery and Specificity of CRISPR/Cas9 Genome Editing Technologies for Human Gene Therapy. Hum. Gene Ther. 2015, 26, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Maeder, M.L.; Gersbach, C.A. Genome-Editing Technologies for Gene and Cell Therapy. Mol. Ther. 2016, 24, 430–446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Lang, Z.; Botella, J.R.; Zhu, J.-K. Genome Editing—Principles and Applications for Functional Genomics Research and Crop Improvement. Crit. Rev. Plant Sci. 2017, 36, 291–309. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome Editing in Rice and Wheat Using the CRISPR/Cas System. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable Base Editing of A• T to G• C in Genomic DNA without DNA Cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Grünewald, J.; Zhou, R.; Garcia, S.P.; Iyer, S.; Lareau, C.A.; Aryee, M.J.; Joung, J.K. Transcriptome-Wide off-Target RNA Editing Induced by CRISPR-Guided DNA Base Editors. Nature 2019, 569, 433–437. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Platt, R.J.; Zhang, F. Therapeutic Genome Editing: Prospects and Challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef]
- Hilton, I.B.; Gersbach, C.A. Enabling Functional Genomics with Genome Engineering. Genome Res. 2015, 25, 1442–1455. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of Targeted Point Mutations in Rice by a Modified CRISPR/Cas9 System. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wen, Y.; Guo, X. CRISPR/Cas9 for Genome Editing: Progress, Implications and Challenges. Hum. Mol. Genet. 2014, 23, R40–R46. [Google Scholar] [CrossRef] [PubMed]
- Vogel, P.; Stafforst, T. Critical Review on Engineering Deaminases for Site-Directed RNA Editing. Curr. Opin. Biotechnol. 2019, 55, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Montiel-Gonzalez, M.F.; Quiroz, J.F.D.; Rosenthal, J.J. Current Strategies for Site-Directed RNA Editing Using ADARs. Methods 2019, 156, 16–24. [Google Scholar] [CrossRef]
- Chen, G.; Katrekar, D.; Mali, P. RNA-Guided Adenosine Deaminases: Advances and Challenges for Therapeutic RNA Editing. Biochemistry 2019, 58, 1947–1957. [Google Scholar] [CrossRef]
- Azad, M.T.; Bhakta, S.; Tsukahara, T. Site-Directed RNA Editing by Adenosine Deaminase Acting on RNA for Correction of the Genetic Code in Gene Therapy. Gene Ther. 2017, 24, 779–786. [Google Scholar] [CrossRef]
- Bhakta, S.; Azad, M.T.A.; Tsukahara, T. Genetic Code Restoration by Artificial RNA Editing of Ochre Stop Codon with ADAR1 Deaminase. Protein Eng. Des. Sel. 2018, 31, 471–478. [Google Scholar] [CrossRef]
- Azad, M.; Thoufic, A.; Qulsum, U.; Tsukahara, T. Comparative Activity of Adenosine Deaminase Acting on RNA (ADARs) Isoforms for Correction of Genetic Code in Gene Therapy. Curr. Gene Ther. 2019, 19, 31–39. [Google Scholar] [CrossRef]
- Vogel, P.; Moschref, M.; Li, Q.; Merkle, T.; Selvasaravanan, K.D.; Li, J.B.; Stafforst, T. Efficient and Precise Editing of Endogenous Transcripts with SNAP-Tagged ADARs. Nat. Methods 2018, 15, 535–538. [Google Scholar] [CrossRef]
- Vallecillo-Viejo, I.C.; Liscovitch-Brauer, N.; Montiel-Gonzalez, M.F.; Eisenberg, E.; Rosenthal, J.J. Abundant Off-Target Edits from Site-Directed RNA Editing Can Be Reduced by Nuclear Localization of the Editing Enzyme. RNA Biol. 2018, 15, 104–114. [Google Scholar] [CrossRef]
- Cox, D.B.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA Editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Katrekar, D.; Chen, G.; Meluzzi, D.; Ganesh, A.; Worlikar, A.; Shih, Y.-R.; Varghese, S.; Mali, P. In Vivo RNA Editing of Point Mutations via RNA-Guided Adenosine Deaminases. Nat. Methods 2019, 16, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Yi, Z.; Zhu, S.; Wang, C.; Cao, Z.; Zhou, Z.; Yuan, P.; Yu, Y.; Tian, F.; Liu, Z. Programmable RNA Editing by Recruiting Endogenous ADAR Using Engineered RNAs. Nat. Biotechnol. 2019, 37, 1059–1069. [Google Scholar] [CrossRef]
- Merkle, T.; Merz, S.; Reautschnig, P.; Blaha, A.; Li, Q.; Vogel, P.; Wettengel, J.; Li, J.B.; Stafforst, T. Precise RNA Editing by Recruiting Endogenous ADARs with Antisense Oligonucleotides. Nat. Biotechnol. 2019, 37, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Franklin, B.; Koob, J.; Kellner, M.J.; Ladha, A.; Joung, J.; Kirchgatterer, P.; Cox, D.B.; Zhang, F. A Cytosine Deaminase for Programmable Single-Base RNA Editing. Science 2019, 365, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Campbell, K.; Rodino-Klapac, L.; Sahenk, Z.; Shilling, C.; Lewis, S.; Bowles, D.; Gray, S.; Li, C.; Galloway, G. Dystrophin Immunity in Duchenne’s Muscular Dystrophy. N. Engl. J. Med. 2010, 363, 1429–1437. [Google Scholar] [CrossRef]
