Translation Regulation by eIF2α Phosphorylation and mTORC1 Signaling Pathways in Non-Communicable Diseases (NCDs)
Abstract
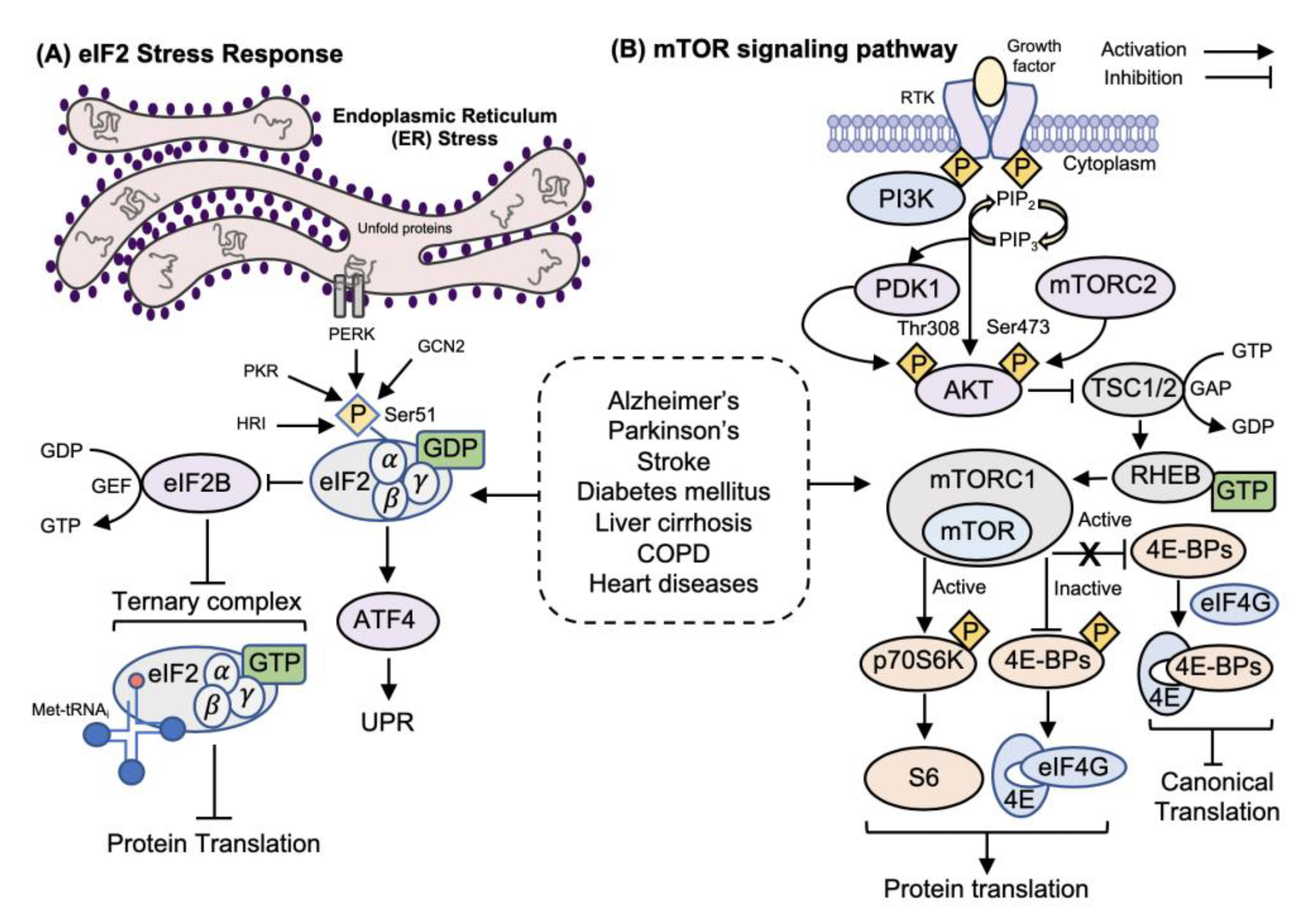
:1. Introduction
1.1. A Brief. Overview of mRNA Translation Initiation in Eukaryotes
1.2. Overview of Regulation of Translation Initiation by the eIF2/ISR and mTORC1 Pathways
1.2.1. eIF2/ISR Regulation of Translation Initiation
1.2.2. mTORC1 Regulation of Translation Initiation
1.2.3. Alternate Mechanisms of mRNA Translation Initiation
2. The eIF2 Stress Response in NCDs
2.1. The eIF2/ISR in Neurodegenerative Disorders
2.2. The eIF2/ISR in Metabolic and Inflammatory Disorders
3. The mTORC1 Signaling Pathway in NCDs
3.1. The mTORC1 Signaling Pathway in Neurodegenerative Disorders
3.2. The mTORC1 Signaling Pathway in Metabolic and Inflammatory Disorders
4. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NCDs | Non-communicable diseases |
| eIF2 | eukaryotic initiation factor 2 |
| mTORC | mammalian target of rapamycin complex |
| 4E-BPs | eukaryotic initiation factor 4E- binding proteins |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| COPD | Chronic obstructive pulmonary disease |
References
- Moriyama, I.M.L.R.; Robb-Smith, A.H.T. History of the Statistical Classification of Diseases and Causes of Death; National Center for Health Statistics: Hyattsville, MD, USA, 2011.
- Budnik, L.T.; Adam, B.; Albin, M.; Banelli, B.; Baur, X.; Belpoggi, F.; Bolognesi, C.; Broberg, K.; Gustavsson, P.; Göen, T.; et al. Diagnosis, monitoring and prevention of exposure-related non-communicable diseases in the living and working environment: DiMoPEx-project is designed to determine the impacts of environmental exposure on human health. J. Occup Med. Toxicol 2018, 13, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Noncommunicable Diseases Country Profiles 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Forouzanfar, M.H.; Afshin, A.; Alexander, L.T.; Anderson, H.R.; Bhutta, Z.A.; Biryukov, S.; Brauer, M.; Burnett, R.; Cercy, K.; Charlson, F.J.; et al. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1659–1724. [Google Scholar] [CrossRef] [Green Version]
- Lawes, C.M.M.; Hoorn, S.V.; Rodgers, A. Global burden of blood-pressure-related disease, 2001. Lancet 2008, 371, 1513–1518. [Google Scholar] [CrossRef]
- Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health. A Report of the Surgeon General: How Tobacco Smoke Causes Disease: What It Means to You; Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health: Atlanta, GA, USA, 2010.
