Knockout of the Transducin-Like Enhancer of Split 6 Gene Affects the Proliferation and Cell Cycle Process of Mouse Spermatogonia
Abstract
1. Introduction
2. Results
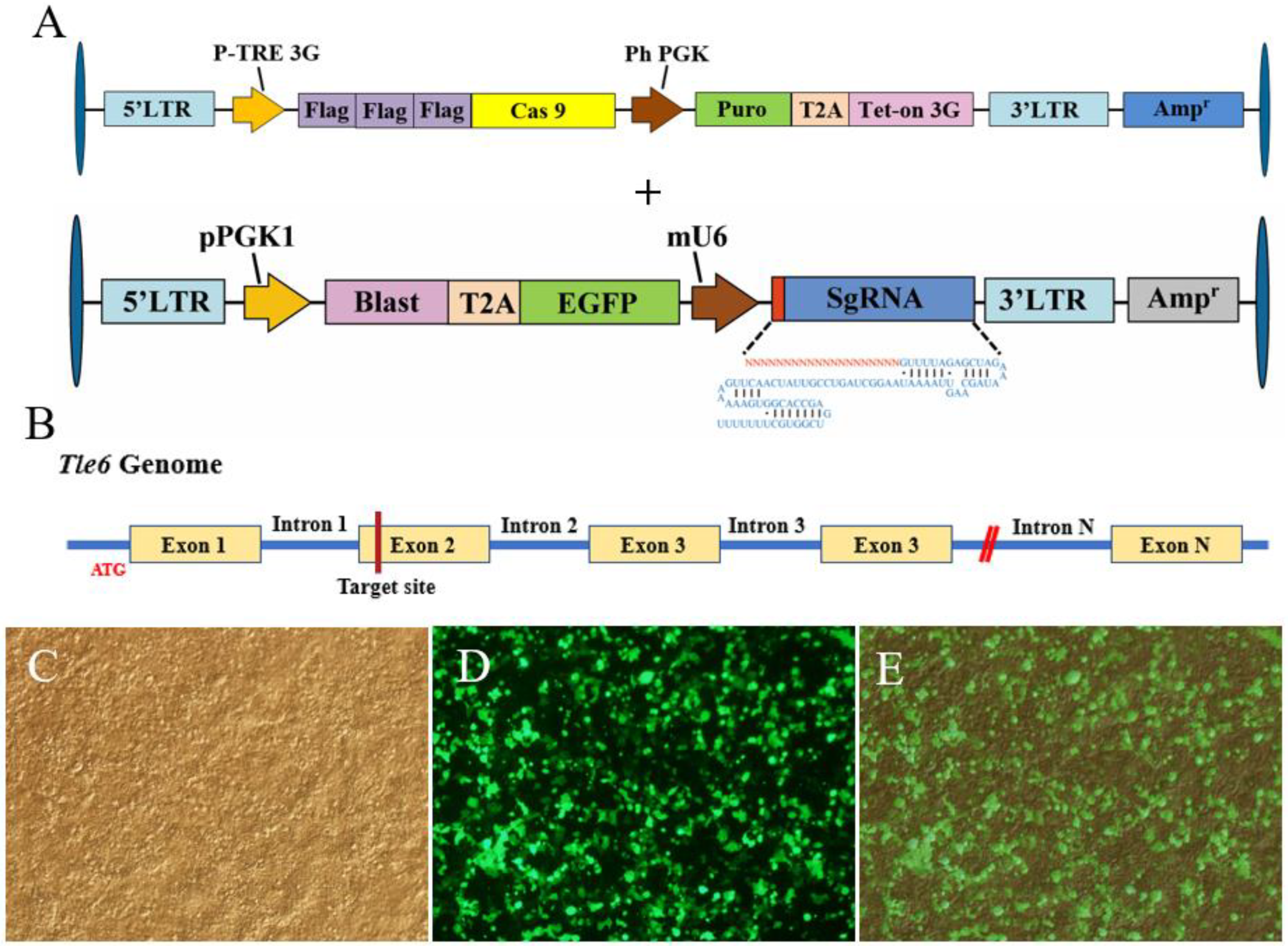
2.1. CRISPR/Cas9-Mediated KO of Tle6 in Spermatogonia
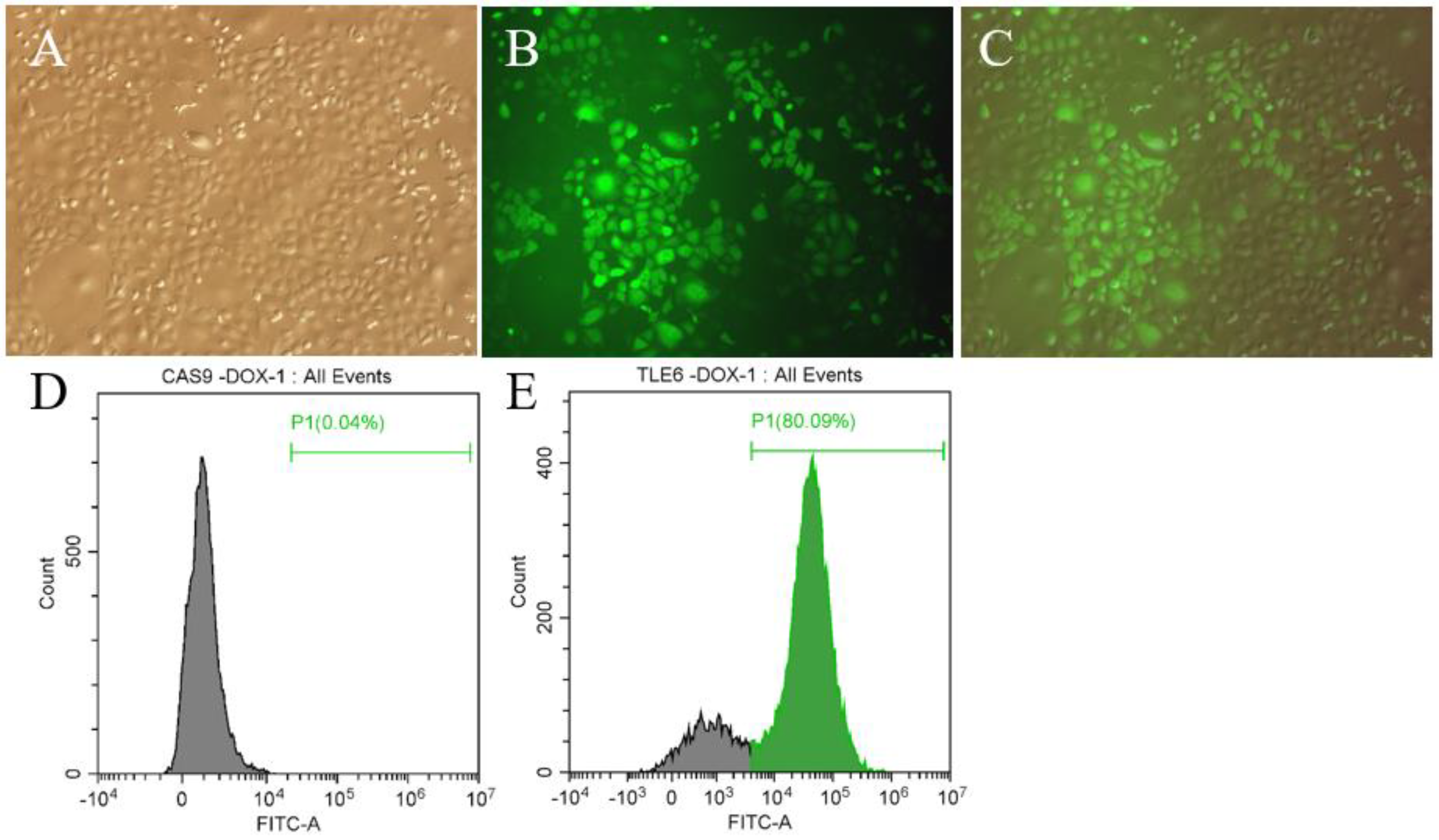
2.2. Detection of the Effect of Tle6 Knockout in Mouse Spermatogonia
2.3. The Effects of the Tle6 Gene Knockout on the Proliferation of Mouse Spermatogonia
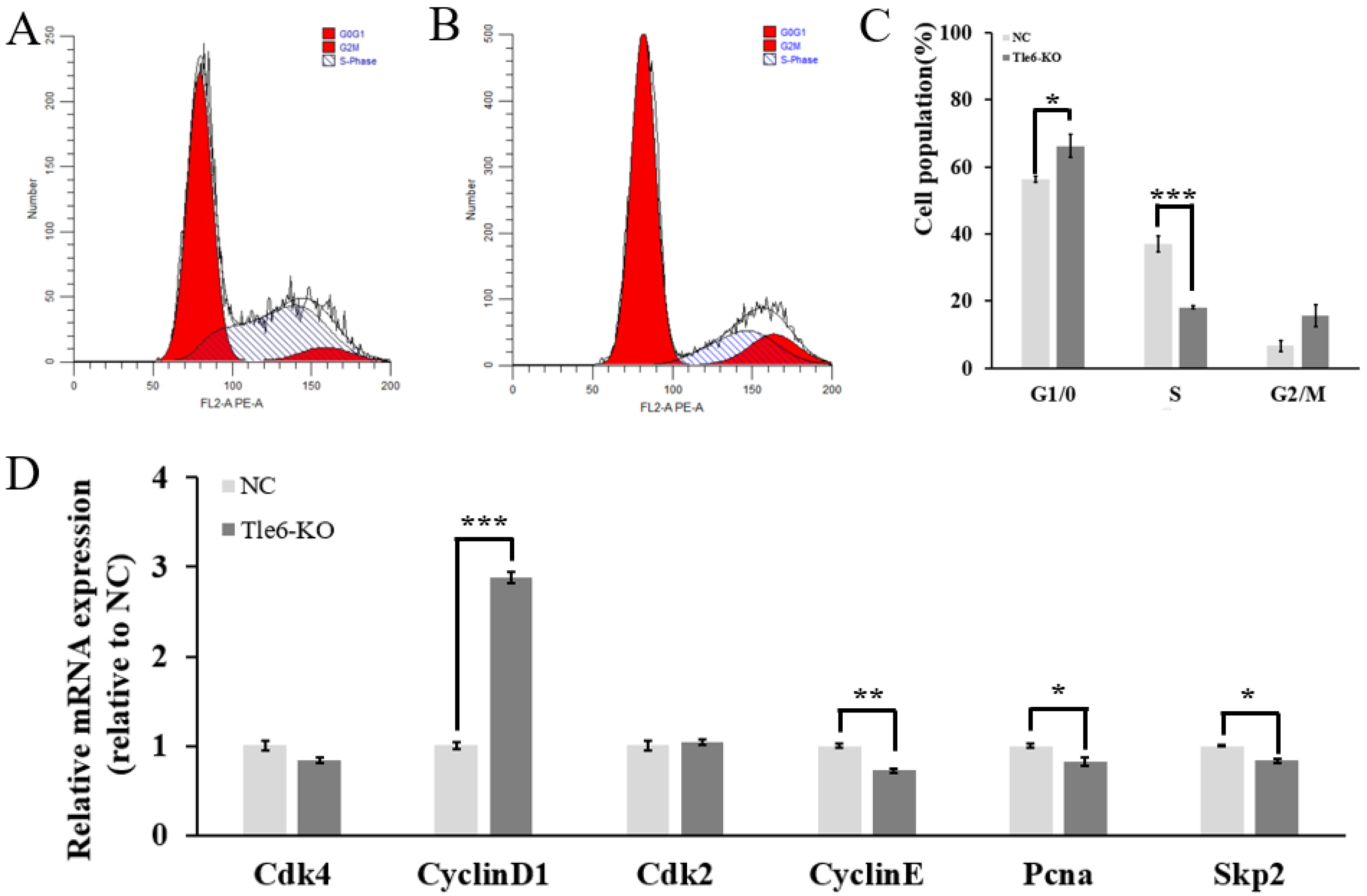
2.4. The Effects of Tle6 Gene Knockout on the Mouse Spermatogonia Cell Cycle
3. Discussion
4. Materials and Methods
4.1. Cell Culture and CRISPR/Cas9-Mediated KO of Tle6 in Spermatogonia
4.2. Target Gene Amplification
4.3. Western Blot Analysis
4.4. Cell Proliferation Analysis
4.5. Cell Cycle and Cell Apoptosis Analysis
4.6. Real-Time Quantity PCR Assay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, W.F.; Wang, Y.G.; Reginato, A.M.; Plotkina, S.; Gridley, T.; Olsen, B.R. Growth defect in Grg5 null mice is associated with reduced Ihh signaling in growth plates. Dev. Dyn. 2002, 224, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, Y.-G.; Reginato, A.M.; Glotzer, D.J.; Fukai, N.; Plotkina, S.; Karsenty, G.; Olsen, B.R. Groucho homologue Grg5 interacts with the transcription factor Runx2–Cbfa1 and modulates its activity during postnatal growth in mice. Dev. Boil. 2004, 270, 364–381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, H.-M.; Jaramillo, E.; Wang, L.; D’Mello, S.R. Histone deacetylase-related protein inhibits AES-mediated neuronal cell death by direct interaction. J. Neurosci. Res. 2008, 86, 2423–2431. