Integrated Omics Analyses Identify Key Pathways Involved in Petiole Rigidity Formation in Sacred Lotus
Abstract
1. Introduction
2. Results
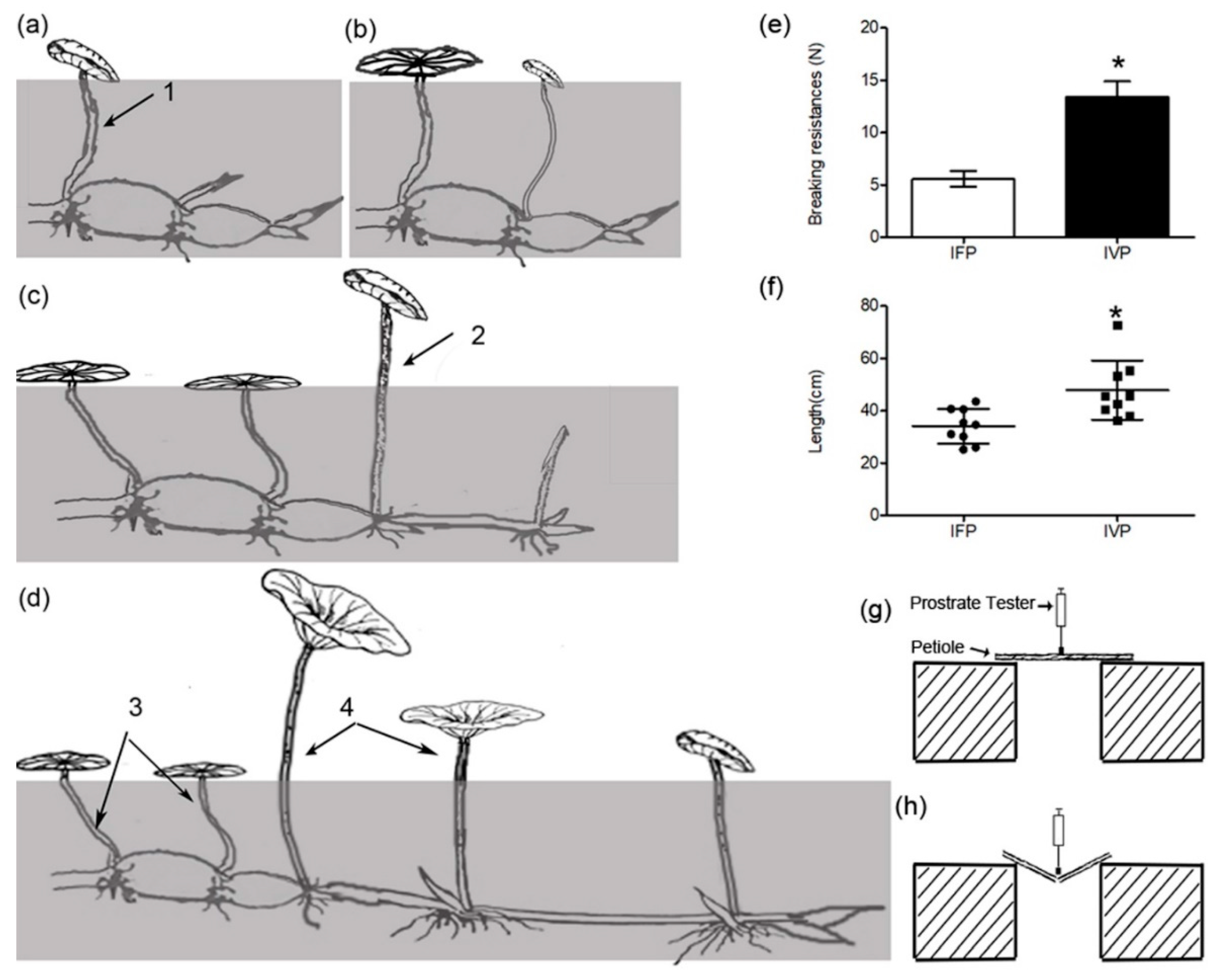
2.1. Phenotypic Evaluation of Lotus Petioles at IFP and IVP Stages
2.2. Anatomic Structure Analysis of Petioles at IFP and IVP Stages
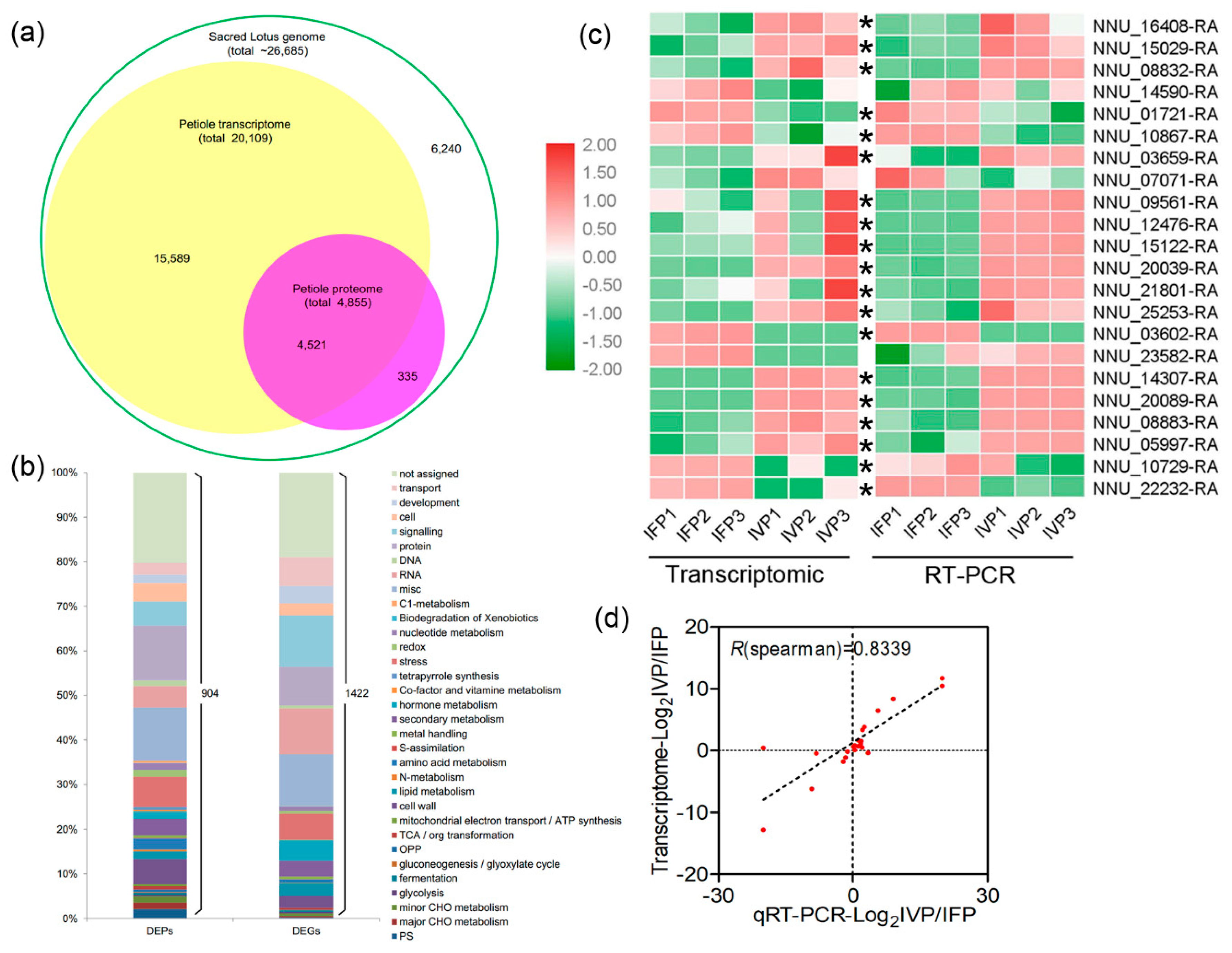
2.3. Overview of Transcriptomic Analysis and Transcriptomic Data Validation
2.4. Overview of Quantitative Proteomics Analysis and Data Validation Using PRM
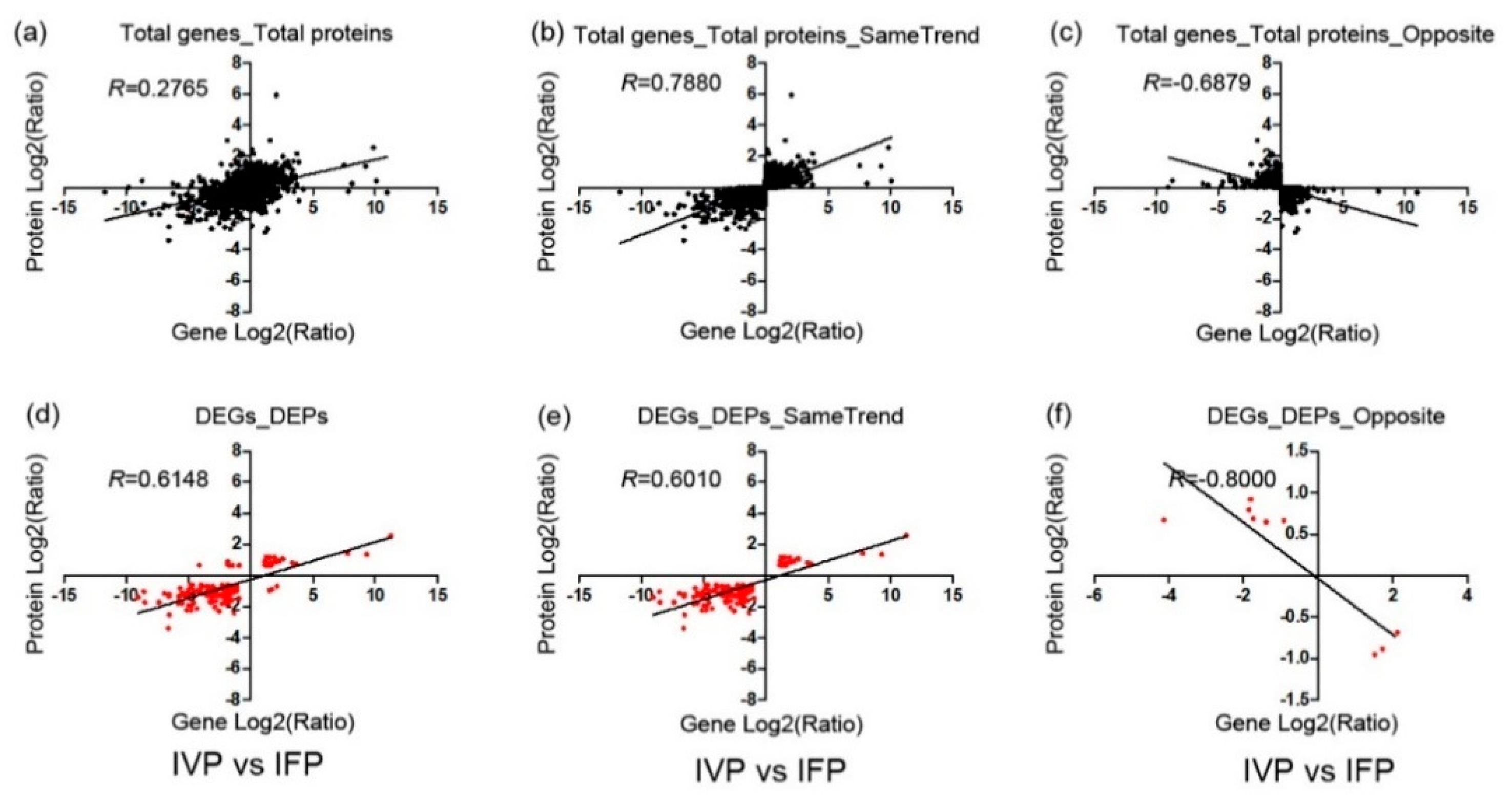
2.5. Integrated Analysis of Transcriptome and Proteome Data
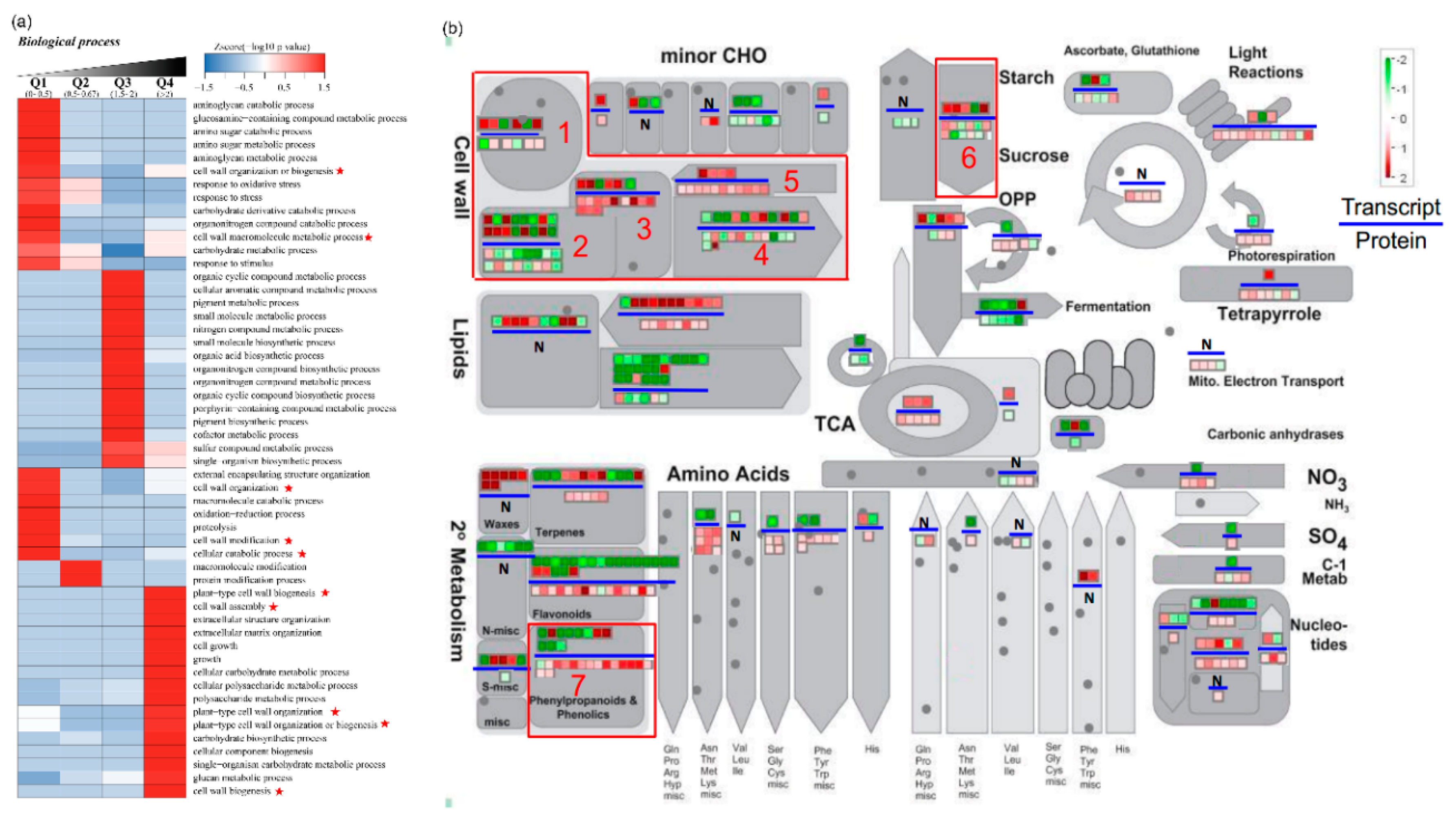
2.6. Functional Enrichment of Quantified DEGs and DEPs
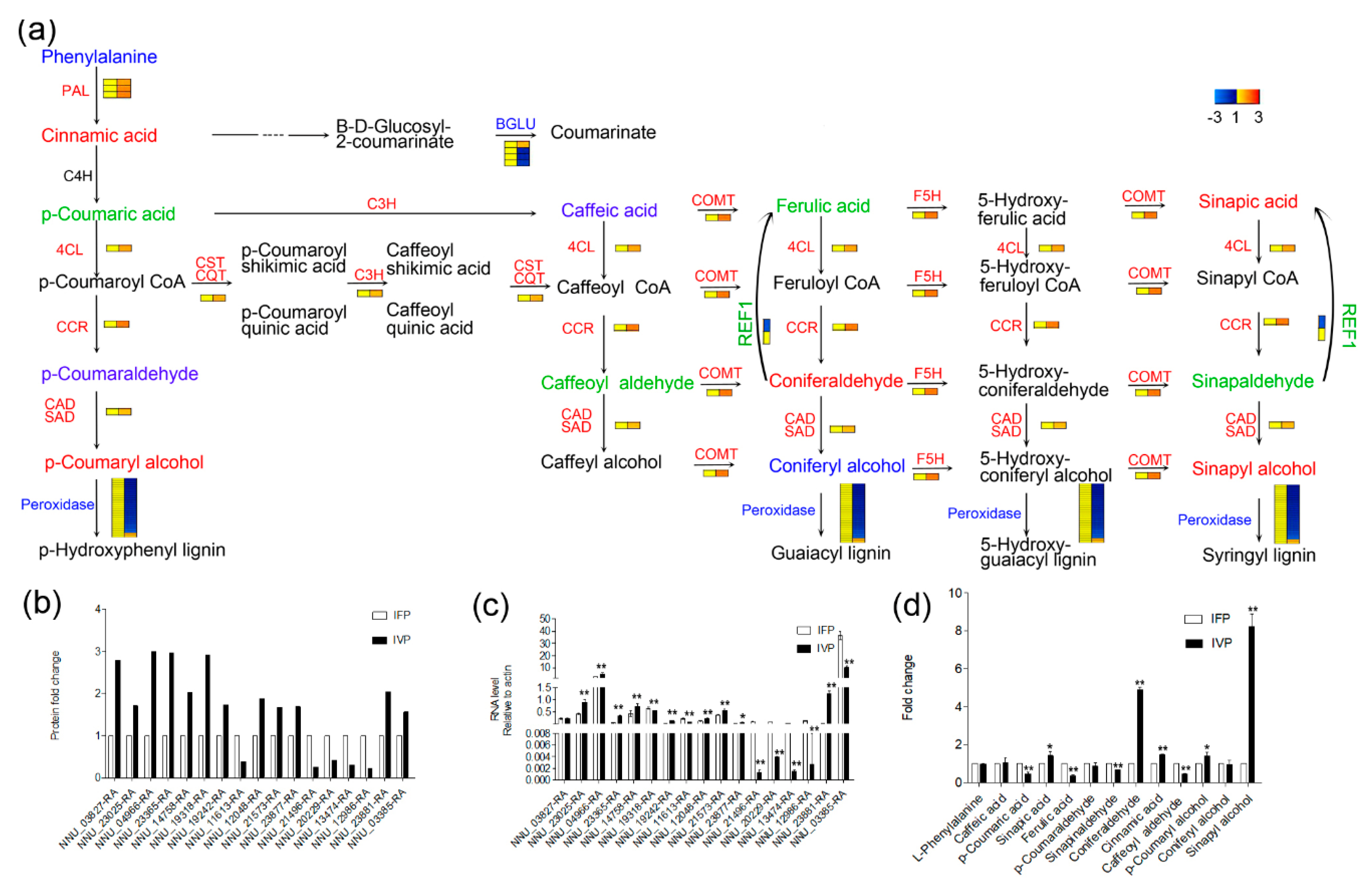
2.7. Polysaccharide and Lignin Biosynthesis Pathway Were Highly Activated in IVP Compared to IFP
3. Discussion
3.1. Genes Involved in Lotus Petioles Formation Exhibit No Synchrony at Transcriptional and Translation Level
3.2. Anatomic Evaluation of Lotus Petioles Indicated Cell Wall Thickness and Cell Differentiation Affect Petiole Rigidity
3.3. Abundance of Lignin Biosynthesis Related Proteins Significantly Higher in IVP Compared to IFP Supports Contribution of Lignin Biosynthesis in Petiole Rigidity
3.4. Abundance of Cell Wall Polysaccharides Biosynthesis Related Proteins Significantly Higher in IVP Than That in IFP
4. Materials and Methods
4.1. Plant Growth and Petiole Collection
4.2. Morphologic Observation
4.2.1. Petiole Microscopic Observation
4.2.2. Breaking Resistance Measurement
4.2.3. Transmission Electron Microscope
4.2.4. Determination of Crude Cellulose and Lignin
4.3. RNA Isolation and Illumina Sequencing
4.3.1. Total RNA Extraction
4.3.2. cDNA Synthesis and Illumina Sequencing
4.3.3. Bioinformatics Analysis
4.4. Proteomics Analysis
4.4.1. Protein Extraction and Trypsin Digestion
4.4.2. TMT Labeling
4.4.3. Quantitative Proteomic Analysis by LC-MS/MS
4.4.4. Database Searching
4.4.5. Protein Annotation, Functional Classification and Enrichment
4.5. Metabolism Analysis
4.5.1. Metabolite Extraction
4.5.2. Metabolite Analysis
4.6. Validation of Transcriptomic and Proteomic Data Using qRT-PCR and PRM Analysis
4.6.1. qRT–PCR
4.6.2. Parallel Reaction Monitoring (PRM)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| ACN | acetonitrile |
| CESA | cellulose synthase |
| DEGs | differentially expressed genes |
| DEPs | differentially expressed proteins |
| DTT | dl-dithiothreitol |
| EDTA | ethylenediaminetetraacetic acid |
| FAA | formaldehyde–acetic acid–alcohol solution |
| FA | formic acid |
| FDR | false discovery rate |
| GA | gibberellin |
| IAM | iodoacetamide |
| IFP | initial floating leaves’ petiole |
| IRX | irregular xylem |
| IVP | initial vertical leaves’ petiole |
| KEGG | Kyoto Gene Ontology and Encyclopedia of Genes and Genomes |
| MFP | mature floating leaves’ petiole |
| MVP | mature vertical leaves’ petiole |
| NCE | normalized collision energy |
| PBS | phosphate buffered solution |
| qRT-PCR | quantitative reverse transcription polymerase chain reaction |
| SUSY | sucrose synthase |
| TEAB | triethylammonium bicarbonate |
| TMT | tandem mass tag |
| UGD | UDP-glucose 6-dehydrogenase |
| UPLC | ultra-performance liquid chromatography system |
| UXS | UDP-glucuronic acid decarboxylase |
References
- Wang, B.; Smith, S.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, S.; Cheng, W.; Fu, Y.; Leng, J.; Yuan, X.; Jiang, N.; Ma, J.; Feng, X. GmILPA1, Encoding an APC8-like Protein, Controls Leaf Petiole Angle in Soybean. Plant Physiol. 2017, 174, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Jo, I.S.; Han, D.U.; Cho, Y.J.; Lee, E.J. Effects of Light, Temperature, and Water Depth on Growth of a Rare Aquatic Plant, Ranunculus kadzusensis. J. Plant Biol. 2010, 53, 88–93. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Zhang, R.; Li, L. Research progress of the plant cell wall signaling. Plant Physiol. J. 2018, 54, 1254–1262. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, Y. Plant Cell Wall Formation and Regulation. Sci. Sin. Vitae 2015, 45, 544–556. [Google Scholar] [CrossRef]