- Rehm, J.; Baliunas, D.; Borges, G.L.G.; Graham, K.; Irving, H.; Kehoe, T.; Parry, C.D.; Patra, J.; Popova, S.; Poznyak, V.; et al. The relation between different dimensions of alcohol consumption and burden of disease: An overview. Addiction 2010, 105, 817–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, A.E.; Coakley, E.H.; Must, A.; Spadano, J.L.; Laird, N.; Dietz, W.H.; Rimm, E.; Colditz, G.A. Impact of Overweight on the Risk of Developing Common Chronic Diseases During a 10-Year Period. Arch. Intern. Med. 2001, 161, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Barouki, R.; Gluckman, P.D.; Grandjean, P.; Hanson, M.; Heindel, J.J. Developmental origins of non-communicable disease: Implications for research and public health. Environ. Health 2012, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Pang, G.; Xie, J.; Chen, Q.; Hu, Z. How functional foods play critical roles in human health. Food Sci. Hum. Wellness 2012, 1, 26–60. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef] [Green Version]
- Bosco, D.A. Translation dysregulation in neurodegenerative disorders. Proc. Natl. Acad. Sci. USA 2018, 115, 12842. [Google Scholar] [CrossRef] [Green Version]
- Zeitz, M.J.; Smyth, J.W. Translating Translation to Mechanisms of Cardiac Hypertrophy. J. Cardiovasc Dev. Dis. 2020, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Meiners, S.; Greene, C.M. Protein quality control in lung disease: It’s all about cloud networking. Eur. Respir. J. 2014, 44, 846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nutter, C.A.; Kuyumcu-Martinez, M.N. Emerging roles of RNA-binding proteins in diabetes and their therapeutic potential in diabetic complications. Wiley Interdiscip Rev. RNA 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Chevet, E.; Harding, H.P. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 2013, 12, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecka, A.; Marcotrigiano, J.; Stepinski, J.; Jankowska-Anyszka, M.; Wyslouch-Cieszynska, A.; Dadlez, M.; Gingras, A.-C.; Mak, P.; Darzynkiewicz, E.; Sonenberg, N.; et al. Biophysical Studies of eIF4E Cap-binding Protein: Recognition of mRNA 5′ Cap Structure and Synthetic Fragments of eIF4G and 4E-BP1 Proteins. J. Mol. Biol. 2002, 319, 615–635. [Google Scholar] [CrossRef]
- Merrick, W.C.; Pavitt, G.D. Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Pain, V.M. Initiation of Protein Synthesis in Eukaryotic Cells. Eur. J. Biochem. 1996, 236, 747–771. [Google Scholar] [CrossRef] [PubMed]
- Pavitt, G.D.; Ramaiah, K.V.; Kimball, S.R.; Hinnebusch, A.G. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998, 12, 514–526. [Google Scholar] [CrossRef] [Green Version]
- Pisareva, V.P.; Pisarev, A.V. eIF5 and eIF5B together stimulate 48S initiation complex formation during ribosomal scanning. Nucleic Acids Res. 2014, 42, 12052–12069. [Google Scholar] [CrossRef]
- Adomavicius, T.; Guaita, M.; Zhou, Y.; Jennings, M.D.; Latif, Z.; Roseman, A.M.; Pavitt, G.D. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 2019, 10, 2136. [Google Scholar] [CrossRef] [Green Version]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef] [Green Version]
- Algire, M.A.; Lorsch, J.R. Where to begin? The mechanism of translation initiation codon selection in eukaryotes. Curr. Opin. Chem. Biol. 2006, 10, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, T.; Pavitt, G.D.; Zhang, F.; Dever, T.E.; Hinnebusch, A.G. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell Biol. 2001, 21, 5018–5030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogorad, A.M.; Lin, K.Y.; Marintchev, A. eIF2B Mechanisms of Action and Regulation: A Thermodynamic View. Biochemistry 2018, 57, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, S.; Miyake, M.; Tsugawa, K.; Oyadomari, M.; Oyadomari, S. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci. Rep. 2016, 6, 32886. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-J. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: Relevance to anemias. Blood 2007, 109, 2693–2699. [Google Scholar] [CrossRef]
- Crosby, J.S.; Lee, K.; London, I.M.; Chen, J.J. Erythroid expression of the heme-regulated eIF-2 alpha kinase. Mol. Cell Biol. 1994, 14, 3906–3914. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Suragani, R.N.V.S.; Wang, F.; Han, A.; Zhao, W.; Andrews, N.C.; Chen, J.-J. The function of heme-regulated eIF2alpha kinase in murine iron homeostasis and macrophage maturation. J. Clin. Investig. 2007, 117, 3296–3305. [Google Scholar] [CrossRef]
- Galluzzi, L.; Brenner, C.; Morselli, E.; Touat, Z.; Kroemer, G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008, 4, e1000018. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Yang, M.; May, W.S. RAX, a Cellular Activator for Double-stranded RNA-dependent Protein Kinase during Stress Signaling. J. Biol. Chem. 1999, 274, 15427–15432. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Furuhashi, M.; Li, P.; Cao, H.; Tuncman, G.; Sonenberg, N.; Gorgun, C.Z.; Hotamisligil, G.S. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell 2010, 140, 338–348. [Google Scholar] [CrossRef] [Green Version]
- Goh, K.C.; deVeer, M.J.; Williams, B.R. The protein kinase PKR is required for p38 MAPK activation and the innate immune response to bacterial endotoxin. EMBO J. 2000, 19, 4292–4297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szegezdi, E.; Logue, S.E.; Gorman, A.M.; Samali, A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006, 7, 880–885. [Google Scholar] [CrossRef] [Green Version]
- Bravo, R.; Parra, V.; Gatica, D.; Rodriguez, A.E.; Torrealba, N.; Paredes, F.; Wang, Z.V.; Zorzano, A.; Hill, J.A.; Jaimovich, E.; et al. Endoplasmic reticulum and the unfolded protein response: Dynamics and metabolic integration. Int. Rev. Cell Mol. Biol. 2013, 301, 215–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Deval, C.; Chaveroux, C.; Maurin, A.-C.; Cherasse, Y.; Parry, L.; Carraro, V.; Milenkovic, D.; Ferrara, M.; Bruhat, A.; Jousse, C.; et al. Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 2009, 276, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Kumanova, M.; Hart, L.S.; Sloane, K.; Zhang, H.; De Panis, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Ron, D.; Koumenis, C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010, 29, 2082–2096. [Google Scholar] [CrossRef] [Green Version]
- Rutkowski, D.T.; Arnold, S.M.; Miller, C.N.; Wu, J.; Li, J.; Gunnison, K.M.; Mori, K.; Sadighi Akha, A.A.; Raden, D.; Kaufman, R.J. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006, 4, e374. [Google Scholar] [CrossRef] [Green Version]
- DuRose, J.B.; Scheuner, D.; Kaufman, R.J.; Rothblum, L.I.; Niwa, M. Phosphorylation of Eukaryotic Translation Initiation Factor 2α Coordinates rRNA Transcription and Translation Inhibition during Endoplasmic Reticulum Stress. Mol. Cell Biol. 2009, 29, 4295. [Google Scholar] [CrossRef] [Green Version]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated Translation Initiation Controls Stress-Induced Gene Expression in Mammalian Cells. Mol. Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef]