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Inukai, T.; Kurosawa, H.; Goi, K.; Inaba, T.; Lenny, N.T.; Downing, J.R.; Stifani, S.; Look, A.T. The E2A-HLF Oncoprotein ActivatesGroucho-Related Genes and SuppressesRunx1. Mol. Cell. Boil. 2001, 21, 5935–5945. [Google Scholar] [CrossRef]
- Li, D.; Roberts, R. WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001, 58, 2085–2097. [Google Scholar] [CrossRef]
- Marcal, N.; Patel, H.; Dong, Z.; Belanger-Jasmin, S.; Hoffman, B.; Helgason, C.D.; Dang, J.; Stifani, S. Antagonistic effects of Grg6 and Groucho/TLE on the transcription repression activity of brain factor 1/FoxG1 and cortical neuron differentiation. Mol. Cell Biol. 2005, 25, 10916–10929. [Google Scholar] [CrossRef]
- Bebbere, D.; Ariu, F.; Bogliolo, L.; Masala, L.; Murrone, O.; Fattorini, M.; Falchi, L.; Ledda, S. Expression of maternally derived KHDC3, NLRP5, OOEP and TLE6 is associated with oocyte developmental competence in the ovine species. BMC Dev. Boil. 2014, 14, 40. [Google Scholar]
- Li, L.; Baibakov, B.; Dean, J. A Subcortical Maternal Complex Essential for Preimplantation Mouse Embryogenesis. Dev. Cell 2008, 15, 416–425. [Google Scholar] [CrossRef]
- Yu, X.-J.; Yi, Z.; Gao, Z.; Qin, D.; Zhai, Y.; Chen, X.; Ou-Yang, Y.; Wang, Z.-B.; Zheng, P.; Zhu, M.-S.; et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat. Commun. 2014, 5, 4887. [Google Scholar] [CrossRef]