- Burton, R.A.; Gidley, M.J.; Fincher, G.B. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 2010, 6, 724–732. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef]
- Wolf, S. Plant cell wall signalling and receptor-like kinases. Biochem. J. 2017, 474, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Gonneau, M.; Desprez, T.; Guillot, A.; Vernhettes, S.; Höfte, H. Catalytic Subunit Stoichiometry within the Cellulose Synthase Complex. Plant Physiol. 2014, 166, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; Hammudi, M.B.; Tien, M. The Arabidopsis Cellulose Synthase Complex: A Proposed Hexamer of CESA Trimers in an Equimolar Stoichiometry. Plant Cell 2014, 26, 4834–4842. [Google Scholar] [CrossRef] [PubMed]
- Grantham, N.; Wurman Rodrich, J.; Terrett, O.; Lyczakowski, J.; Stott, K.; Iuga, D.; Simmons, T.; Durand-Tardif, M.; Brown, S.P.; Dupree, R.; et al. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat. Plants 2017, 3, 859–865. [Google Scholar] [CrossRef]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef]
- Zhong, R.; Demura, T.; Yu, L. SND1, a NAC Domain Transcription Factor, Is a Key Regulator of Secondary Wall Synthesis in Fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-C.; Ko, J.-H.; Han, K.-H. Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol. 2012, 78, 489–501. [Google Scholar] [CrossRef]
- Sato, M.; Tsutsumi, M.; Ohtsubo, A.; Nishii, K.; Kuwabara, A.; Nagata, T. Temperature-dependent changes of cell shape during heterophyllous leaf formation in Ludwigia arcuata (Onagraceae). Planta 2008, 228, 27–36. [Google Scholar] [CrossRef]
- Jackson, M.B. Ethylene-promoted Elongation: An Adaptation to Submergence Stress. Ann. Bot. 2008, 101, 229–248. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Keuskamp, D.H.; Kooke, R.; Voesenek, L.A.C.J.; Pierik, R. Interactions between Auxin, Microtubules and XTHsMediate Green Shade- Induced Petiole Elongation in Arabidopsis. PLoS ONE 2013, 9, e90587. [Google Scholar] [CrossRef]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Manisseri, C.; Fagerstrom, A.; Peck, M.L.; Vega-Sanchez, M.E.; Williams, B.; Chiniquy, D.M.; Saha, P.; Pattathil, S.; Conlin, B.; et al. Cell Wall Composition and Candidate Biosynthesis Gene Expression During Rice Development. Plant Cell Physiol. 2016, 57, 2058–2075. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Song, D.; Sun, J.; Shen, J.; Li, L. Formation of wood secondary cell wall may involve two type cellulose synthase complexes in Populus. Plant Mol. Biol. 2017, 93, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Li, W.; Schmidt, W. Complementary Proteome and Transcriptome Profiling in Phosphate-Deficient Arabidopsis Roots Reveals Multiple Levels of Gene Regulation. Mol. Cell. Proteom. 2012, 11, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Busch, W.; Benfey, P. Omics meet networks—Using systems approaches to infer regulatory networks in plants. Curr. Opin. Plant Biol. 2010, 13, 126–131. [Google Scholar] [CrossRef]
- Jamet, E.; Roujol, D.; San-Clemente, H.; Irshad, M.; Soubigou-Taconnat, L.; Renou, J.P.; Pont-Lezica, R. Cell wall biogenesis of Arabidopsis thaliana elongating cells: Transcriptomics complements proteomics. BMC Genom. 2009, 10, 505. [Google Scholar] [CrossRef]
- Minic, Z.; Jamet, E.; San-Clemente, H.; Pelletier, S.; Renou, J.P.; Rihouey, C.; Okinyo, D.P.; Proux, C.; Lerouge, P.; Jouanin, L. Transcriptomic analysis of Arabidopsis developing stems: A close-up on cell wall genes. BMC Plant Biol. 2009, 9, 6. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, Z.; Zhang, X.; Liu, Z.; Li, G.; Wang, S.; Ding, Y. Complementary Proteome and Transcriptome Profiling in Developing Grains of a Notched-Belly Rice Mutant Reveals Key Pathways Involved in Chalkiness Formation. Plant Cell Physiol. 2017, 58, 560–573. [Google Scholar] [CrossRef]
- Li, J.; Ren, L.; Gao, Z.; Jiang, M.; Liu, Y.; Zhou, L.; He, Y.; Chen, H. Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Cell Environ. 2017, 40, 3069–3087. [Google Scholar] [CrossRef] [PubMed]