- Vattem, K.M.; Wek, R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11269–11274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.D.; Harding, H.P.; Ron, D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004, 167, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Burke, J.E.; Williams, R.L. Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS). Adv. Biol. Regul. 2013, 53, 97–110. [Google Scholar] [CrossRef] [Green Version]
- Denley, A.; Gymnopoulos, M.; Kang, S.; Mitchell, C.; Vogt, P.K. Requirement of Phosphatidylinositol(3,4,5)Trisphosphate in Phosphatidylinositol 3-Kinase-Induced Oncogenic Transformation. Mol. Cancer Res. 2009, 7, 1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef] [Green Version]
- Tee, A.R.; Fingar, D.C.; Manning, B.D.; Kwiatkowski, D.J.; Cantley, L.C.; Blenis, J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 13571–13576. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Zhang, Y.; Arrazola, P.; Hino, O.; Kobayashi, T.; Yeung, R.S.; Ru, B.; Pan, D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 2002, 4, 699–704. [Google Scholar] [CrossRef]
- Li, Y.; Inoki, K.; Guan, K.-L. Biochemical and Functional Characterizations of Small GTPase Rheb and TSC2 GAP Activity. Mol. Cell Biol. 2004, 24, 7965. [Google Scholar] [CrossRef] [Green Version]
- Inoki, K.; Li, Y.; Zhu, T.; Wu, J.; Guan, K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002, 4, 648–657. [Google Scholar] [CrossRef]
- Hannan, K.M.; Sanij, E.; Hein, N.; Hannan, R.D.; Pearson, R.B. Signaling to the ribosome in cancer--It is more than just mTORC1. Iubmb Life 2011, 63, 79–85. [Google Scholar] [CrossRef]
- Iadevaia, V.; Caldarola, S.; Tino, E.; Amaldi, F.; Loreni, F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5’-terminal oligopyrimidine (TOP) mRNAs. RNA 2008, 14, 1730–1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnuson, B.; Ekim, B.; Fingar, D.C. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012, 441, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gingras, A.C.; Gygi, S.P.; Raught, B.; Polakiewicz, R.D.; Abraham, R.T.; Hoekstra, M.F.; Aebersold, R.; Sonenberg, N. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999, 13, 1422–1437. [Google Scholar] [CrossRef]
- Piazzi, M.; Bavelloni, A.; Gallo, A.; Faenza, I.; Blalock, W.L. Signal Transduction in Ribosome Biogenesis: A Recipe to Avoid Disaster. Int. J. Mol. Sci. 2019, 20, 2718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauvin, C.; Koka, V.; Nouschi, A.; Mieulet, V.; Hoareau-Aveilla, C.; Dreazen, A.; Cagnard, N.; Carpentier, W.; Kiss, T.; Meyuhas, O.; et al. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 2014, 33, 474–483. [Google Scholar] [CrossRef]
- Grüner, S.; Peter, D.; Weber, R.; Wohlbold, L.; Chung, M.-Y.; Weichenrieder, O.; Valkov, E.; Igreja, C.; Izaurralde, E. The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol. Cell 2016, 64, 467–479. [Google Scholar] [CrossRef] [Green Version]
- Haneke, K.; Schott, J.; Lindner, D.; Hollensen, A.K.; Damgaard, C.K.; Mongis, C.; Knop, M.; Palm, W.; Ruggieri, A.; Stoecklin, G. CDK1 couples proliferation with protein synthesis. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef] [Green Version]
- Monnier, A.; Bellé, R.; Morales, J.; Cormier, P.; Boulben, S.; Mulner-Lorillon, O. Evidence for regulation of protein synthesis at the elongation step by CDK1/cyclin B phosphorylation. Nucleic Acids Res. 2001, 29, 1453–1457. [Google Scholar] [CrossRef] [Green Version]
- Shuda, M.; Velásquez, C.; Cheng, E.; Cordek, D.G.; Kwun, H.J.; Chang, Y.; Moore, P.S. CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc. Natl. Acad. Sci. USA 2015, 112, 5875. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Valle, F.; Badura, M.L.; Braunstein, S.; Narasimhan, M.; Schneider, R.J. Mitotic raptor promotes mTORC1 activity, G(2)/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol. Cell Biol. 2010, 30, 3151–3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coldwell, M.; Cowan, J.; Vlasak, M.; Mead, A.; Willett, M.; Perry, L.; Morley, S. Phosphorylation of eIF4GII and 4E-BP1 in response to nocodazole treatment: A reappraisal of translation initiation during mitosis. Cell Cycle 2013, 12, 3615–3628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stonyte, V.; Boye, E.; Grallert, B. Regulation of global translation during the cell cycle. J. Cell Sci. 2018, 131, jcs220327. [Google Scholar] [CrossRef] [Green Version]
- Anda, S.; Grallert, B. Cell-Cycle-Dependent Regulation of Translation: New Interpretations of Old Observations in Light of New Approaches. BioEssays 2019, 41, 1900022. [Google Scholar] [CrossRef] [PubMed]
- Walters, B.; Thompson, S.R. Cap-Independent Translational Control of Carcinogenesis. Front. Oncol 2016, 6, 128. [Google Scholar] [CrossRef] [Green Version]
- Stoneley, M.; Willis, A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene 2004, 23, 3200–3207. [Google Scholar] [CrossRef] [Green Version]
- Hellen, C.U.; Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001, 15, 1593–1612. [Google Scholar] [CrossRef] [Green Version]
- Mokrejs, M.; Masek, T.; Vopálensky, V.; Hlubucek, P.; Delbos, P.; Pospísek, M. IRESite--a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 2010, 38, D131–D136. [Google Scholar] [CrossRef] [Green Version]
- Baird, S.D.; Lewis, S.M.; Turcotte, M.; Holcik, M. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007, 35, 4664–4677. [Google Scholar] [CrossRef] [Green Version]
- Komar, A.A.; Hatzoglou, M. Internal Ribosome Entry Sites in Cellular mRNAs: Mystery of Their Existence. J. Biol. Chem. 2005, 280, 23425–23428. [Google Scholar] [CrossRef] [Green Version]
- King, H.A.; Cobbold, L.C.; Willis, A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010, 38, 1581–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, S.R. So you want to know if your message has an IRES? Wiley Interdiscip. Rev. RNA 2012, 3, 697–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Parra, C.; Ernlund, A.; Alard, A.; Ruggles, K.; Ueberheide, B.; Schneider, R.J. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 2018, 9, 3068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imataka, H.; Olsen, H.S.; Sonenberg, N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997, 16, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; McCormick, F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 2006, 25, 4008–4019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, S.; Poksay, K.S.; Arnold, K.S.; Innerarity, T.L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997, 11, 321–333. [Google Scholar] [CrossRef] [Green Version]
- Lewis, S.M.; Cerquozzi, S.; Graber, T.E.; Ungureanu, N.H.; Andrews, M.; Holcik, M. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res. 2008, 36, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Nevins, T.A.; Harder, Z.M.; Korneluk, R.G.; Holčík, M. Distinct Regulation of Internal Ribosome Entry Site-mediated Translation following Cellular Stress Is Mediated by Apoptotic Fragments of eIF4G Translation Initiation Factor Family Members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 2003, 278, 3572–3579. [Google Scholar] [CrossRef] [Green Version]