- Alazami, A.M.; Awad, S.M.; Coskun, S.; Al-Hassan, S.; Hijazi, H.; Abdulwahab, F.M.; Poizat, C.; Alkuraya, F.S. TLE6 mutation causes the earliest known human embryonic lethality. Genome Boil. 2015, 16, 240. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93–115. [Google Scholar] [CrossRef]
- Tchakarska, G.; Sola, B. The double dealing of cyclin D1. Cell Cycle 2019, 19, 163–178. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Li, J.; Mao, Q. Construction of a Single Lentiviral Vector Containing Tetracycline-Inducible Alb-uPA for Transduction of uPA Expression in Murine Hepatocytes. PLoS ONE 2013, 8, e61412. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bai, Y.; Zhu, C.; Feng, M.; Pan, B.; Zhang, S.; Zhan, X.; Chen, H.; Wang, B.; Li, J. Establishment of A Reversibly Inducible Porcine Granulosa Cell Line. Cells 2020, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liu, K.; Wei, H.; Li, L.; Zhang, S. Generation of porcine fetal fibroblasts expressing the tetracycline-inducible Cas9 gene by somatic cell nuclear transfer. Mol. Med. Rep. 2016, 14, 2527–2533. [Google Scholar] [CrossRef][Green Version]
- Chen, P.-C.; Kuraguchi, M.; Velasquez, J.; Wang, Y.; Yang, K.; Edwards, R.; Gillen, D.; Edelmann, W.; Kucherlapati, R.; Lipkin, S.M. Novel Roles for MLH3 Deficiency and TLE6-Like Amplification in DNA Mismatch Repair-Deficient Gastrointestinal Tumorigenesis and Progression. PLoS Genet. 2008, 4, e1000092. [Google Scholar] [CrossRef]
- Verginelli, F.; Perin, A.; Dali, R.; Fung, K.H.; Lo, R.; Longatti, P.; Guiot, M.-C.; Del Maestro, R.F.; Rossi, S.; Di Porzio, U.; et al. Transcription factors FOXG1 and Groucho/TLE promote glioblastoma growth. Nat. Commun. 2013, 4, 2956. [Google Scholar] [CrossRef]