- Casas-Vila, N.; Bluhm, A.; Sayols, S.; Dinges, N.; Dejung, M.; Altenhein, T.; Kappei, D.; Altenhein, B.; Roignant, J.Y.; Butter, F. The developmental proteome of Drosophila melanogaster. Genome Res. 2017, 27, 1273–1285. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Vanburen, R.; Liu, Y.; Yang, M.; Han, Y.; Li, L.-T.; Zhang, Q.; Kim, M.-J.; Schatz, M.C.; Campbell, M.; et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 2013, 14, R41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nyong, A.T.; Shi, T.; Yang, P. The complexity of alternative splicing and landscape of tissue-specific expression in lotus (Nelumbo nucifera) unveiled by Illumina- and single-molecule real-time-based RNA-sequencing. DNA Res. 2019, 26, 301–311. [Google Scholar] [CrossRef]
- Yang, M.; Xu, L.; Liu, Y.; Yang, P. RNA-Seq Uncovers SNPs and Alternative Splicing Events in Asian Lotus (Nelumbo nucifera). PLoS ONE 2015, 10, e0125702. [Google Scholar] [CrossRef]
- Chen, D.; Melton, L.D.; Zujovic, Z.; Harris, P.J. Developmental changes in collenchyma cell-wall polysaccharides in celery (Apium graveolens L.) petioles. BMC Plant Biol. 2019, 19, 81. [Google Scholar] [CrossRef]
- Khan, G.A.; Persson, S. Cell Wall Biology: Dual Control of Cellulose Synthase Guidance. Curr. Biol. 2020, 30, R232–R234. [Google Scholar] [CrossRef]
- Endler, A.; Persson, S. Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 2011, 4, 199–211. [Google Scholar] [CrossRef]
- Kuang, B.; Zhao, X.; Zhou, C.; Zeng, W.; Ren, J.; Ebert, B.; Beahan, C.T.; Deng, X.; Zeng, Q.; Zhou, G.; et al. Role of UDP-Glucuronic Acid Decarboxylase in Xylan Biosynthesis in Arabidopsis. Mol. Plant 2016, 9, 1119–1131. [Google Scholar] [CrossRef]
- Xu, P.; Kong, Y.; Li, X.; Li, L. Identification of molecular processes needed for vascular formation through transcriptome analysis of different vascular systems. BMC Genom. 2013, 14, 217. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, D.; Sun, J.; Wang, X.; Li, L. PtrHB7, a class III HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol. Plant 2013, 6, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, J.; Xu, P.; Zhang, R.; Li, L. Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 2014, 164, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, H.; Zhang, B.; Fei, B.; Burgert, I. Cell wall structure and formation of maturing fibres of moso bamboo (Phyllostachys pubescens) increase buckling resistance. J. R. Soc. Interface 2012, 9, 988–996. [Google Scholar] [CrossRef] [PubMed]
- La Camera, S.; Gouzerh, G.; Dhondt, S.; Hoffmann, L.; Fritig, B.; Legrand, M.; Heitz, T. Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2004, 198, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Pollet, B.; Legrand, M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 2007, 19, 148–162. [Google Scholar] [CrossRef]
- Lan, W.; Lu, F.; Regner, M.; Zhu, Y.; Rencoret, J.; Ralph, S.A.; Zakai, U.I.; Morreel, K.; Boerjan, W.; Ralph, J. Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 2015, 167, 1284–1295. [Google Scholar] [CrossRef]
- McFarlane, H.E.; Doring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef]
- Guerriero, G.; Fugelstad, J.; Bulone, V. What do we really know about cellulose biosynthesis in higher plants? J. Integr. Plant Biol. 2010, 52, 161–175. [Google Scholar] [CrossRef]
- Richmond, T.A.; Somerville, C.R. The cellulose synthase superfamily. Plant Physiol. 2000, 124, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Elliott, J.E.; Williamson, R.E. Features of the primary wall CESA complex in wild type and cellulose-deficient mutants of Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 2627–2637. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Chan, W.C.W. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Desprez, T.; Juraniec, M.; Crowell, E.F.; Jouy, H.; Pochylova, Z.; Parcy, F.; Hofte, H.; Gonneau, M.; Vernhettes, S. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 15572–15577. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Shen, J.; Li, L. Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol. 2010, 187, 777–790. [Google Scholar] [CrossRef]