- Marash, L.; Liberman, N.; Henis-Korenblit, S.; Sivan, G.; Reem, E.; Elroy-Stein, O.; Kimchi, A. DAP5 Promotes Cap-Independent Translation of Bcl-2 and CDK1 to Facilitate Cell Survival during Mitosis. Mol. Cell 2008, 30, 447–459. [Google Scholar] [CrossRef]
- Virgili, G.; Frank, F.; Feoktistova, K.; Sawicki, M.; Sonenberg, N.; Fraser, C.S.; Nagar, B. Structural analysis of the DAP5 MIF4G domain and its interaction with eIF4A. Structure 2013, 21, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Stoneley, M.; Chappell, S.A.; Jopling, C.L.; Dickens, M.; MacFarlane, M.; Willis, A.E. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell Biol. 2000, 20, 1162–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coldwell, M.J.; Mitchell, S.A.; Stoneley, M.; MacFarlane, M.; Willis, A.E. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 2000, 19, 899–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weingarten-Gabbay, S.; Khan, D.; Liberman, N.; Yoffe, Y.; Bialik, S.; Das, S.; Oren, M.; Kimchi, A. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 2014, 33, 611–618. [Google Scholar] [CrossRef]
- Holcik, M.; Lefebvre, C.; Yeh, C.; Chow, T.; Korneluk, R.G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1999, 1, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriyarachchi, D.; Cerquozzi, S.; Cheung, H.H.; Holcík, M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem 2004, 279, 17148–17157. [Google Scholar] [CrossRef] [Green Version]
- Hundsdoerfer, P.; Thoma, C.; Hentze, M.W. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. USA 2005, 102, 13421–13426. [Google Scholar] [CrossRef] [Green Version]
- Henis-Korenblit, S.; Strumpf, N.L.; Goldstaub, D.; Kimchi, A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell Biol. 2000, 20, 496–506. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Valle, F.; Braunstein, S.; Zavadil, J.; Formenti, S.C.; Schneider, R.J. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 2008, 181, 293–307. [Google Scholar] [CrossRef] [Green Version]
- Badura, M.; Braunstein, S.; Zavadil, J.; Schneider, R.J. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc. Natl. Acad. Sci. USA 2012, 109, 18767–18772. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, H.; Takahashi, K.; Yamamoto, T.; Iwasaki, M.; Narita, M.; Nakamura, M.; Rand, T.A.; Nakagawa, M.; Watanabe, A.; Yamanaka, S. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2017, 114, 340–345. [Google Scholar] [CrossRef] [Green Version]
- Yoffe, Y.; David, M.; Kalaora, R.; Povodovski, L.; Friedlander, G.; Feldmesser, E.; Ainbinder, E.; Saada, A.; Bialik, S.; Kimchi, A. Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev. 2016, 30, 1991–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.S.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, B.-J.; van Hoef, V.; Jobava, R.; Elroy-Stein, O.; Valasek, L.S.; Cargnello, M.; Gao, X.-H.; Krokowski, D.; Merrick, W.C.; Kimball, S.R.; et al. A Unique ISR Program Determines Cellular Responses to Chronic Stress. Mol. Cell 2017, 68, 885–900.e886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques-Ramos, A.; Candeias, M.M.; Menezes, J.; Lacerda, R.; Willcocks, M.; Teixeira, A.; Locker, N.; Romão, L. Cap-independent translation ensures mTOR expression and function upon protein synthesis inhibition. RNA 2017, 23, 1712–1728. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.; Xu, W.; Reed, J.C. Cell death and endoplasmic reticulum stress: Disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008, 7, 1013–1030. [Google Scholar] [CrossRef]
- Holcik, M.; Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 318–327. [Google Scholar] [CrossRef]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef]
- Sharma, D.K.; Bressler, K.; Patel, H.; Balasingam, N.; Thakor, N. Role of Eukaryotic Initiation Factors during Cellular Stress and Cancer Progression. J. Nucleic Acids 2016, 2016, 8235121. [Google Scholar] [CrossRef] [Green Version]
- Moreno, J.A.; Radford, H.; Peretti, D.; Steinert, J.R.; Verity, N.; Martin, M.G.; Halliday, M.; Morgan, J.; Dinsdale, D.; Ortori, C.A.; et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature 2012, 485, 507–511. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.W.A.-A.L.; Jia, H.; Martin, I.; Dawson, T.M.; Dawson, V.L. Protein Translation in Parkinson’s Disease. In Parkinson’s Disease Molecular Mechanism Underlying Pathology, 1rd ed.; Verstreken, P., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 281–286. [Google Scholar] [CrossRef]
- Gerakis, Y.; Hetz, C. Emerging roles of ER stress in the etiology and pathogenesis of Alzheimer’s disease. FEBS J. 2018, 285, 995–1011. [Google Scholar] [CrossRef] [Green Version]
- Costa-Mattioli, M.; Gobert, D.; Stern, E.; Gamache, K.; Colina, R.; Cuello, C.; Sossin, W.; Kaufman, R.; Pelletier, J.; Rosenblum, K.; et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 2007, 129, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Querfurth, H.W.; LaFerla, F.M. Alzheimer’s Disease. N. Engl. J. Med. 2010, 362, 329–344. [Google Scholar] [CrossRef] [Green Version]
- Chang, R.C.C.; Wong, A.K.Y.; Ng, H.-K.; Hugon, J. Phosphorylation of eukaryotic initiation factor-2α (eIF2α) is associated with neuronal degeneration in Alzheimer’s disease. NeuroReport 2002, 13, 2429–2432. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.A.; Halliday, M.; Molloy, C.; Radford, H.; Verity, N.; Axten, J.M.; Ortori, C.A.; Willis, A.E.; Fischer, P.M.; Barrett, D.A.; et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 2013, 5, 206ra138. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hong, S.; Shepardson, N.E.; Walsh, D.M.; Shankar, G.M.; Selkoe, D. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron 2009, 62, 788–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radford, H.; Moreno, J.A.; Verity, N.; Halliday, M.; Mallucci, G.R. PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol. 2015, 130, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Bellucci, A.; Navarria, L.; Zaltieri, M.; Falarti, E.; Bodei, S.; Sigala, S.; Battistin, L.; Spillantini, M.; Missale, C.; Spano, P. Induction of the unfolded protein response by α-synuclein in experimental models of Parkinson’s disease. J. Neurochem. 2011, 116, 588–605. [Google Scholar] [CrossRef]
- Liu, M.; Qin, L.; Wang, L.; Tan, J.; Zhang, H.; Tang, J.; Shen, X.; Tan, L.; Wang, C. α-synuclein induces apoptosis of astrocytes by causing dysfunction of the endoplasmic reticulum-Golgi compartment. Mol. Med. Rep. 2018, 18, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Hoozemans, J.J.; van Haastert, E.S.; Eikelenboom, P.; de Vos, R.A.; Rozemuller, J.M.; Scheper, W. Activation of the unfolded protein response in Parkinson’s disease. Biochem. Biophys. Res. Commun. 2007, 354, 707–711. [Google Scholar] [CrossRef]
- Mercado, G.; Castillo, V.; Soto, P.; López, N.; Axten, J.M.; Sardi, S.P.; Hoozemans, J.J.M.; Hetz, C. Targeting PERK signaling with the small molecule GSK2606414 prevents neurodegeneration in a model of Parkinson’s disease. Neurobiol. Dis. 2018, 112, 136–148. [Google Scholar] [CrossRef]
