Heterologous Expression of Poplar WRKY18/35 Paralogs in Arabidopsis Reveals Their Antagonistic Regulation on Pathogen Resistance and Abiotic Stress Tolerance via Variable Hormonal Pathways
Abstract
1. Introduction
2. Results
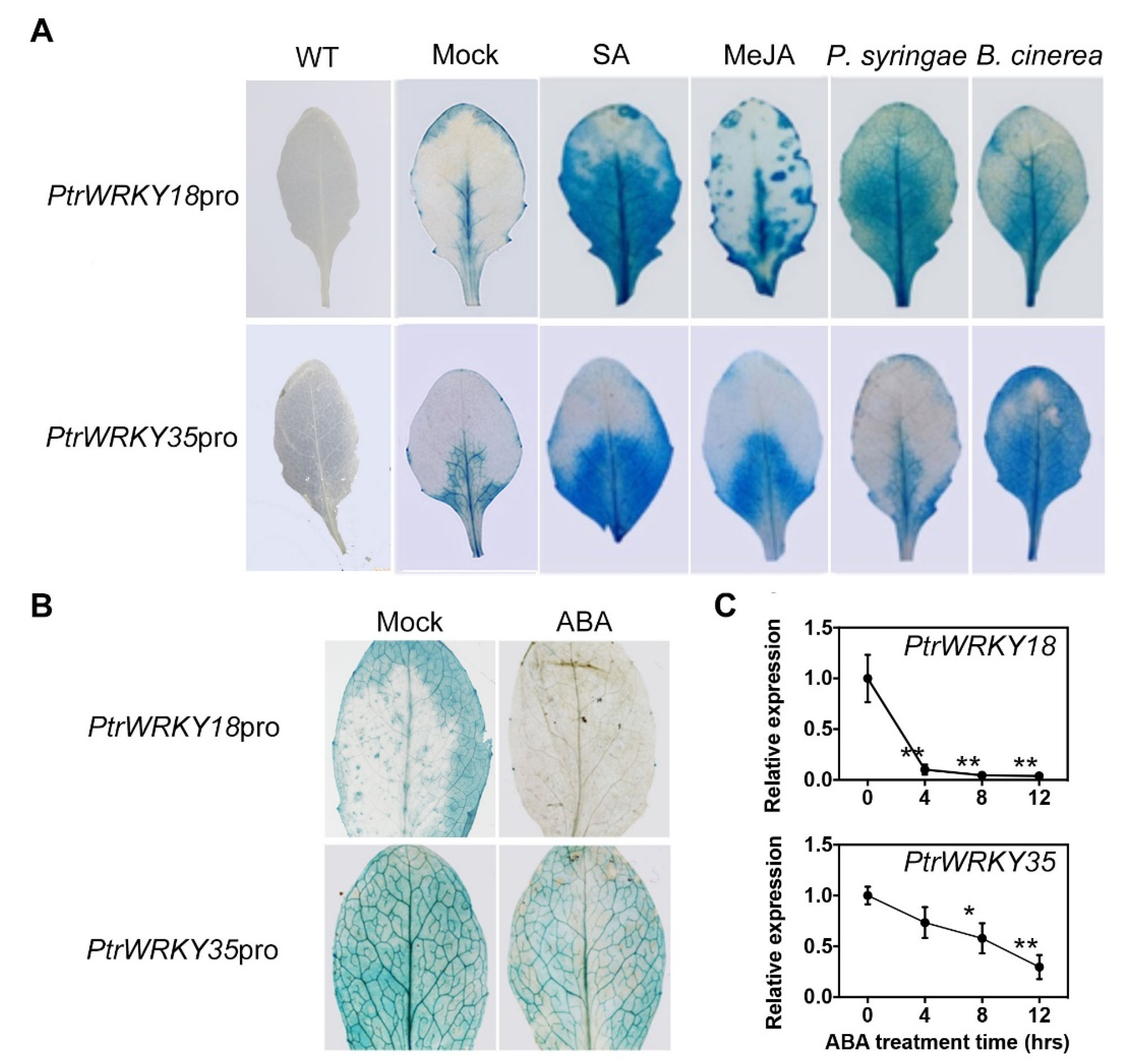
2.1. The Expression of PtrWRKY18 and PtrWRKY35 Is Induced by Various Hormones and Biotic Stresses
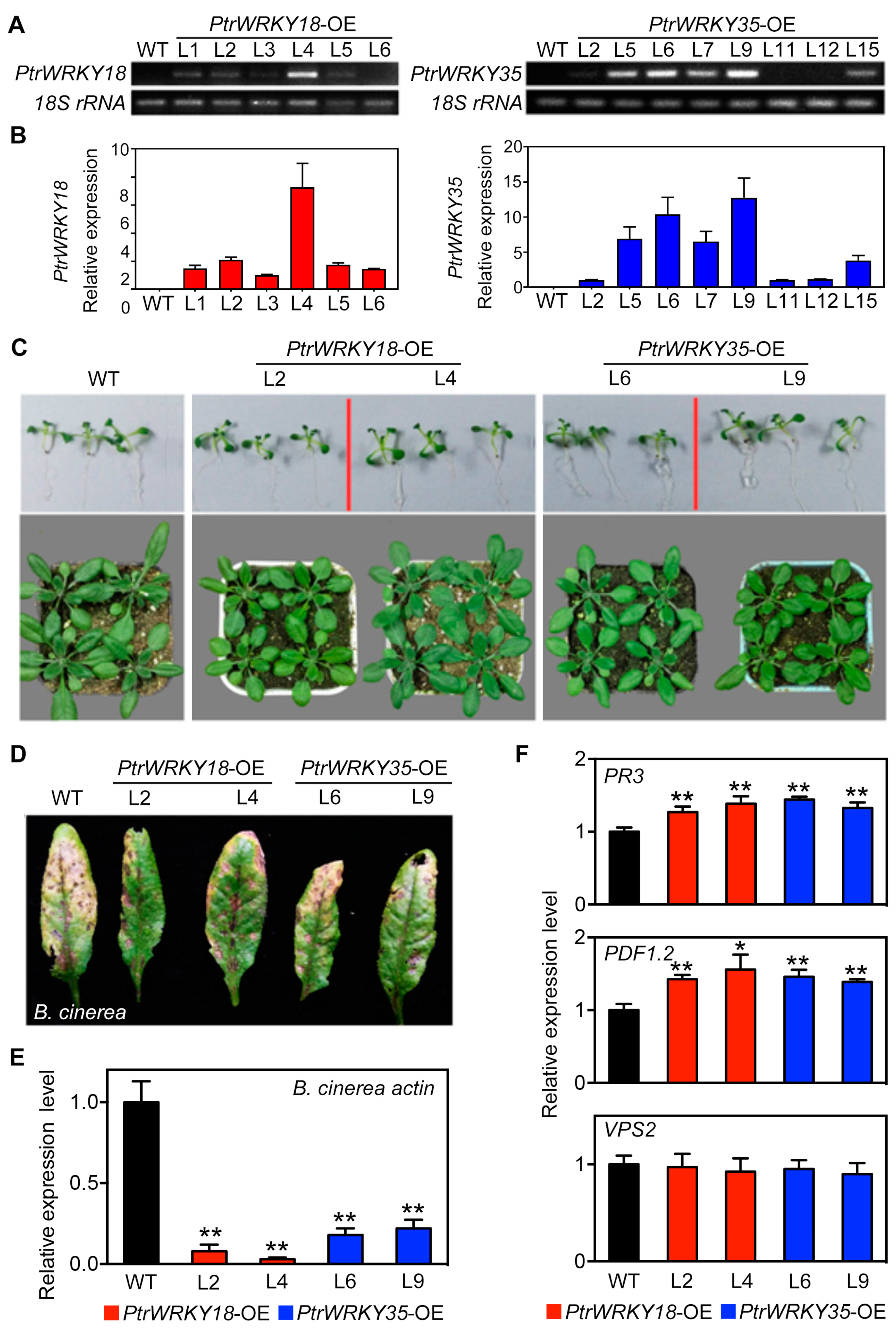
2.2. PtrWRKY18 and PtrWRKY35 Play Positive Roles in Resistance Against B. cinerea
2.3. PtrWRKY18 and PtrWRKY35 Play Negative Roles in Resistance Against P. syringae
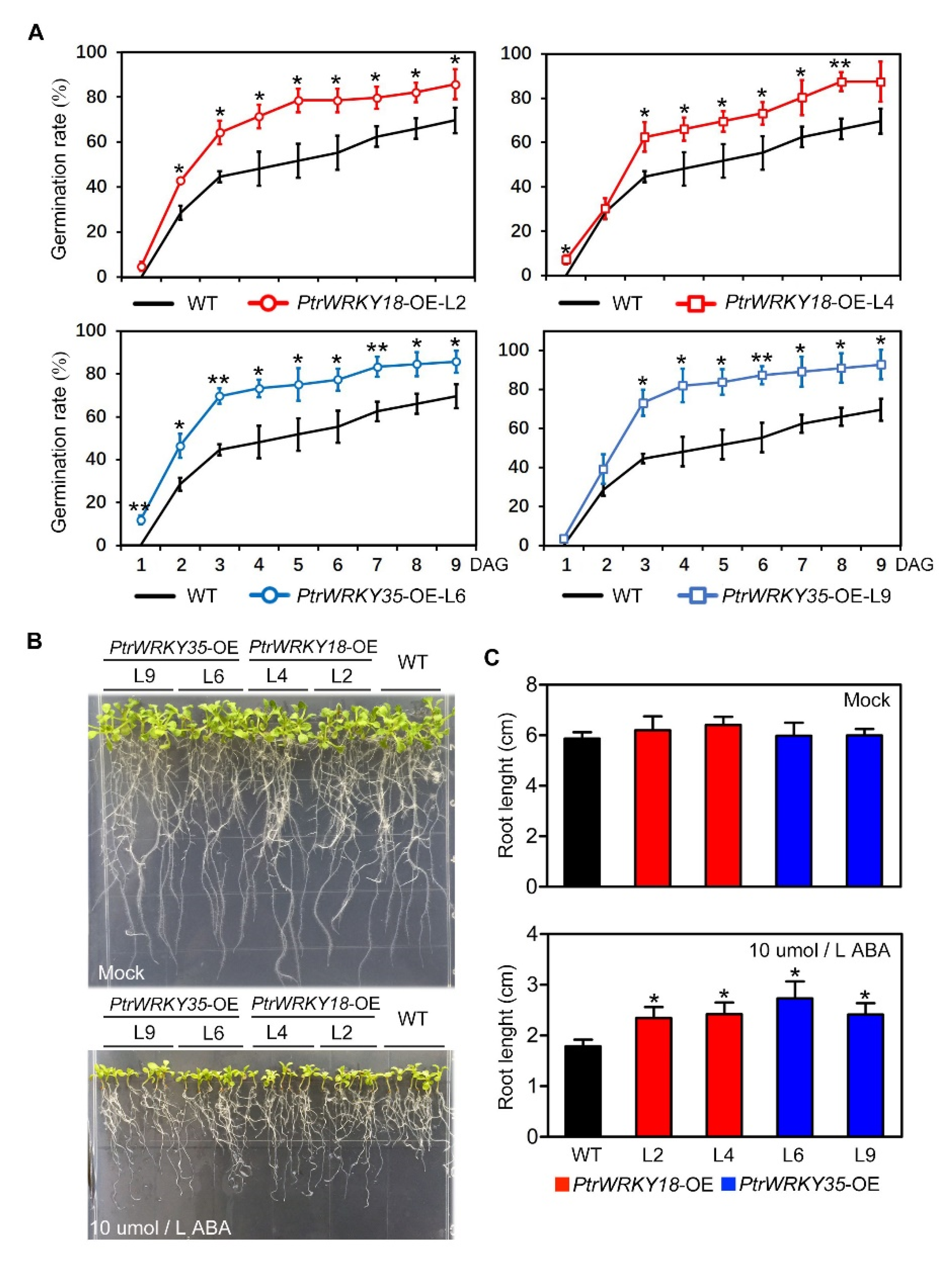
2.4. Overexpression of PtrWRKY18 and PtrWRKY35 Reduced Sensitivity to ABA in Transgenic Arabidopsis
2.5. Overexpression of PtrWRKY18 and PtrWRKY35 Reduced Drought Tolerance in Arabidopsis
3. Discussion
3.1. SA, JA, and ABA All Play Important Roles in Regulating the Expression of PtrWRKY18 and PtrWRKY35
3.2. PtrWRKY18 and PtrWRKY35 Play Antagonistic Roles in JA and SA Signaling Pathway
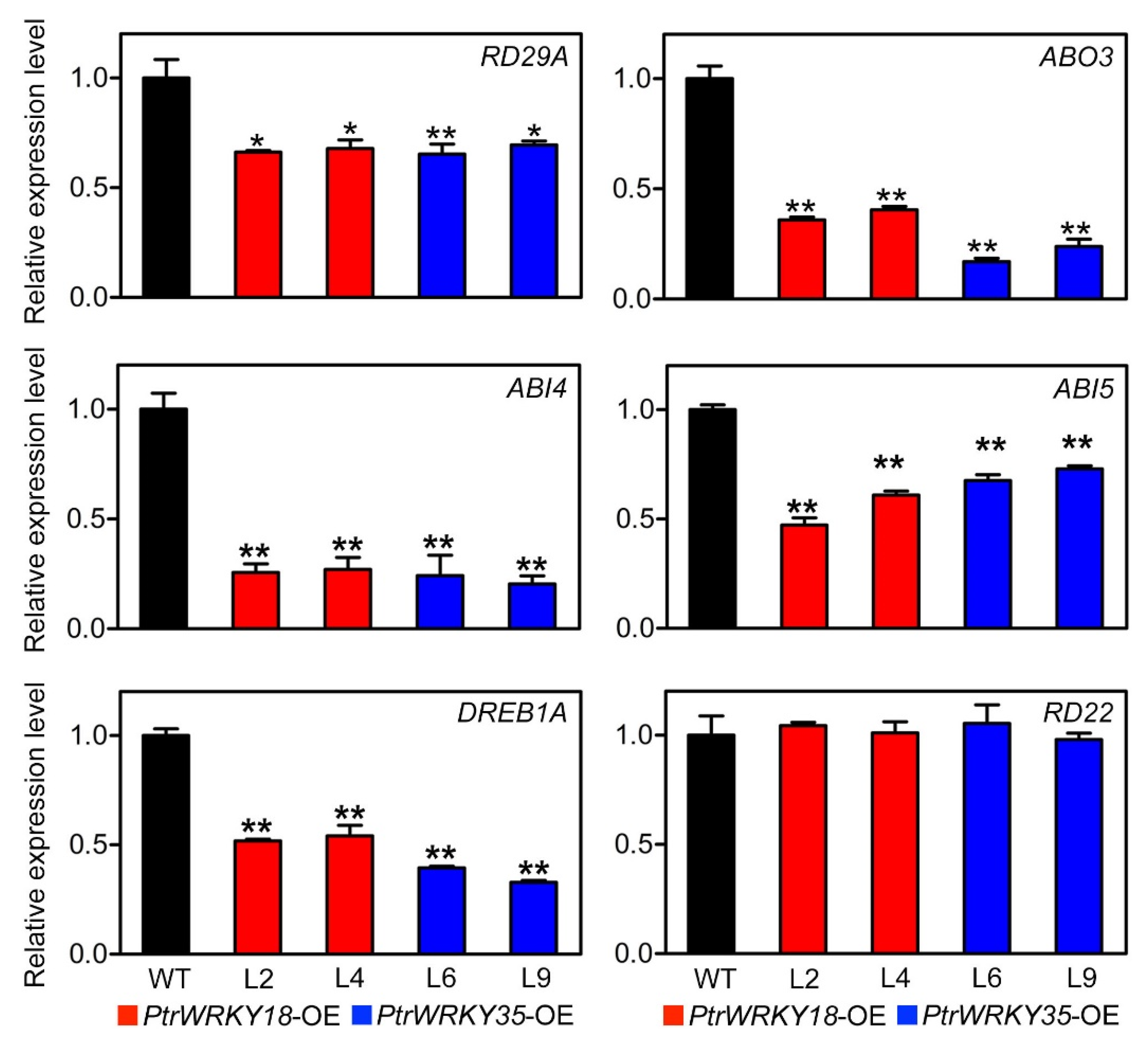
3.3. PtrWRKY18 and PtrWRKY35 Play Roles in ABA Signal Perception, and Can Regulate the Tolerance to Osmotic Stress through Stomatal Movement
4. Materials and Methods
4.1. Plant Material and Treatments
4.2. GUS Staining
4.3. Chlorophyll Content Detection
4.4. Growth State Detection of P. syringae in Plants
4.5. Stomatal Movement Assay Response to ABA