- Mutwil, M.; Debolt, S.; Persson, S. Cellulose synthesis: A complex complex. Curr. Opin. Plant Biol. 2008, 11, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, H.; Sun, J.; Li, L. PtrKOR1 is required for secondary cell wall cellulose biosynthesis in Populus. Tree Physiol. 2014, 34, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Sun, J.; Li, L. PtrCel9A6, an endo-1,4-beta-glucanase, is required for cell wall formation during xylem differentiation in Populus. Mol. Plant 2013, 6, 1904–1917. [Google Scholar] [CrossRef]
- Kim, W.C.; Ko, J.H.; Kim, J.Y.; Kim, J.; Bae, H.J.; Han, K.H. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 2013, 73, 26–36. [Google Scholar] [CrossRef]
- Coleman, H.D.; Yan, J.; Mansfield, S.D. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. USA 2009, 106, 13118–13123. [Google Scholar] [CrossRef]
- Klinghammer, M.; Tenhaken, R. Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J. Exp. Bot. 2007, 58, 3609–3621. [Google Scholar] [CrossRef]
- Ebert, B.; Rautengarten, C.; Guo, X.; Xiong, G.; Stonebloom, S.; Smith-Moritz, A.M.; Herter, T.; Chan, L.J.; Adams, P.D.; Petzold, C.J.; et al. Identification and Characterization of a Golgi-Localized UDP-Xylose Transporter Family from Arabidopsis. Plant Cell 2015, 27, 1218–1227. [Google Scholar] [CrossRef]
- Jensen, J.K.; Johnson, N.R.; Wilkerson, C.G. Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J. 2014, 80, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhen, L.; Tan, X.; Li, L.; Wang, X. The involvement of hexokinase in the coordinated regulation of glucose and gibberellin on cell wall invertase and sucrose synthesis in grape berry. Mol. Biol. Rep. 2014, 41, 7899–7910. [Google Scholar] [CrossRef]
- Frankova, L.; Fry, S.C. Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. J. Exp. Bot. 2013, 64, 3519–3550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Q. Preliminary study of the ecotypes of genetic resources of tropical lotus. Landsc. Plants 2006, 20, 82–85. [Google Scholar]
- Yang, M.; Zhu, L.; Xu, L.; Pan, C.; Liu, Y. Comparative transcriptomic analysis of the regulation of flowering in temperate and tropical lotus (Nelumbo nucifera) by RNA-Seq. Ann. Appl. Biol. 2014, 165, 73–95. [Google Scholar] [CrossRef]
- Hai, L.; Guo, H.; Xiao, S.; Jiang, G.; Zhang, X.; Yan, C.; Xin, Z.; Jia, J. Quantitative trait loci (QTL) of stem strength and related traits in a doubled-haploid population of wheat (Triticum aestivum L.). Euphytica 2005, 141, 1–9. [Google Scholar] [CrossRef]
- Yadav, S.; Singh, U.M.; Naik, S.M.; Venkateshwarlu, C.; Ramayya, P.J.; Raman, K.A.; Sandhu, N.; Kumar, A. Molecular Mapping of QTLs Associated with Lodging Resistance in Dry Direct-Seeded Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1431. [Google Scholar] [CrossRef]
- Lin, S.Y.; Dence, C.W. Methods in Lignin Chemistry; Springer: Berlin, Germany, 1992. [Google Scholar]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Guerreiro, A.C.; Benevento, M.; Lehmann, R.; van Breukelen, B.; Post, H.; Giansanti, P.; Maarten Altelaar, A.F.; Axmann, I.M.; Heck, A.J. Daily rhythms in the cyanobacterium synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol. Cell. Proteom. 2014, 13, 2042–2055. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Aoki-Kinoshita, K.F.; Kanehisa, M. Gene Annotation and Pathway Mapping in KEGG; Humana Press Inc.: Totowa, NJ, USA, 2007; Volume 396. [Google Scholar]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Vizcaino, J.A.; Cote, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef] [PubMed]


| ID | Name | Log2(IVP/IFP) | Log2(IVP/IFP) |
|---|---|---|---|
| cell wall precursor synthesis | |||
| NNU_03659-RA | UDP-glucose 6-dehydrogenase 4-like | 0.9 | Null |
| NNU_04520-RA | UDP-glucose 6-dehydrogenase 1 | 0.7 | Null |
| NNU_07386-RA | UDP-glucose 6-dehydrogenase 1-like | 0.9 | Null |
| a NNU_08832-RA | probable rhamnose biosynthetic enzyme 1 | 1.2 | 1.3 |
| NNU_10172-RA | probable rhamnose biosynthetic enzyme 1 | 0.8 | Null |
| NNU_12302-RA | UDP-glucuronic acid decarboxylase 6 | 0.8 | Null |
| NNU_15122-RA | UDP-glucuronic acid decarboxylase 6 | 1.2 | Null |
| NNU_25253-RA | hypothetical protein OsI_05369 | 0.6 | Null |
| NNU_26559-RA | probable rhamnose biosynthetic enzyme 1 | 1.0 | Null |