- Mingozzi, F.; Meulenberg, J.J.; Hui, D.J.; Basner-Tschakarjan, E.; Hasbrouck, N.C.; Edmonson, S.A.; Hutnick, N.A.; Betts, M.R.; Kastelein, J.J.; Stroes, E.S. AAV-1–Mediated Gene Transfer to Skeletal Muscle in Humans Results in Dose-Dependent Activation of Capsid-Specific T Cells. Blood J. Am. Soc. Hematol. 2009, 114, 2077–2086. [Google Scholar] [CrossRef]
- Montiel-González, M.F.; Vallecillo-Viejo, I.C.; Rosenthal, J.J. An Efficient System for Selectively Altering Genetic Information within MRNAs. Nucleic Acids Res. 2016, 44, e157. [Google Scholar] [CrossRef]
- Seth Chhabra, E.; Moore, N.; Furcht, C.; Holthaus, A.M.; Liu, J.; Liu, T.; Schellenberger, V.; Kulman, J.; Salas, J.; Peters, R. Evaluation of Enhanced In Vitro Plasma Stability of a Novel Long Acting Recombinant FVIIIFc-VWF-XTEN Fusion Protein. Blood. 2015, 126, 2279. [Google Scholar] [CrossRef]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of Catalytically Inactive Cas9 to FokI Nuclease Improves the Specificity of Genome Modification. Nat. Biotechnol. 2014, 32, 577. [Google Scholar] [CrossRef]
- Eggington, J.M.; Greene, T.; Bass, B.L. Predicting Sites of ADAR Editing in Double-Stranded RNA. Nat. Commun. 2011, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.; Spingola, M.; Peabody, D.S. Altering the RNA Binding Specificity of a Translational Repressor. J. Biol. Chem. 1994, 269, 9006–9010. [Google Scholar]
- Peabody, D.S.; Lim, F. Complementation of RNA Binding Site Mutations in MS2 Coat Protein Heterodimers. Nucleic Acids Res. 1996, 24, 2352–2359. [Google Scholar] [CrossRef] [PubMed]
- Tsai, B.P.; Wang, X.; Huang, L.; Waterman, M.L. Quantitative Profiling of in Vivo-Assembled RNA-Protein Complexes Using a Novel Integrated Proteomic Approach. Mol. Cell. Proteomics 2011, 10. [Google Scholar] [CrossRef]
- Hook, B.; Bernstein, D.; Zhang, B.; Wickens, M. RNA–Protein Interactions in the Yeast Three-Hybrid System: Affinity, Sensitivity, and Enhanced Library Screening. RNA 2005, 11, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Cokol, M.; Nair, R.; Rost, B. Finding Nuclear Localization Signals. EMBO Rep. 2000, 1, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Conti, E.; Uy, M.; Leighton, L.; Blobel, G.; Kuriyan, J. Crystallographic Analysis of the Recognition of a Nuclear Localization Signal by the Nuclear Import Factor Karyopherin α. Cell 1998, 94, 193–204. [Google Scholar] [CrossRef]
- Peabody, D.S. The RNA Binding Site of Bacteriophage MS2 Coat Protein. EMBO J. 1993, 12, 595–600. [Google Scholar] [CrossRef]
- Johansson, H.E.; Liljas, L.; Uhlenbeck, O.C. RNA Recognition by the MS2 Phage Coat Protein. In Seminars in Virology; Elsevier: Amsterdam, The Netherlands, 1997; Volume 8, pp. 176–185. [Google Scholar]
- Horn, W.T.; Convery, M.A.; Stonehouse, N.J.; Adams, C.J.; Liljas, L.; Phillips, S.E.; Stockley, P.G. The Crystal Structure of a High Affinity RNA Stem-Loop Complexed with the Bacteriophage MS2 Capsid: Further Challenges in the Modeling of Ligand–RNA Interactions. RNA 2004, 10, 1776–1782. [Google Scholar] [CrossRef]
- Arenal, A.; Pimentel, R.; García, C.; Pimentel, E.; Aleström, P. The SV40 T Antigen Nuclear Localization Sequence Enhances Nuclear Import of Vector DNA in Embryos of a Crustacean (Litopenaeus Schmitti). Gene 2004, 337, 71–77. [Google Scholar] [CrossRef]
- Rinkevich, F.D.; Schweitzer, P.A.; Scott, J.G. Antisense Sequencing Improves the Accuracy and Precision of A-to-I Editing Measurements Using the Peak Height Ratio Method. BMC Res. Notes 2012, 5, 63. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tohama, T.; Sakari, M.; Tsukahara, T. Development of a Single Construct System for Site-Directed RNA Editing Using MS2-ADAR. Int. J. Mol. Sci. 2020, 21, 4943. https://doi.org/10.3390/ijms21144943
Tohama T, Sakari M, Tsukahara T. Development of a Single Construct System for Site-Directed RNA Editing Using MS2-ADAR. International Journal of Molecular Sciences. 2020; 21(14):4943. https://doi.org/10.3390/ijms21144943
Chicago/Turabian StyleTohama, Tetsuto, Matomo Sakari, and Toshifumi Tsukahara. 2020. "Development of a Single Construct System for Site-Directed RNA Editing Using MS2-ADAR" International Journal of Molecular Sciences 21, no. 14: 4943. https://doi.org/10.3390/ijms21144943
APA StyleTohama, T., Sakari, M., & Tsukahara, T. (2020). Development of a Single Construct System for Site-Directed RNA Editing Using MS2-ADAR. International Journal of Molecular Sciences, 21(14), 4943. https://doi.org/10.3390/ijms21144943





