Two-Player Game in a Complex Landscape: 26S Proteasome, PKA, and Intracellular Calcium Concentration Modulate Mammalian Sperm Capacitation by Creating an Integrated Dialogue—A Computational Analysis
Abstract
1. Introduction
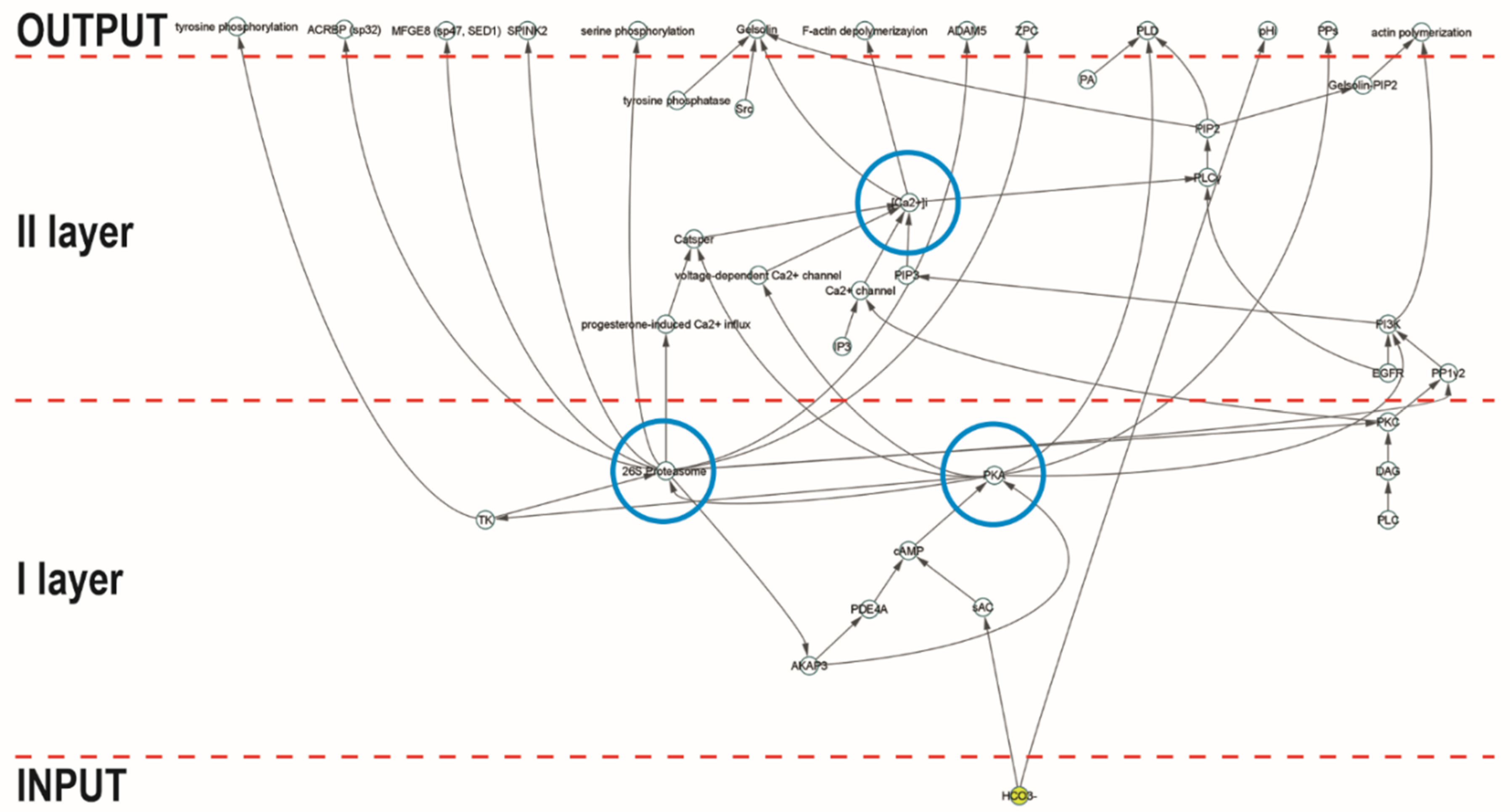
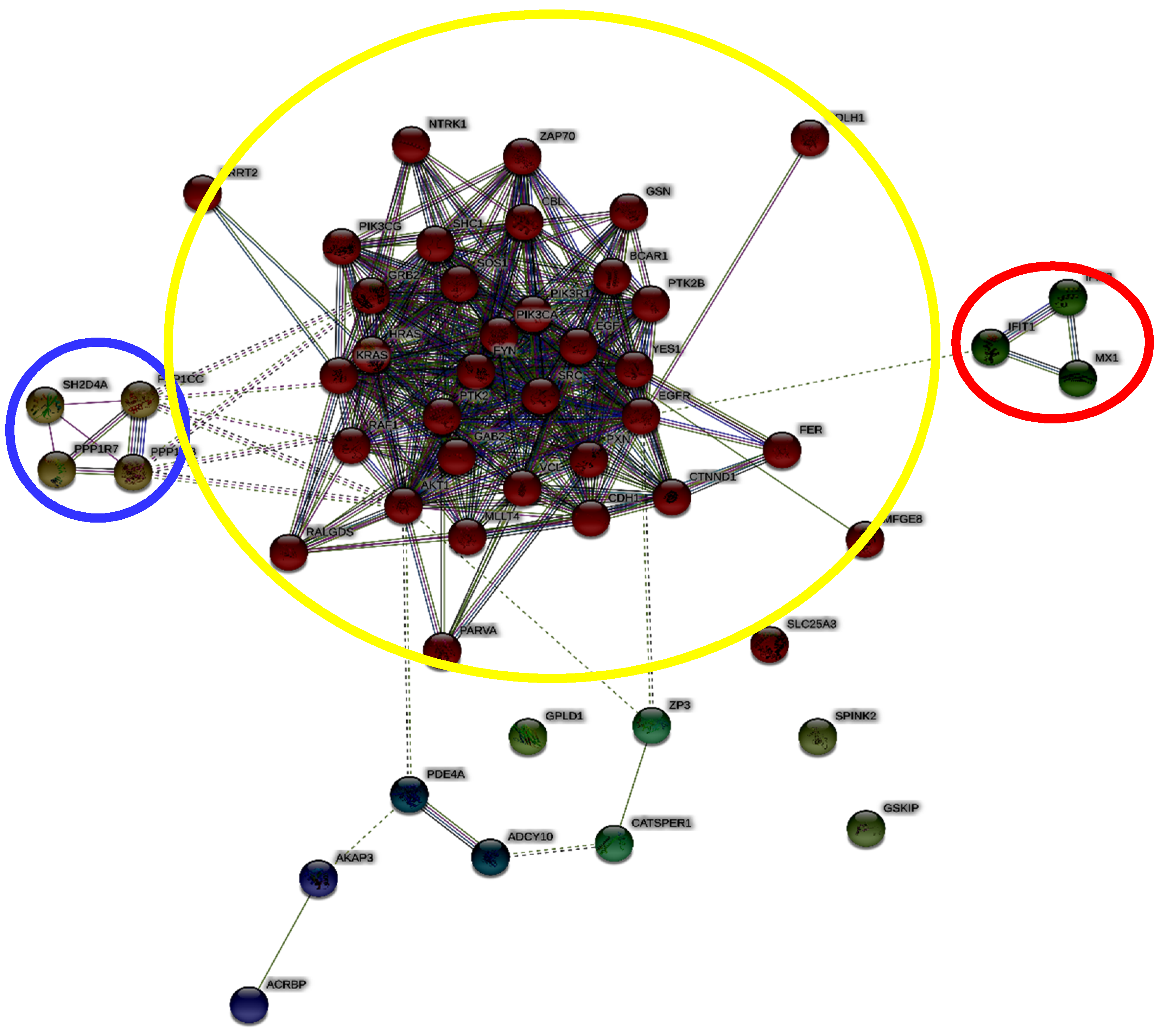
2. Results and Discussion
3. Materials and Methods
3.1. Data Collection, Network Creation, and General Analysis
3.2. Identification of 26SN Hubs
3.3. Identification of Bottleneck Nodes within 26SN
3.4. Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Suarez, S.S. Formation of a Reservoir of Sperm in the Oviduct. Reprod. Domest. Anim. 2002, 37, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Bernabò, N.; Greco, L.; Ordinelli, A.; Mattioli, M.; Barboni, B. Capacitation-Related Lipid Remodeling of Mammalian Spermatozoa Membrane Determines the Final Fate of Male Gametes: A Computational Biology Study. OMICS A J. Integr. Boil. 2015, 19, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Ickowicz, D.; Finkelstein, M.; Breitbart, H. Mechanism of sperm capacitation and the acrosome reaction: Role of protein kinases. Asian J. Androl. 