4.6. Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The Plant NADPH Oxidase RBOHD Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Zhu, Z.; Shi, J.; Cao, J.; He, M.; Wang, Y. VpWRKY3, a biotic and abiotic stress-related transcription factor from the Chinese wild Vitis pseudoreticulata. Plant Cell Rep. 2012, 31, 2109–2120. [Google Scholar] [CrossRef]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef]
- Oh, S.-K.; Yi, S.Y.; Yu, S.H.; Moon, J.; Park, J.M.; Choi, D. CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Mol. Cells 2006, 22, 58–64. [Google Scholar]
- Karim, A.; Jiang, Y.; Guo, L.; Ling, Z.; Ye, S.; Duan, Y.; Li, C.; Luo, K. Isolation and characterization of a subgroup IIa WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 2015, 35, 1129–1139. [Google Scholar] [CrossRef]
- Zheng, Z.; Mosher, S.L.; Fan, B.; Klessig, D.F.; Chen, Z. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Vinod, K.; Zheng, Z.; Fan, B.; Chen, Z. Roles of Arabidopsis WRKY3 and WRKY4 Transcription Factors in Plant Responses to Pathogens. BMC Plant Biol. 2008, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.-K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Liu, Z.-Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.-T.; Liang, S.; Zhang, X.-F.; Wang, X.-F.; Zhang, D.-P. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 2012, 63, 6371–6392. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.-L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and Functional Analyses of the Rice WRKY Gene Superfamily Reveal Positive and Negative Regulators of Abscisic Acid Signaling in Aleurone Cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Huang, Y.; Feng, C.-Z.; Ye, Q.; Wu, W.-H.; Chen, Y.-F. Arabidopsis WRKY6 Transcription Factor Acts as a Positive Regulator of Abscisic Acid Signaling during Seed Germination and Early Seedling Development. PLoS Genet. 2016, 12, e1005833. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef]