| NNU_00650-RA | probable arabinose 5-phosphate isomerase | Null | 1.4 |
| NNU_21054-RA | bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 | Null | 1.9 |
| cell wall proteins | |||
| NNU_01080-RA | glucomannan 4-beta-mannosyltransferase 2-like | 0.9 | Null |
| NNU_25605-RA | fasciclin-like arabinogalactan protein 17 | 0.7 | Null |
| a NNU_11213-RA | fasciclin-like arabinogalactan protein 13 | −1.6 | −3.2 |
| NNU_12269-RA | fasciclin-like arabinogalactan protein 4 | Null | 1.4 |
| NNU_15965-RA | fasciclin-like arabinogalactan protein 7 | Null | 1.8 |
| NNU_06301-RA | leucine-rich repeat extensin-like protein 6 | Null | −8.3 |
| NNU_16861-RA | leucine-rich repeat extensin-like protein 4 | Null | 3.0 |
| NNU_24457-RA | leucine-rich repeat extensin-like protein 4 | Null | 3.0 |
| NNU_25277-RA | glucomannan 4-beta-mannosyltransferase 9 | Null | 1.7 |
| cell wall degradation | |||
| NNU_05055-RA | probable polygalacturonase | 0.9 | Null |
| NNU_11467-RA | hypothetical protein PHAVU_009G016100 g | 2.2 | Null |
| NNU_11761-RA | probable polygalacturonase | 1.3 | Null |
| NNU_23253-RA | probable pectate lyase 18 | 0.7 | Null |
| NNU_23813-RA | GDSL esterase/lipase At5g14450 isoform X3 | 0.9 | Null |
| NNU_00300-RA | probable polygalacturonase isoform X1 | −0.7 | Null |
| NNU_05224-RA | probable rhamnogalacturonate lyase B isoform X1 | −0.8 | Null |
| a NNU_10867-RA | alpha-L-arabinofuranosidase 1-like | −1.2 | −2.1 |
| NNU_11580-RA | polygalacturonase inhibitor-like | −0.9 | Null |
| NNU_13918-RA | putative beta-D-xylosidase | −0.9 | Null |
| NNU_22026-RA | probable pectate lyase 18 | −2.9 | Null |
| NNU_07943-RA | probable polygalacturonase | Null | −2.3 |
| NNU_11529-RA | probable polygalacturonase isoform X2 | Null | −1.9 |
| NNU_11581-RA | polygalacturonase inhibitor-like | Null | −9.0 |
| NNU_18600-RA | lysosomal beta glucosidase-like isoform X4 | Null | −1.0 |
| NNU_19090-RA | polygalacturonase inhibitor-like | Null | −2.8 |
| NNU_13976-RA | probable polygalacturonase | Null | 1.3 |
| NNU_23205-RA | probable polygalacturonase isoform X1 | Null | 1.3 |
| NNU_23792-RA | probable polygalacturonase non-catalytic subunit JP650 | Null | 1.8 |
| NNU_26608-RA | alpha-L-fucosidase 1-like | Null | 1.1 |
| cell wall modification | |||
| a NNU_15029-RA | xyloglucan endotransglucosylase/hydrolase protein 22-like | 1.1 | 1.9 |
| a NNU_16408-RA | probable xyloglucan endotransglucosylase/hydrolase protein 8 | 0.9 | 1.6 |
| NNU_17562-RA | expansin-A13-like | 0.7 | Null |
| NNU_25629-RA | probable xyloglucan endotransglucosylase/hydrolase protein 6 | 1.0 | Null |
| NNU_12232-RA | expansin-A8-like | −1.1 | Null |
| NNU_12958-RA | expansin-A4 | −0.8 | Null |
| NNU_24404-RA | expansin-A8-like | −1.1 | Null |
| NNU_24832-RA | probable xyloglucan endotransglucosylase/hydrolase protein 23 | −0.8 | Null |
| NNU_20658-RA | pectinesterase-like | 0.9 | Null |
| a NNU_01721-RA | probable pectinesterase/pectinesterase inhibitor 51 | −1.1 | −1.6 |
| NNU_05006-RA | pectinesterase | −1.9 | Null |
| NNU_05086-RA | pectinesterase | −1.9 | Null |
| NNU_08272-RA | protein notum homolog | −1.1 | Null |
| NNU_11705-RA | pectinesterase-like | −1.5 | Null |
| NNU_15192-RA | pectinesterase-like | −0.8 | Null |
| NNU_18238-RA | pectinesterase 2-like | −0.7 | Null |
| NNU_05158-RA | putative expansin-A17 isoform X1 | Null | −8.6 |
| NNU_05160-RA | putative expansin-A17 | Null | −4.6 |
| NNU_23652-RA | expansin-A15-like | Null | −2.9 |
| NNU_15361-RA | brassinosteroid-regulated protein BRU1-like | Null | 1.6 |
| NNU_16495-RA | xyloglucan endotransglucosylase/hydrolase protein 9 | Null | 2.3 |
| NNU_24956-RA | probable xyloglucan endotransglucosylase/hydrolase protein 33 | Null | 4.4 |
| NNU_12324-RA | probable pectinesterase/pectinesterase inhibitor 41 | Null | −2.6 |
| NNU_14002-RA | pectinesterase-like | Null | −1.7 |
| NNU_14557-RA | pectinesterase 2-like | Null | −5.6 |
| NNU_24388-RA | probable pectinesterase/pectinesterase inhibitor 41 | Null | −1.4 |
| NNU_05007-RA | pectinesterase/pectinesterase inhibitor PPE8B | Null | 2.3 |
| NNU_18245-RA | probable pectinesterase/pectinesterase inhibitor 41 | Null | 3.3 |