- Celardo, I.; Costa, A.C.; Lehmann, S.; Jones, C.; Wood, N.; Mencacci, N.E.; Mallucci, G.R.; Loh, S.H.Y.; Martins, L.M. Mitofusin-mediated ER stress triggers neurodegeneration in pink1/parkin models of Parkinson’s disease. Cell Death Dis. 2016, 7, e2271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Aimé, P.; Dai, D.; Ramalingam, N.; Crary, J.F.; Burke, R.E.; Greene, L.A.; Levy, O.A. Guanabenz promotes neuronal survival via enhancement of ATF4 and parkin expression in models of Parkinson disease. Exp. Neurol. 2018, 303, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, W. Impaired capacity to restore proteostasis in the aged brain after ischemia: Implications for translational brain ischemia research. Neurochem. Int. 2019, 127, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Krause, G.S.; Tiffany, B.R. Suppression of protein synthesis in the reperfused brain. Stroke 1993, 24, 747–755. [Google Scholar] [CrossRef] [Green Version]
- DeGracia, D.J.; Montie, H.L. Cerebral ischemia and the unfolded protein response. J. Neurochem. 2004, 91, 1–8. [Google Scholar] [CrossRef]
- White, B.C.; Sullivan, J.M.; DeGracia, D.J.; O’Neil, B.J.; Neumar, R.W.; Grossman, L.I.; Rafols, J.A.; Krause, G.S. Brain ischemia and reperfusion: Molecular mechanisms of neuronal injury. J. Neurol. Sci. 2000, 179, 1–33. [Google Scholar] [CrossRef]
- Kumar, R.; Azam, S.; Sullivan, J.M.; Owen, C.; Cavener, D.R.; Zhang, P.; Ron, D.; Harding, H.P.; Chen, J.J.; Han, A.; et al. Brain ischemia and reperfusion activates the eukaryotic initiation factor 2alpha kinase, PERK. J. Neurochem. 2001, 77, 1418–1421. [Google Scholar] [CrossRef] [Green Version]
- Doutheil, J.; Althausen, S.; Gissel, C.; Paschen, W. Activation of MYD116 (gadd34) expression following transient forebrain ischemia of rat: Implications for a role of disturbances of endoplasmic reticulum calcium homeostasis. Mol. Brain Res. 1999, 63, 225–232. [Google Scholar] [CrossRef]
- Imai, H.; Harland, J.; McCulloch, J.; Graham, D.I.; Brown, S.M.; Macrae, I.M. Specific expression of the cell cycle regulation proteins, GADD34 and PCNA, in the peri-infarct zone after focal cerebral ischaemia in the rat. Eur. J. Neurosci. 2002, 15, 1929–1936. [Google Scholar] [CrossRef]
- Paschen, W.; Hayashi, T.; Saito, A.; Chan, P.H. GADD34 protein levels increase after transient ischemia in the cortex but not in the CA1 subfield: Implications for post-ischemic recovery of protein synthesis in ischemia-resistant cells. J. Neurochem. 2004, 90, 694–701. [Google Scholar] [CrossRef]
- McCaig, D.; Imai, H.; Gallagher, L.; Graham, D.I.; Harland, J.; Moira Brown, S.; Mhairi Macrae, I. Evolution of GADD34 expression after focal cerebral ischaemia. Brain Res. 2005, 1034, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.W.; Chen, T.Y.; Hung, C.Y.; Tai, S.H.; Huang, S.Y.; Chang, C.C.; Hung, H.Y.; Lee, E.J. Melatonin protects brain against ischemia/reperfusion injury by attenuating endoplasmic reticulum stress. Int. J. Mol. Med. 2018, 42, 182–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghemrawi, R.; Battaglia-Hsu, S.-F.; Arnold, C. Endoplasmic Reticulum Stress in Metabolic Disorders. Cells 2018, 7, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheuner, D.; Vander Mierde, D.; Song, B.; Flamez, D.; Creemers, J.W.; Tsukamoto, K.; Ribick, M.; Schuit, F.C.; Kaufman, R.J. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 2005, 11, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Baird, T.D.; Wek, R.C. Eukaryotic Initiation Factor 2 Phosphorylation and Translational Control in Metabolism. Adv. Nutr. 2012, 3, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Harding, H.P.; Zeng, H.; Zhang, Y.; Jungries, R.; Chung, P.; Plesken, H.; Sabatini, D.D.; Ron, D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/-mice reveals a role for translational control in secretory cell survival. Mol. Cell 2001, 7, 1153–1163. [Google Scholar] [CrossRef]
- Delépine, M.; Nicolino, M.; Barrett, T.; Golamaully, M.; Lathrop, G.M.; Julier, C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000, 25, 406–409. [Google Scholar] [CrossRef]
- Scheuner, D.; Song, B.; McEwen, E.; Liu, C.; Laybutt, R.; Gillespie, P.; Saunders, T.; Bonner-Weir, S.; Kaufman, R.J. Translational Control Is Required for the Unfolded Protein Response and In Vivo Glucose Homeostasis. Mol. Cell 2001, 7, 1165–1176. [Google Scholar] [CrossRef]
- Han, A.P.; Yu, C.; Lu, L.; Fujiwara, Y.; Browne, C.; Chin, G.; Fleming, M.; Leboulch, P.; Orkin, S.H.; Chen, J.J. Heme-regulated eIF2α kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001, 20, 6909–6918. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; McGrath, B.C.; Reinert, J.; Olsen, D.S.; Lei, L.; Gill, S.; Wek, S.A.; Vattem, K.M.; Wek, R.C.; Kimball, S.R.; et al. The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell Biol. 2002, 22, 6681–6688. [Google Scholar] [CrossRef] [Green Version]
- Abraham, N.; Stojdl, D.F.; Duncan, P.I.; Méthot, N.; Ishii, T.; Dubé, M.; Vanderhyden, B.C.; Atkins, H.L.; Gray, D.A.; McBurney, M.W.; et al. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 1999, 274, 5953–5962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skelly, R.H.; Schuppin, G.T.; Ishihara, H.; Oka, Y.; Rhodes, C.J. Glucose-regulated translational control of proinsulin biosynthesis with that of the proinsulin endopeptidases PC2 and PC3 in the insulin-producing MIN6 cell line. Diabetes 1996, 45, 37–43. [Google Scholar] [CrossRef]
- Ginès, P.; Graupera, I.; Lammert, F.; Angeli, P.; Caballeria, L.; Krag, A.; Guha, I.N.; Murad, S.D.; Castera, L. Screening for liver fibrosis in the general population: A call for action. Lancet Gastroenterol. Hepatol. 2016, 1, 256–260. [Google Scholar] [CrossRef]
- Dasarathy, S.; Hatzoglou, M. Hyperammonemia and proteostasis in cirrhosis. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Davuluri, G.; Krokowski, D.; Guan, B.J.; Kumar, A.; Thapaliya, S.; Singh, D.; Hatzoglou, M.; Dasarathy, S. Metabolic adaptation of skeletal muscle to hyperammonemia drives the beneficial effects of l-leucine in cirrhosis. J. Hepatol. 2016, 65, 929–937. [Google Scholar] [CrossRef] [Green Version]
- Tsien, C.; Davuluri, G.; Singh, D.; Allawy, A.; Ten Have, G.A.M.; Thapaliya, S.; Schulze, J.M.; Barnes, D.; McCullough, A.J.; Engelen, M.P.K.J.; et al. Metabolic and molecular responses to leucine-enriched branched chain amino acid supplementation in the skeletal muscle of alcoholic cirrhosis. Hepatology 2015, 61, 2018–2029. [Google Scholar] [CrossRef] [PubMed]
- Leweling, H.; Breitkreutz, R.; Behne, F.; Staedt, U.; Striebel, J.P.; Holm, E. Hyperammonemia-induced depletion of glutamate and branched-chain amino acids in muscle and plasma. J. Hepatol. 1996, 25, 756–762. [Google Scholar] [CrossRef]
- Kumar, A.; Davuluri, G.; Silva, R.N.E.; Engelen, M.; Ten Have, G.A.M.; Prayson, R.; Deutz, N.E.P.; Dasarathy, S. Ammonia lowering reverses sarcopenia of cirrhosis by restoring skeletal muscle proteostasis. Hepatology 2017, 65, 2045–2058. [Google Scholar] [CrossRef] [Green Version]