- Wang, H.; Avci, U.; Nakashima, J.; Hahn, M.G.; Chen, F.; Dixon, R.A. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 2010, 107, 22338–22343. [Google Scholar] [CrossRef]
- Jiang, Y.; Duan, Y.; Yin, J.; Ye, S.; Zhu, J.; Zhang, F.; Lu, W.; Fan, D.; Luo, K. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 2014, 65, 6629–6644. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Guo, L.; Ma, X.; Zhao, X.; Jiao, B.; Li, C.; Luo, K. The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 2017, 37, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; De Vos, R.C.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Boil. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Clouse, S.D. Brassinosteroid/Abscisic Acid Antagonism in Balancing Growth and Stress. Dev. Cell 2016, 38, 118–120. [Google Scholar] [CrossRef]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.P. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Amagai, A.; Honda, Y.; Ishikawa, S.; Hara, Y.; Kuwamura, M.; Shinozawa, A.; Sugiyama, N.; Ishihama, Y.; Takezawa, D.; Sakata, Y.; et al. Phosphoproteomic profiling reveals ABA—Responsive phosphosignaling pathways in Physcomitrella patens. Plant J. 2018, 94, 699–708. [Google Scholar] [CrossRef]
- Stevenson, S.R.; Kamisugi, Y.; Trinh, C.H.; Schmutz, J.; Jenkins, J.; Grimwood, J.; Muchero, W.; Tuskan, G.A.; Rensing, S.A.; Lang, D.; et al. Genetic Analysis of Physcomitrella patens Identifies ABSCISIC ACID NON-RESPONSIVE, a Regulator of ABA Responses Unique to Basal Land Plants and Required for Desiccation Tolerance. Plant Cell 2016, 28, 1310–1327. [Google Scholar] [CrossRef]
- Luan, S. Signalling drought in guard cells. Plant Cell Environ. 2002, 25, 229–237. [Google Scholar] [CrossRef]
- Hovenden, M.J.; Schoor, J.K.V. Nature vs nurture in the leaf morphology of Southern beech, Nothofagus cunninghamii (Nothofagaceae). New Phytol. 2004, 161, 585–594. [Google Scholar] [CrossRef]
- Wei, T.; Deng, K.; Liu, N.; Gao, Y.; Yang, M.; Zhang, Y.; Zheng, X.; Wang, C.; Song, W.; Chen, C. Ectopic expression of DREB transcription factor, AtDREB1A, confers tolerance to drought in transgenic Salvia miltiorrhiza. Plant Cell Physiol. 2016, 57, 1593–1609. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Hu, L.; Yuan, D.; Xu, J.; Gao, W.; He, L.; Yang, X.; Zhang, X. Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: Evidence for stress response functions in SE development. Plant Biotechnol. J. 2013, 12, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a WRKY Transcription Factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Pesch, M.; Hülskamp, M. One, two, three…models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 2009, 12, 587–592. [Google Scholar] [CrossRef]