| NNU_18519-RA | L-ascorbate oxidase homolog | Null | 1.7 |
| cellulose synthase | |||
| NNU_09561-RA | probable cellulose synthase A catalytic subunit 5 | 2.4 | Null |
| NNU_12044-RA | cellulose synthase A catalytic subunit 7 | 1.3 | Null |
| NNU_21632-RA | probable cellulose synthase A catalytic subunit 1 | 1.2 | Null |
| NNU_07451-RA | protein COBRA-like isoform X1 | 1.4 | Null |
| NNU_07455-RA | COBRA-like protein 4 | 1.3 | Null |
| NNU_13962-RA | COBRA-like protein 7 | 1.3 | Null |
| NNU_21801-RA | endoglucanase 9-like | 1.1 | Null |
| NNU_18140-RA | cellulose synthase-like protein G3 | Null | 1.5 |
| NNU_20039-RA | cellulose synthase A catalytic subunit 2 | Null | 1.8 |
| NNU_10059-RA | endoglucanase 12-like | Null | −5.2 |
| NNU_07071-RA | xyloglucan glycosyltransferase 4 isoform X1 | Null | 3.4 |
| hemicellulose synthesis | |||
| NNU_01719-RA | putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 | 1.2 | Null |
| a NNU_10542-RA | xyloglucan galactosyltransferase KATAMARI1-like | 0.7 | −1.7 |
| NNU_12476-RA | probable beta-1,4-xylosyltransferase IRX9 isoform X1 | 2.0 | Null |
| NNU_13626-RA | probable beta-1,4-xylosyltransferase IRX14H | 1.5 | Null |
| lignin biosynthesis | |||
| NNU_03759-RA | laccase-4-like | 0.7 | Null |
| NNU_03827-RA | phenylalanine ammonia-lyase | 1.5 | Null |
| NNU_04966-RA | caffeic acid 3-O-methyltransferase 1 | 1.6 | Null |
| NNU_06036-RA | isoflavone reductase homolog | 0.6 | Null |
| NNU_12048-RA | shikimate O-hydroxycinnamoyltransferase-like | 0.9 | Null |
| NNU_12868-RA | phenylalanine ammonia-lyase-like | 1.1 | Null |
| NNU_13598-RA | caffeoyl-CoA O-methyltransferase-like | 1.0 | Null |
| NNU_14758-RA | 4-coumarate--CoA ligase 2-like | 1.0 | Null |
| NNU_17055-RA | caffeoyl-CoA O-methyltransferase | 1.5 | Null |
| NNU_18746-RA | laccase-17-like | 0.6 | Null |
| NNU_19318-RA | cinnamoyl-CoA reductase 1-like | 1.5 | Null |
| NNU_21321-RA | phenylalanine ammonia-lyase | 1.4 | Null |
| NNU_23025-RA | cytochrome P450 98A2 | 0.8 | Null |
| NNU_23365-RA | cytochrome P450 84A1-like | 1.6 | Null |
| a NNU_23877-RA | probable cinnamyl alcohol dehydrogenase 6 | 0.8 | 3.6 |
| NNU_24517-RA | cinnamoyl-CoA reductase 2 | −0.8 | Null |
| NNU_04106-RA | phenylalanine ammonia-lyase-like | Null | −2.1 |
| NNU_05129-RA | phenylalanine ammonia-lyase-like | Null | −3.6 |
| NNU_07568-RA | cinnamoyl-CoA reductase 2 isoform X2 | Null | −1.4 |
| NNU_11085-RA | laccase-17-like | Null | −5.6 |
| NNU_12126-RA | cinnamoyl-CoA reductase 1-like | Null | −5.0 |
| NNU_18647-RA | laccase-7-like | Null | −7.3 |
| NNU_08076-RA | cytochrome P450 84A1-like | Null | 1.8 |
| NNU_22838-RA | cinnamoyl-CoA reductase 2-like | Null | 2.1 |
| major CHO metabolism | |||
| NNU_02421-RA | maltose excess protein 1-like, chloroplastic | 0.6 | Null |
| NNU_04943-RA | alkaline/neutral invertase CINV2 | 1.0 | Null |
| a NNU_17880-RA | probable fructokinase-1 | 1.2 | 1.6 |
| a NNU_18248-RA | beta-fructofuranosidase, soluble isoenzyme I-like | 0.9 | 2.3 |
| NNU_19077-RA | sucrose synthase | 1.0 | Null |
| NNU_04529-RA | alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic | −0.8 | Null |
| NNU_05767-RA | sucrose synthase 2-like | −1.1 | Null |
| a NNU_11941-RA | beta-fructofuranosidase, insoluble isoenzyme CWINV3-like isoform X1 | −0.9 | −2.7 |
| NNU_13572-RA | pentatricopeptide repeat-containing protein At2g04860 | −1.7 | Null |
| NNU_18912-RA | phosphoglucan phosphatase DSP4, amyloplastic-like | −0.7 | Null |
| NNU_08846-RA | probable fructokinase-7 | Null | 1.8 |
| NNU_09096-RA | alkaline/neutral invertase CINV2 | Null | 1.3 |
| Protein Accession | Protein Name | VvsF Ratio | Regulated Type | p-Value |
|---|---|---|---|---|
| Biosynthesis of amino acids | ||||
| NNU_09156-RA | S-adenosylmethionine synthase 5 | 2.5 | Up | 0 |
| NNU_25903-RA | S-adenosylmethionine synthase 2 | 2.3 | Up | 0 |
| NNU_07815-RA | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 2-like | 2.3 | Up | 0.001 |
| NNU_06116-RA | glutamine synthetase leaf isozyme, chloroplastic | 2.2 | Up | 0 |
| NNU_15496-RA | pyrroline-5-carboxylate reductase isoform X1 | 2.2 | Up | 0.002 |