- Welte, K.H. G-CSF: Filgrastim, lenograstim and biosimilars. Expert Opin. Boil. Ther. 2014, 14, 983–993. [Google Scholar] [CrossRef]
- Miyamoto, M.; Natsume, H.; Satoh, I.; Ohtake, K.; Yamaguchi, M.; Kobayashi, D.; Sugibayashi, K.; Morimoto, Y. Effect of poly-l-arginine on the nasal absorption of FITC-dextran of different molecular weights and recombinant human granulocyte colony-stimulating factor (rhG-CSF) in rats. Int. J. Pharm. 2001, 226, 127–138. [Google Scholar] [CrossRef]
- Kotzur, T.; Benavides-Garcia, R.; Mecklenburg, J.; Sanchez, J.R.; Reilly, M.A.; Hermann, B.P. Granulocyte colony-stimulating factor (G-CSF) promotes spermatogenic regeneration from surviving spermatogonia after high-dose alkylating chemotherapy. Reprod. Boil. Endocrinol. 2017, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Begay, V.; Baumeier, C.; Zimmermann, K.; Heuser, A.; Leutz, A. The C/EBPbeta LIP isoform rescues loss of C/EBPbeta function in the mouse. Sci. Rep. 2018, 8, 8417. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.B.; Porse, B.T. C/EBPalpha: A tumour suppressor in multiple tissues? Biochim. Biophys. Acta. 2006, 1766, 88–103. [Google Scholar] [PubMed]
- Chen, S.S.; Chen, J.F.; Johnson, P.F.; Muppala, V.; Lee, Y.H. C/EBPbeta, when expressed from the C/EBPalpha gene locus, can functionally replace C/EBPalpha in liver but not in adipose tissue. Mol. Cell Biol. 2000, 20, 7292–7299. [Google Scholar] [CrossRef]
- Jones, L.C.; Lin, M.-L.; Chen, S.-S.; Krug, U.; Hofmann, W.; Lee, S.; Lee, Y.-H.; Koeffler, H.P. Expression of C/EBPbeta from the C/ebpalpha gene locus is sufficient for normal hematopoiesis in vivo. Blood 2002, 99, 2032–2036. [Google Scholar] [CrossRef]