- Miao, Y.; Zentgraf, U. The Antagonist Function of Arabidopsis WRKY53 and ESR/ESP in Leaf Senescence Is Modulated by the Jasmonic and Salicylic Acid Equilibrium. Plant Cell 2007, 19, 819–830. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Wei, W.; Hu, Y.; Han, Y.-T.; Zhang, K.; Zhao, F.-L.; Feng, J.-Y. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 2016, 105, 129–144. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 Is a Key Transcriptional Regulator of Hormonal and Metabolic Responses toward Botrytis cinerea Infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.-Q.; Chen, Z. Protein–Protein Interactions in the Regulation of WRKY Transcription Factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and Functional Interactions between Pathogen-Induced Arabidopsis WRKY18, WRKY40, and WRKY60 Transcription Factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef]
- Shen, Q.-H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ülker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear Activity of MLA Immune Receptors Links Isolate-Specific and Basal Disease-Resistance Responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef]
- Kim, K.-C.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 Transcription Factors Interact with Histone Deacetylase 19 in Basal Defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef]
- Schwessinger, B.; Bahar, O.; Thomas, N.; Holton, N.; Nekrasov, V.; Ruan, D.; Canlas, P.E.; Daudi, A.; Petzold, J.; Singan, V.R.; et al. Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PloS Pathog. 2015, 11, e1004872. [Google Scholar]
- Wang, M.; Vannozzi, A.; Wang, G.; Liang, Y.-H.; Tornielli, G.B.; Zenoni, S.; Cavallini, E.; Pezzotti, M.; Cheng, Z.-M. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 2014, 1, 14016. [Google Scholar] [CrossRef]