| NNU_16927-RA | S-adenosylmethionine synthase 1 | 1.9 | Up | 0 |
| NNU_22690-RA | phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplastic-like | 1.9 | Up | 0 |
| NNU_02525-RA | glutamate synthase 1 | 1.8 | Up | 0 |
| NNU_16800-RA | serine acetyltransferase 5-like | 1.8 | Up | 0.003 |
| NNU_18211-RA | 3-phosphoshikimate 1-carboxyvinyltransferase 2 | 1.7 | Up | 0.001 |
| NNU_21805-RA | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 1.7 | Up | 0 |
| NNU_18019-RA | serine hydroxymethyltransferase 4 | 1.7 | Up | 0 |
| NNU_01496-RA | glutamate synthase 1 | 1.7 | Up | 0.031 |
| NNU_13370-RA | phospho-2-dehydro-3-deoxyheptonate aldolase 2, chloroplastic-like | 1.7 | Up | 0 |
| NNU_04572-RA | chorismate mutase 3, chloroplastic-like | 1.7 | Up | 0 |
| NNU_16636-RA | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | 1.6 | Up | 0 |
| NNU_17273-RA | probable fructose-bisphosphate aldolase 3, chloroplastic | 1.6 | Up | 0 |
| NNU_18207-RA | transketolase, chloroplastic | 1.6 | Up | 0 |
| NNU_20134-RA | shikimate kinase, chloroplastic isoform X1 | 1.6 | Up | 0 |
| NNU_26622-RA | indole-3-glycerol phosphate synthase, chloroplastic-like isoform X1 | 1.5 | Up | 0.015 |
| NNU_01941-RA | aspartokinase 2, chloroplastic-like isoform X1 | 1.5 | Up | 0 |
| NNU_14707-RA | D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like | 1.5 | Up | 0 |
| NNU_13158-RA | chorismate synthase, chloroplastic isoform X1 | 1.5 | Up | 0.015 |
| NNU_20724-RA | glutamine synthetase cytosolic isozyme 1 | 1.5 | Up | 0 |
| NNU_21817-RA | phosphoserine aminotransferase 1, chloroplastic-like | 1.5 | Up | 0 |
| Flavonoid biosynthesis | ||||
| NNU_16100-RA | naringenin, 2-oxoglutarate 3-dioxygenase-like | 4.4 | Up | 0 |
| NNU_24753-RA | flavonol synthase/flavanone 3-hydroxylase-like | 2.7 | Up | 0.002 |
| NNU_19543-RA | flavonol synthase/flavanone 3-hydroxylase | 2.0 | Up | 0 |
| NNU_12048-RA | shikimate O-hydroxycinnamoyltransferase-like | 1.9 | Up | 0 |
| NNU_04498-RA | flavonoid 3′-monooxygenase-like | 1.7 | Up | 0 |
| NNU_23025-RA | cytochrome P450 98A2 | 1.7 | Up | 0 |
| NNU_08856-RA | leucoanthocyanidin dioxygenase-like | 1.6 | Up | 0 |
| Phenylpropanoid biosynthesis | ||||
| NNU_04966-RA | caffeic acid 3-O-methyltransferase 1 | 3.0 | Up | 0 |
| NNU_23365-RA | cytochrome P450 84A1-like | 3.0 | Up | 0 |
| NNU_19318-RA | cinnamoyl-CoA reductase 1-like | 2.9 | Up | 0.001 |
| NNU_03827-RA | phenylalanine ammonia–lyase | 2.8 | Up | 0 |
| NNU_21321-RA | phenylalanine ammonia–lyase | 2.6 | Up | 0.001 |
| NNU_12868-RA | phenylalanine ammonia–lyase-like | 2.2 | Up | 0.001 |
| NNU_23881-RA | peroxidase 64 | 2.0 | Up | 0 |
| NNU_14758-RA | 4-coumarate--CoA ligase 2-like | 2.0 | Up | 0 |
| NNU_12048-RA | shikimate O-hydroxycinnamoyltransferase-like | 1.9 | Up | 0 |
| NNU_23025-RA | cytochrome P450 98A2 | 1.7 | Up | 0 |
| NNU_23877-RA | probable cinnamyl alcohol dehydrogenase 6 | 1.7 | Up | 0.001 |
| NNU_21573-RA | caffeoylshikimate esterase | 1.7 | Up | 0 |
| NNU_03385-RA | peroxidase 42-like | 1.6 | Up | 0.006 |
| NNU_13717-RA | peroxidase 73-like | 0.6 | Down | 0.001 |
| NNU_16422-RA | peroxidase 27-like | 0.6 | Down | 0 |
| NNU_04050-RA | peroxidase P7-like | 0.6 | Down | 0 |
| NNU_20058-RA | peroxidase 12-like | 0.6 | Down | 0 |
| NNU_18799-RA | peroxidase 4-like | 0.6 | Down | 0 |
| NNU_23337-RA | cationic peroxidase 1-like | 0.6 | Down | 0 |
| NNU_04265-RA | peroxidase 27-like | 0.6 | Down | 0.012 |
| NNU_04268-RA | peroxidase 3-like | 0.5 | Down | 0 |
| NNU_22685-RA | peroxidase 17-like | 0.5 | Down | 0 |
| NNU_13190-RA | peroxidase 21-like | 0.5 | Down | 0 |
| NNU_20096-RA | cationic peroxidase 1-like | 0.5 | Down | 0 |
| NNU_02934-RA | peroxidase N-like | 0.5 | Down | 0 |
| NNU_13360-RA | peroxidase 17-like | 0.5 | Down | 0.003 |
| NNU_24553-RA | peroxidase 47-like | 0.5 | Down | 0.001 |
| NNU_12989-RA | peroxidase 2-like | 0.5 | Down | 0 |
| NNU_06410-RA | peroxidase 12-like | 0.5 | Down | 0 |
| NNU_20132-RA | peroxidase 3-like | 0.5 | Down | 0.004 |
| NNU_01736-RA | peroxidase 43-like isoform X1 | 0.4 | Down | 0 |
| NNU_20229-RA | peroxidase N1-like | 0.4 | Down | 0 |