- Hoffman, B.G.; Zavaglia, B.; Beach, M.; Helgason, C.D. Expression of Groucho/TLE proteins during pancreas development. BMC Dev. Boil. 2008, 8, 81. [Google Scholar] [CrossRef]
- Duncan, F.E.; Padilla-Banks, E.; Bernhardt, M.L.; Ord, T.S.; Jefferson, W.N.; Moss, S.B.; Williams, C.J. Transducin-Like Enhancer of Split-6 (TLE6) Is a Substrate of Protein Kinase A Activity During Mouse Oocyte Maturation1. Boil. Reprod. 2014, 90, 63. [Google Scholar] [CrossRef]
- Zhu, K.; Yan, L.-Y.; Zhang, X.; Lu, X.; Wang, T.; Liu, X.; Qiao, J.; Li, L. Identification of a human subcortical maternal complex. Mol. Hum. Reprod. 2014, 21, 320–329. [Google Scholar] [CrossRef]
- Swanton, C. Cell-cycle targeted therapies. Lancet Oncol. 2004, 5, 27–36. [Google Scholar] [CrossRef]
- Bochis, O.V.; Irimie, A.; Pichler, M.; Neagoe, I.B. The Role of Skp2 and its Substrate CDKN1B (p27) in Colorectal Cancer. J. Gastrointest. Liver Dis. 2015, 24, 225–234. [Google Scholar] [CrossRef]
- Malumbres, M.; Barbacid, M. To cycle or not to cycle: A critical decision in cancer. Nat. Rev. Cancer 2001, 1, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Cao, L.; Sun, Z.; Xu, J.; Tang, L.; Chen, W.; Luo, J.; Yang, F.; Wang, Y.; Guan, X. Skp2 is over-expressed in breast cancer and promotes breast cancer cell proliferation. Cell Cycle 2016, 15, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, E.; Bartnicki, F.; Fujisawa, R.; Bonarek, P.; Hermanowicz, P.; Tsurimoto, T.; Muszyńska, K.; Strzalka, W. Inhibition of DNA replication by an anti-PCNA aptamer/PCNA complex. Nucleic Acids Res. 2018, 46, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.C.