- Ye, S.; Jiang, Y.; Duan, Y.; Karim, A.; Fan, D.; Yang, L.; Zhao, X.; Yin, J.; Luo, K.; Li, C. Constitutive expression of the poplar WRKY transcription factor PtoWRKY60 enhances resistance to Dothiorella gregaria Sacc. in transgenic plants. Tree Physiol. 2014, 34, 1118–1129. [Google Scholar] [CrossRef]
- Adachi, H.; Nakano, T.; Miyagawa, N.; Ishihama, N.; Yoshioka, M.; Katou, Y.; Yaeno, T.; Shirasu, K.; Yoshioka, H. WRKY Transcription Factors Phosphorylated by MAPK Regulate a Plant Immune NADPH Oxidase in Nicotiana benthamiana. Plant Cell 2015, 27, 2645–2663. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D.; et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 2014, 150, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, E.; Lai, Y.; Liu, Z.-Q.; Wang, X.; Qiu, A.-L.; Zhang, T.-X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.-W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.-K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, G.; Yu, D. Activated Expression of WRKY57 Confers Drought Tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The Cotton WRKY Transcription Factor GhWRKY17 Functions in Drought and Salt Stress in Transgenic Nicotiana benthamiana Through ABA Signaling and the Modulation of Reactive Oxygen Species Production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Jiang, Y.; Guo, L.; Liu, R.; Jiao, B.; Zhao, X.; Ling, Z.; Luo, K. Overexpression of Poplar PtrWRKY89 in Transgenic Arabidopsis Leads to a Reduction of Disease Resistance by Regulating Defense-Related Genes in Salicylate- and Jasmonate-Dependent Signaling. PLoS ONE 2016, 11, e0149137. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, L.; Li, C.; Jiang, Y.; Luo, K.; Xu, C. Heterologous Expression of Poplar WRKY18/35 Paralogs in Arabidopsis Reveals Their Antagonistic Regulation on Pathogen Resistance and Abiotic Stress Tolerance via Variable Hormonal Pathways. Int. J. Mol. Sci. 2020, 21, 5440. https://doi.org/10.3390/ijms21155440
Guo L, Li C, Jiang Y, Luo K, Xu C. Heterologous Expression of Poplar WRKY18/35 Paralogs in Arabidopsis Reveals Their Antagonistic Regulation on Pathogen Resistance and Abiotic Stress Tolerance via Variable Hormonal Pathways. International Journal of Molecular Sciences. 2020; 21(15):5440. https://doi.org/10.3390/ijms21155440
Chicago/Turabian StyleGuo, Li, Chaofeng Li, Yuanzhong Jiang, Keming Luo, and Changzheng Xu. 2020. "Heterologous Expression of Poplar WRKY18/35 Paralogs in Arabidopsis Reveals Their Antagonistic Regulation on Pathogen Resistance and Abiotic Stress Tolerance via Variable Hormonal Pathways" International Journal of Molecular Sciences 21, no. 15: 5440. https://doi.org/10.3390/ijms21155440
APA StyleGuo, L., Li, C., Jiang, Y., Luo, K., & Xu, C. (2020). Heterologous Expression of Poplar WRKY18/35 Paralogs in Arabidopsis Reveals Their Antagonistic Regulation on Pathogen Resistance and Abiotic Stress Tolerance via Variable Hormonal Pathways. International Journal of Molecular Sciences, 21(15), 5440. https://doi.org/10.3390/ijms21155440