| NNU_02064-RA | peroxidase 51 | 0.4 | Down | 0 |
| NNU_11196-RA | peroxidase P7-like | 0.4 | Down | 0 |
| NNU_00369-RA | peroxidase 57-like | 0.3 | Down | 0 |
| NNU_13474-RA | peroxidase 10 | 0.3 | Down | 0 |
| NNU_21496-RA | aldehyde dehydrogenase family 2 member C4-like | 0.3 | Down | 0 |
| NNU_04048-RA | peroxidase P7-like | 0.2 | Down | 0.003 |
| NNU_12986-RA | peroxidase P7-like | 0.2 | Down | 0 |
| Amino sugar and nucleotide sugar metabolism | ||||
| NNU_05331-RA | glucose-1-phosphate adenylyltransferase large subunit 1-like | 0.7 | Down | 0.012 |
| NNU_12353-RA | beta-hexosaminidase 1 isoform X1 | 0.6 | Down | 0.001 |
| NNU_09609-RA | basic endochitinase-like | 0.6 | Down | 0 |
| NNU_10055-RA | beta-hexosaminidase 3-like | 0.6 | Down | 0 |
| NNU_06174-RA | glucose-1-phosphate adenylyltransferase large subunit 3, chloroplastic/amyloplastic | 0.6 | Down | 0 |
| NNU_10728-RA | acidic endochitinase-like | 0.5 | Down | 0 |
| NNU_12150-RA | G-type lectin S-receptor-like serine/threonine–protein kinase At1g11300 | 0.5 | Down | 0 |
| NNU_17908-RA | endochitinase PR4-like | 0.5 | Down | 0 |
| NNU_17907-RA | endochitinase PR4-like | 0.4 | Down | 0 |
| NNU_10867-RA | alpha-l-arabinofuranosidase 1-like | 0.4 | Down | 0 |
| NNU_20770-RA | acidic endochitinase-like | 0.4 | Down | 0 |
| NNU_14306-RA | basic endochitinase-like | 0.4 | Down | 0 |
| NNU_22938-RA | chitinase 5-like, partial | 0.3 | Down | 0 |
| NNU_11632-RA | acidic endochitinase-like | 0.3 | Down | 0 |
| NNU_09610-RA | endochitinase A-like | 0.3 | Down | 0 |
| NNU_17910-RA | endochitinase PR4-like | 0.3 | Down | 0 |
| NNU_12151-RA | acidic mammalian chitinase-like | 0.3 | Down | 0 |
| NNU_22939-RA | chitinase 5-like | 0.2 | Down | 0 |
| NNU_24291-RA | acidic endochitinase-like, partial | 0.2 | Down | 0 |
| Starch and sucrose metabolism | ||||
| NNU_24463-RA | glucan endo-1,3-beta-glucosidase 6-like | 0.7 | Down | 0.001 |
| NNU_05331-RA | glucose-1-phosphate adenylyltransferase large subunit 1-like | 0.7 | Down | 0.012 |
| NNU_26480-RA | probable sucrose-phosphate synthase 1 isoform X1 | 0.6 | Down | 0 |
| NNU_04529-RA | alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic | 0.6 | Down | 0 |
| NNU_06174-RA | glucose-1-phosphate adenylyltransferase large subunit 3, chloroplastic/amyloplastic | 0.6 | Down | 0 |
| NNU_11941-RA | beta-fructofuranosidase, insoluble isoenzyme CWINV3-like isoform X1 | 0.5 | Down | 0.001 |
| NNU_07396-RA | beta-glucosidase 40-like | 0.5 | Down | 0 |
| NNU_11617-RA | beta-glucosidase 12-like | 0.5 | Down | 0 |
| NNU_05767-RA | sucrose synthase 2-like | 0.5 | Down | 0 |
| NNU_11613-RA | beta-glucosidase 12-like | 0.4 | Down | 0 |
| NNU_13572-RA | pentatricopeptide repeat-containing protein At2g04860 | 0.3 | Down | 0 |
| Glycosphingolipid biosynthesis | ||||
| NNU_12353-RA | beta-hexosaminidase 1 isoform X1 | 0.6 | Down | 0.001 |
| NNU_10055-RA | beta-hexosaminidase 3-like | 0.6 | Down | 0 |
| NNU_08403-RA | alpha-galactosidase-like | 0.6 | Down | 0 |
| NNU_24417-RA | alpha-galactosidase isoform X1 | 0.5 | Down | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Hameed, I.; Cao, D.; He, D.; Yang, P. Integrated Omics Analyses Identify Key Pathways Involved in Petiole Rigidity Formation in Sacred Lotus. Int. J. Mol. Sci. 2020, 21, 5087. https://doi.org/10.3390/ijms21145087
Li M, Hameed I, Cao D, He D, Yang P. Integrated Omics Analyses Identify Key Pathways Involved in Petiole Rigidity Formation in Sacred Lotus. International Journal of Molecular Sciences. 2020; 21(14):5087. https://doi.org/10.3390/ijms21145087
Chicago/Turabian StyleLi, Ming, Ishfaq Hameed, Dingding Cao, Dongli He, and Pingfang Yang. 2020. "Integrated Omics Analyses Identify Key Pathways Involved in Petiole Rigidity Formation in Sacred Lotus" International Journal of Molecular Sciences 21, no. 14: 5087. https://doi.org/10.3390/ijms21145087
APA StyleLi, M., Hameed, I., Cao, D., He, D., & Yang, P. (2020). Integrated Omics Analyses Identify Key Pathways Involved in Petiole Rigidity Formation in Sacred Lotus. International Journal of Molecular Sciences, 21(14), 5087. https://doi.org/10.3390/ijms21145087