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 2013, 343, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Xue, X.; Fu, J. Effect of OLIG1 on the development of oligodendrocytes and myelination in a neonatal rat PVL model induced by hypoxia-ischemia. Mol. Med. Rep. 2014, 11, 2379–2386. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, M.; Bai, Y.; Chen, Y.; Wang, K. Knockout of the Transducin-Like Enhancer of Split 6 Gene Affects the Proliferation and Cell Cycle Process of Mouse Spermatogonia. Int. J. Mol. Sci. 2020, 21, 5827. https://doi.org/10.3390/ijms21165827
Feng M, Bai Y, Chen Y, Wang K. Knockout of the Transducin-Like Enhancer of Split 6 Gene Affects the Proliferation and Cell Cycle Process of Mouse Spermatogonia. International Journal of Molecular Sciences. 2020; 21(16):5827. https://doi.org/10.3390/ijms21165827
Chicago/Turabian StyleFeng, Meiying, Yinshan Bai, Yun Chen, and Kai Wang. 2020. "Knockout of the Transducin-Like Enhancer of Split 6 Gene Affects the Proliferation and Cell Cycle Process of Mouse Spermatogonia" International Journal of Molecular Sciences 21, no. 16: 5827. https://doi.org/10.3390/ijms21165827
APA StyleFeng, M., Bai, Y., Chen, Y., & Wang, K. (2020). Knockout of the Transducin-Like Enhancer of Split 6 Gene Affects the Proliferation and Cell Cycle Process of Mouse Spermatogonia. International Journal of Molecular Sciences, 21(16), 5827. https://doi.org/10.3390/ijms21165827