- Tran, I.; Ji, C.; Ni, I.; Min, T.; Tang, D.; Vij, N. Role of Cigarette Smoke-Induced Aggresome Formation in Chronic Obstructive Pulmonary Disease-Emphysema Pathogenesis. Am. J. Respir. Cell Mol. Biol. 2015, 53, 159–173. [Google Scholar] [CrossRef]
- Min, T.; Bodas, M.; Mazur, S.; Vij, N. Critical role of proteostasis-imbalance in pathogenesis of COPD and severe emphysema. J. Mol. Med. 2011, 89, 577–593. [Google Scholar] [CrossRef] [Green Version]
- Kenche, H.; Baty, C.J.; Vedagiri, K.; Shapiro, S.D.; Blumental-Perry, A. Cigarette smoking affects oxidative protein folding in endoplasmic reticulum by modifying protein disulfide isomerase. FASEB J. 2012, 27, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Steiling, K.; van den Berge, M.; Hijazi, K.; Florido, R.; Campbell, J.; Liu, G.; Xiao, J.; Zhang, X.; Duclos, G.; Drizik, E.; et al. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am. J. Respir. Crit. Care Med. 2013, 187, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, M.; Komoike, Y. Experimental Evidence Shows Salubrinal, an eIF2α Dephosphorylation Inhibitor, Reduces Xenotoxicant-Induced Cellular Damage. Int. J. Mol. Sci. 2015, 16, 16275–16287. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, X.; Fassett, J.; Kwak, D.; Liu, X.; Hu, X.; Falls, T.J.; Bell, J.C.; Li, H.; Bitterman, P.; et al. Double-stranded RNA-dependent protein kinase deficiency protects the heart from systolic overload-induced congestive heart failure. Circulation 2014, 129, 1397–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Xu, X.; Fassett, J.; Kwak, D.; Liu, X.; Hu, X.; Wang, H.; Guo, H.; Xu, D.; Yan, S.; et al. Loss of the eukaryotic initiation factor 2α kinase general control nonderepressible 2 protects mice from pressure overload-induced congestive heart failure without affecting ventricular hypertrophy. Hypertension 2014, 63, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Kwak, D.; Lu, Z.; Xu, X.; Fassett, J.; Wang, H.; Wei, Y.; Cavener, D.R.; Hu, X.; Hall, J.; et al. Endoplasmic reticulum stress sensor protein kinase R-like endoplasmic reticulum kinase (PERK) protects against pressure overload-induced heart failure and lung remodeling. Hypertension 2014, 64, 738–744. [Google Scholar] [CrossRef] [Green Version]
- Neuber, C.; Uebeler, J.; Schulze, T.; Sotoud, H.; El-Armouche, A.; Eschenhagen, T. Guanabenz interferes with ER stress and exerts protective effects in cardiac myocytes. PLoS ONE 2014, 9, e98893. [Google Scholar] [CrossRef]
- Hoeffer, C.A.; Klann, E. mTOR signaling: At the crossroads of plasticity, memory and disease. Trends Neurosci. 2010, 33, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Alafuzoff, I.; Soininen, H.; Winblad, B.; Pei, J.J. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer’s disease brain. FEBS J. 2005, 272, 4211–4220. [Google Scholar] [CrossRef]
- Li, X.; An, W.L.; Alafuzoff, I.; Soininen, H.; Winblad, B.; Pei, J.J. Phosphorylated eukaryotic translation factor 4E is elevated in Alzheimer brain. Neuroreport 2004, 15, 2237–2240. [Google Scholar] [CrossRef]
- An, W.L.; Cowburn, R.F.; Li, L.; Braak, H.; Alafuzoff, I.; Iqbal, K.; Iqbal, I.G.; Winblad, B.; Pei, J.J. Up-regulation of phosphorylated/activated p70 S6 kinase and its relationship to neurofibrillary pathology in Alzheimer’s disease. Am. J. Pathol. 2003, 163, 591–607. [Google Scholar] [CrossRef]
- Caccamo, A.; Majumder, S.; Richardson, A.; Strong, R.; Oddo, S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: Effects on cognitive impairments. J. Biol. Chem. 2010, 285, 13107–13120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaudoin, M.E.; Poirel, V.-J.; Krushel, L.A. Regulating amyloid precursor protein synthesis through an internal ribosomal entry site. Nucleic Acids Res. 2008, 36, 6835–6847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, T.; Hoeffer, C.A.; Capetillo-Zarate, E.; Yu, F.; Wong, H.; Lin, M.T.; Tampellini, D.; Klann, E.; Blitzer, R.D.; Gouras, G.K. Dysregulation of the mTOR Pathway Mediates Impairment of Synaptic Plasticity in a Mouse Model of Alzheimer’s Disease. PLoS ONE 2010, 5, e12845. [Google Scholar] [CrossRef]
- Bové, J.; Martínez-Vicente, M.; Vila, M. Fighting neurodegeneration with rapamycin: Mechanistic insights. Nat. Rev. Neurosci. 2011, 12, 437–452. [Google Scholar] [CrossRef]
- Ducker, G.S.; Atreya, C.E.; Simko, J.P.; Hom, Y.K.; Matli, M.R.; Benes, C.H.; Hann, B.; Nakakura, E.K.; Bergsland, E.K.; Donner, D.B.; et al. Incomplete inhibition of phosphorylation of 4E-BP1 as a mechanism of primary resistance to ATP-competitive mTOR inhibitors. Oncogene 2014, 33, 1590–1600. [Google Scholar] [CrossRef] [Green Version]
- Crino, P.B. The mTOR signalling cascade: Paving new roads to cure neurological disease. Nat. Rev. Neurol. 2016, 12, 379–392. [Google Scholar] [CrossRef]
- Tramutola, A.; Lanzillotta, C.; Di Domenico, F. Targeting mTOR to reduce Alzheimer-related cognitive decline: From current hits to future therapies. Expert Rev. Neurother. 2017, 17, 33–45. [Google Scholar] [CrossRef]
- Crews, L.; Spencer, B.; Desplats, P.; Patrick, C.; Paulino, A.; Rockenstein, E.; Hansen, L.; Adame, A.; Galasko, D.; Masliah, E. Selective Molecular Alterations in the Autophagy Pathway in Patients with Lewy Body Disease and in Models of α-Synucleinopathy. PLoS ONE 2010, 5, e9313. [Google Scholar] [CrossRef] [Green Version]
- Magalhaes, J.; Gegg, M.E.; Migdalska-Richards, A.; Doherty, M.K.; Whitfield, P.D.; Schapira, A.H. Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: Relevance to Parkinson disease. Hum. Mol. Genet. 2016, 25, 3432–3445. [Google Scholar] [CrossRef]
- Imai, Y.; Gehrke, S.; Wang, H.-Q.; Takahashi, R.; Hasegawa, K.; Oota, E.; Lu, B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008, 27, 2432–2443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, I.; Abalde-Atristain, L.; Kim, J.W.; Dawson, T.M.; Dawson, V.L. Abberant protein synthesis in G2019S LRRK2 Drosophila Parkinson disease-related phenotypes. Fly 2014, 8, 165–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, X.; Jiang, B.; Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 2016, 15, 781–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chartier-Harlin, M.-C.; Dachsel, J.C.; Vilariño-Güell, C.; Lincoln, S.J.; Leprêtre, F.; Hulihan, M.M.; Kachergus, J.; Milnerwood, A.J.; Tapia, L.; Song, M.-S.; et al. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am. J. Hum. Genet. 2011, 89, 398–406. [Google Scholar] [CrossRef] [Green Version]
- Tucci, A.; Charlesworth, G.; Sheerin, U.-M.; Plagnol, V.; Wood, N.W.; Hardy, J. Study of the genetic variability in a Parkinson’s Disease gene: EIF4G1. Neurosci. Lett. 2012, 518, 19–22. [Google Scholar] [CrossRef] [Green Version]
- Koukouraki, P.; Doxakis, E. Constitutive translation of human α-synuclein is mediated by the 5’-untranslated region. Open Biol. 2016, 6, 160022. [Google Scholar] [CrossRef] [Green Version]
- Masini, D.; Bonito-Oliva, A.; Bertho, M.; Fisone, G. Inhibition of mTORC1 Signaling Reverts Cognitive and Affective Deficits in a Mouse Model of Parkinson’s Disease. Front. Neurol. 2018, 9, 208. [Google Scholar] [CrossRef]
- Perez-Alvarez, M.J.; Villa Gonzalez, M.; Benito-Cuesta, I.; Wandosell, F.G. Role of mTORC1 Controlling Proteostasis after Brain Ischemia. Front. Neurosci. 2018, 12, 60. [Google Scholar] [CrossRef]
- Zare Mehrjerdi, F.; Aboutaleb, N.; Habibey, R.; Ajami, M.; Soleimani, M.; Arabian, M.; Niknazar, S.; Hossein Davoodi, S.; Pazoki-Toroudi, H. Increased phosphorylation of mTOR is involved in remote ischemic preconditioning of hippocampus in mice. Brain Res. 2013, 1526, 94–101. [Google Scholar] [CrossRef]
- Xie, R.; Wang, P.; Ji, X.; Zhao, H. Ischemic post-conditioning facilitates brain recovery after stroke by promoting Akt/mTOR activity in nude rats. J. Neurochem. 2013, 127, 723–732. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wu, X.; Pu, J.; Luo, P.; Ma, W.; Wang, J.; Wei, J.; Wang, Y.; Fei, Z. Lycium barbarum polysaccharide protects against oxygen glucose deprivation/reoxygenation-induced apoptosis and autophagic cell death via the PI3K/Akt/mTOR signaling pathway in primary cultured hippocampal neurons. Biochem. Biophys. Res. Commun. 2018, 495, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, M.-H.; Qin, C.; Fan, W.-H.; Tian, D.-S.; Liu, J.-L. Fingolimod suppresses neuronal autophagy through the mTOR/p70S6K pathway and alleviates ischemic brain damage in mice. PLoS ONE 2017, 12, e0188748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, C.; Zhang, J.; Li, B.; Ding, Z.; Cheng, W.; Gao, F.; Zhang, Y.; Xu, Y.; Zhang, S. Cornin protects SH-SY5Y cells against oxygen and glucose deprivation-induced autophagy through the PI3K/Akt/mTOR pathway. Mol. Med. Rep. 2018, 17, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.; Brunmair, B.; Brehm, A.; Artwohl, M.; Szendroedi, J.; Nowotny, P.; Roth, E.; Fürnsinn, C.; Promintzer, M.; Anderwald, C.; et al. The Mammalian Target of Rapamycin Pathway Regulates Nutrient-Sensitive Glucose Uptake in Man. Diabetes 2007, 56, 1600. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, F.; Krebs, M.; Dombrowski, L.; Brehm, A.; Bernroider, E.; Roth, E.; Nowotny, P.; Waldhäusl, W.; Marette, A.; Roden, M. Overactivation of S6 Kinase 1 as a Cause of Human Insulin Resistance During Increased Amino Acid Availability. Diabetes 2005, 54, 2674. [Google Scholar] [CrossRef] [Green Version]
- Catania, C.; Binder, E.; Cota, D. mTORC1 signaling in energy balance and metabolic disease. Int. J. Obes. 2011, 35, 751–761. [Google Scholar] [CrossRef] [Green Version]
- Um, S.H.; Frigerio, F.; Watanabe, M.; Picard, F.; Joaquin, M.; Sticker, M.; Fumagalli, S.; Allegrini, P.R.; Kozma, S.C.; Auwerx, J.; et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004, 431, 200–205. [Google Scholar] [CrossRef]
- Le Bacquer, O.; Petroulakis, E.; Paglialunga, S.; Poulin, F.; Richard, D.; Cianflone, K.; Sonenberg, N. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J. Clin. Investig. 2007, 117, 387–396. [Google Scholar] [CrossRef]
- Tsai, S.-Y.; Rodriguez, A.A.; Dastidar, S.G.; Del Greco, E.; Carr, K.L.; Sitzmann, J.M.; Academia, E.C.; Viray, C.M.; Martinez, L.L.; Kaplowitz, B.S.; et al. Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice. Cell Rep. 2016, 16, 1903–1914. [Google Scholar] [CrossRef] [Green Version]
- Mariappan, M.M.; Feliers, D.; Mummidi, S.; Choudhury, G.G.; Kasinath, B.S. High Glucose, High Insulin, and Their Combination Rapidly Induce Laminin-β1 Synthesis by Regulation of mRNA Translation in Renal Epithelial Cells. Diabetes 2007, 56, 476. [Google Scholar] [CrossRef] [Green Version]
- Da, L.; Cao, T.; Sun, X.; Jin, S.; Di, X.; Huang, X.; Yang, X.; Carmichael, G.G.; Taylor, H.S.; Diano, S.; et al. Hepatic TET3 contributes to type-2 diabetes by inducing the HNF4α fetal isoform. Nat. Commun. 2020, 11, 342. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Sun, X.; Zhang, R.; Cao, T.; Cai, S.-Y.; Boyer, J.L.; Zhang, X.; Li, D.; Huang, Y. A Positive Feedback Loop of TET3 and TGF-β1 Promotes Liver Fibrosis. Cell Rep. 2020, 30, 1310–1318.e1315. [Google Scholar] [CrossRef] [PubMed]
- Tandon, P.; Low, G.; Mourtzakis, M.; Zenith, L.; Myers, R.P.; Abraldes, J.G.; Shaheen, A.A.; Qamar, H.; Mansoor, N.; Carbonneau, M.; et al. A Model to Identify Sarcopenia in Patients With Cirrhosis. Clin. Gastroenterol. Hepatol. 2016, 14, 1473–1480.e1473. [Google Scholar] [CrossRef] [PubMed]
- Preedy, V.R.; Peters, T.J.; Patel, V.B.; Miell, J.P. Chronic alcoholic myopathy: Transcription and translational alterations. FASEB J. 1994, 8, 1146–1151. [Google Scholar] [CrossRef] [PubMed]
- Paek, K.Y.; Kim, C.S.; Park, S.M.; Kim, J.H.; Jang, S.K. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 2008, 82, 12082–12093. [Google Scholar] [CrossRef] [Green Version]
- Lang, C.H.; Frost, R.A.; Deshpande, N.; Kumar, V.; Vary, T.C.; Jefferson, L.S.; Kimball, S.R. Alcohol impairs leucine-mediated phosphorylation of 4E-BP1, S6K1, eIF4G, and mTOR in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2003, 285, E1205–E1215. [Google Scholar] [CrossRef]
- Levine, A.J.; Feng, Z.; Mak, T.W.; You, H.; Jin, S. Coordination and communication between the p53 and IGF-1–AKT–TOR signal transduction pathways. Genes Dev. 2006, 20, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Barreiro, E.; Gea, J. Respiratory and Limb Muscle Dysfunction in COPD. Copd J. Chronic Obstr. Pulm. Dis. 2015, 12, 413–426. [Google Scholar] [CrossRef]
- Pérez-Baos, S.; Prieto-Potin, I.; Román-Blas, J.A.; Sánchez-Pernaute, O.; Largo, R.; Herrero-Beaumont, G. Mediators and Patterns of Muscle Loss in Chronic Systemic Inflammation. Front. Physiol. 2018, 9, 409. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, W.; Guo, Q.; Wang, Y.; Ma, L.; Zhang, X. Insulin-Like Growth Factor-1 Signaling in Lung Development and Inflammatory Lung Diseases. Biomed. Res. Int. 2018, 2018, 6057589. [Google Scholar] [CrossRef] [Green Version]
- Ye, M.; Yu, H.; Yu, W.; Zhang, G.; Xiao, L.; Zheng, X.; Wu, J. Evaluation of the Significance of Circulating Insulin-Like Growth Factor-1 and C-Reactive Protein in Patients with Chronic Obstructive Pulmonary Disease. J. Int. Med. Res. 2012, 40, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Coşkun, F.E.E.; Uzaslan, E.; Ediger, D.; Karadağ, M.; Gözü, O. Evaluation of thyroid hormone levels and somatomedin-C (IGF-1) in patients with chronic obstructive pulmonary disease (COPD) and relation with the severity of the disease. Tuberk Toraks 2009, 57, 369–375. [Google Scholar] [PubMed]
- Debigaré, R.; Marquis, K.; Côté, C.H.; Tremblay, R.R.; Michaud, A.; LeBlanc, P.; Maltais, F. Catabolic/Anabolic Balance and Muscle Wasting in Patients With COPD. Chest 2003, 124, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogiatzis, I.; Simoes, D.C.M.; Stratakos, G.; Kourepini, E.; Terzis, G.; Manta, P.; Athanasopoulos, D.; Roussos, C.; Wagner, P.D.; Zakynthinos, S. Effect of pulmonary rehabilitation on muscle remodelling in cachectic patients with COPD. Eur. Respir. J. 2010, 36, 301. [Google Scholar] [CrossRef] [Green Version]
- Houssaini, A.; Breau, M.; Kebe, K.; Abid, S.; Marcos, E.; Lipskaia, L.; Rideau, D.; Parpaleix, A.; Huang, J.; Amsellem, V.; et al. mTOR pathway activation drives lung cell senescence and emphysema. JCI Insight 2018, 3, e93203. [Google Scholar] [CrossRef] [Green Version]
- Doucet, M.; Russell, A.P.; Léger, B.; Debigaré, R.; Joanisse, D.R.; Caron, M.-A.; LeBlanc, P.; Maltais, F. Muscle Atrophy and Hypertrophy Signaling in Patients with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2007, 176, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Pasini, E.; Flati, V.; Comini, L.; Olivares, A.; Bertella, E.; Corsetti, G.; Vitacca, M. Mammalian Target of Rapamycin: Is It Relevant to COPD Pathogenesis or Treatment? Copd J. Chronic Obstr. Pulm. Dis. 2019, 16, 89–92. [Google Scholar] [CrossRef]
- Giraud, S.; Greco, A.; Brink, M.; Diaz, J.-J.; Delafontaine, P. Translation Initiation of the Insulin-like Growth Factor I Receptor mRNA Is Mediated by an Internal Ribosome Entry Site. J. Biol. Chem. 2001, 276, 5668–5675. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Contu, R.; Latronico, M.V.G.; Zhang, J.; Rizzi, R.; Catalucci, D.; Miyamoto, S.; Huang, K.; Ceci, M.; Gu, Y.; et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J. Clin. Investig. 2010, 120, 2805–2816. [Google Scholar] [CrossRef]
- Connolly, E.P.; Thuillier, V.; Rouy, D.; Bouétard, G.; Schneider, R.J. Inhibition of Cap-initiation complexes linked to a novel mechanism of eIF4G depletion in acute myocardial ischemia. Cell Death Differ. 2006, 13, 1586–1594. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Cao, Y.; Nie, J.; Liu, H.; Lu, S.; Hu, X.; Zhu, J.; Zhao, X.; Chen, J.; Chen, X.; et al. Genetic and pharmacological inhibition of Rheb1-mTORC1 signaling exerts cardioprotection against adverse cardiac remodeling in mice. Am. J. Pathol. 2013, 182, 2005–2014. [Google Scholar] [CrossRef]
- Sciarretta, S.; Zhai, P.; Shao, D.; Maejima, Y.; Robbins, J.; Volpe, M.; Condorelli, G.; Sadoshima, J. Rheb is a critical regulator of autophagy during myocardial ischemia: Pathophysiological implications in obesity and metabolic syndrome. Circulation 2012, 125, 1134–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buss, S.J.; Muenz, S.; Riffel, J.H.; Malekar, P.; Hagenmueller, M.; Weiss, C.S.; Bea, F.; Bekeredjian, R.; Schinke-Braun, M.; Izumo, S.; et al. Beneficial Effects of Mammalian Target of Rapamycin Inhibition on Left Ventricular Remodeling After Myocardial Infarction. J. Am. Coll. Cardiol. 2009, 54, 2435–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, R.; Van Diest, P.J.; De Jong, J.S.; Van Der Groep, P.; Van Der Valk, P.; Van Der Wall, E. Hypoxia-inducible factor-1α is associated with angiogenesis, and expression of bFGF, PDGF-BB, and EGFR in invasive breast cancer. Histopathology 2005, 46, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.J.D.; Kappel, A.; Goodall, G.J. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell 2002, 13, 1792–1801. [Google Scholar] [CrossRef] [PubMed]
- Stein, I.; Itin, A.; Einat, P.; Skaliter, R.; Grossman, Z.; Keshet, E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: Implications for translation under hypoxia. Mol. Cell Biol. 1998, 18, 3112–3119. [Google Scholar] [CrossRef] [Green Version]
- Hantelys, F.; Godet, A.-C.; David, F.; Tatin, F.; Renaud-Gabardos, E.; Pujol, F.; Diallo, L.H.; Ader, I.; Ligat, L.; Henras, A.K.; et al. Vasohibin1, a new mouse cardiomyocyte IRES trans-acting factor that regulates translation in early hypoxia. Elife 2019, 8, e50094. [Google Scholar] [CrossRef]
- Gao, G.; Chen, W.; Yan, M.; Liu, J.; Luo, H.; Wang, C.; Yang, P. Rapamycin regulates the balance between cardiomyocyte apoptosis and autophagy in chronic heart failure by inhibiting mTOR signaling. Int. J. Mol. Med. 2020, 45, 195–209. [Google Scholar] [CrossRef] [Green Version]
- Buszczak, M.; Signer, R.A.J.; Morrison, S.J. Cellular differences in protein synthesis regulate tissue homeostasis. Cell 2014, 159, 242–251. [Google Scholar] [CrossRef] [Green Version]
- Polymenis, M.; Aramayo, R. Translate to divide: Control of the cell cycle by protein synthesis. Microb. Cell 2015, 2, 94–104. [Google Scholar] [CrossRef]
- Katz, A.; Orellana, O. Protein synthesis and the stress response. In Cell-Free Protein Synthesis; InTech: Rijeka, Croatia, 2012. [Google Scholar]
- van der Knaap, J.A.; Verrijzer, C.P. Undercover: Gene control by metabolites and metabolic enzymes. Genes Dev. 2016, 30, 2345–2369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, A.R.; Gsponer, J.; Foster, L.J. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst Biol. 2013, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Saelens, X.; Kalai, M.; Vandenabeele, P. Translation Inhibition in Apoptosis: CASPASE-DEPENDENT PKR ACTIVATION AND eIF2-α PHOSPHORYLATION. J. Biol. Chem. 2001, 276, 41620–41628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.-G.; Han, J.; Kim, J.-H.; Kim, M.-J.; Park, J.-W.; Song, B.; Cha, H.-J.; Choi, H.-S.; Chung, H.-T.; Lee, I.-K.; et al. eIF2α phosphorylation is required to prevent hepatocyte death and liver fibrosis in mice challenged with a high fructose diet. Nutr. Metab. 2017, 14, 48. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Pires, K.M.P.; Whitehead, K.J.; Olsen, C.D.; Wayment, B.; Zhang, Y.C.; Bugger, H.; Ilkun, O.; Litwin, S.E.; Thomas, G.; et al. Mechanistic Target of Rapamycin (Mtor) Is Essential for Murine Embryonic Heart Development and Growth. PLoS ONE 2013, 8, e54221. [Google Scholar] [CrossRef]
- Guo, Q.; Zhang, Y.; Zhang, S.; Jin, J.; Pang, S.; Wu, X.; Zhang, W.; Bi, X.; Zhang, Y.; Zhang, Q.; et al. Genome-wide translational reprogramming of genes important for myocyte functions in overload-induced heart failure. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165649. [Google Scholar] [CrossRef]
- Buffington, S.A.; Huang, W.; Costa-Mattioli, M. Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 2014, 37, 17–38. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rios-Fuller, T.J.; Mahe, M.; Walters, B.; Abbadi, D.; Pérez-Baos, S.; Gadi, A.; Andrews, J.J.; Katsara, O.; Vincent, C.T.; Schneider, R.J. Translation Regulation by eIF2α Phosphorylation and mTORC1 Signaling Pathways in Non-Communicable Diseases (NCDs). Int. J. Mol. Sci. 2020, 21, 5301. https://doi.org/10.3390/ijms21155301
Rios-Fuller TJ, Mahe M, Walters B, Abbadi D, Pérez-Baos S, Gadi A, Andrews JJ, Katsara O, Vincent CT, Schneider RJ. Translation Regulation by eIF2α Phosphorylation and mTORC1 Signaling Pathways in Non-Communicable Diseases (NCDs). International Journal of Molecular Sciences. 2020; 21(15):5301. https://doi.org/10.3390/ijms21155301
Chicago/Turabian StyleRios-Fuller, Tiffany J., Melanie Mahe, Beth Walters, Dounia Abbadi, Sandra Pérez-Baos, Abhilash Gadi, John J. Andrews, Olga Katsara, C. Theresa Vincent, and Robert J. Schneider. 2020. "Translation Regulation by eIF2α Phosphorylation and mTORC1 Signaling Pathways in Non-Communicable Diseases (NCDs)" International Journal of Molecular Sciences 21, no. 15: 5301. https://doi.org/10.3390/ijms21155301
APA StyleRios-Fuller, T. J., Mahe, M., Walters, B., Abbadi, D., Pérez-Baos, S., Gadi, A., Andrews, J. J., Katsara, O., Vincent, C. T., & Schneider, R. J. (2020). Translation Regulation by eIF2α Phosphorylation and mTORC1 Signaling Pathways in Non-Communicable Diseases (NCDs). International Journal of Molecular Sciences, 21(15), 5301. https://doi.org/10.3390/ijms21155301