2012, 14, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.C.P.; Luque, G.M.; Balestrini, P.A.; Marín-Briggiler, C.I.; Romarowski, A.; Buffone, M.G. Molecular Basis of Human Sperm Capacitation. Front. Cell Dev. Boil. 2018, 6, 1–23. [Google Scholar] [CrossRef]
- Signorelli, J.; Diaz, E.S.; Morales, P. Kinases, phosphatases and proteases during sperm capacitation. Cell Tissue Res. 2012, 349, 765–782. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Boil. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Kerns, K.C.; Morales, P.; Sutovsky, P. Regulation of Sperm Capacitation by the 26S Proteasome: An Emerging New Paradigm in Spermatology. Boil. Reprod. 2016, 94, 117. [Google Scholar] [CrossRef]
- Navon, A.; Ciechanover, A. The 26 S Proteasome: From Basic Mechanisms to Drug Targeting. J. Boil. Chem. 2009, 284, 33713–33718. [Google Scholar] [CrossRef]
- Morales, P.; Kong, M.; Pizarro, E.; Pasten, C. Participation of the sperm proteasome in human fertilization. Hum. Reprod. 2003, 18, 1010–1017. [Google Scholar] [CrossRef]
- Bard, J.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Ciechanover, A.; Iwai, K. The Ubiquitin System: From Basic Mechanisms to the Patient Bed. IUBMB Life 2004, 56, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.; Sutovsky, P. The sperm proteasome during sperm capacitation and fertilization. J. Reprod. Immunol. 2009, 83, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Zapata-Carmona, H.; Barón, L.; Zuñiga, L.M.; Díaz, E.S.; Kong, M.; Drobnis, E.Z.; Sutovsky, P.; Morales, P. The activation of the chymotrypsin-like activity of the proteasome is regulated by soluble adenyl cyclase/cAMP/protein kinase A pathway and required for human sperm capacitation. Mol. Hum. Reprod. 2019, 25, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Balbach, M.; Beckert, V.; Hansen, J.N.; Wachten, D. Shedding light on the role of cAMP in mammalian sperm physiology. Mol. Cell. Endocrinol. 2018, 468, 111–120. [Google Scholar] [CrossRef]
- Kong, M.; Diaz, E.S.; Morales, P. Participation of the Human Sperm Proteasome in the Capacitation Process and Its Regulation by Protein Kinase A and Tyrosine Kinase1. Boil. Reprod. 2009, 80, 1026–1035. [Google Scholar] [CrossRef]
- Zhang, F.; Hu, Y.; Huang, P.; Toleman, C.A.; Paterson, A.J.; Kudlow, J.E. Proteasome Function Is Regulated by Cyclic AMP-dependent Protein Kinase through Phosphorylation of Rpt6. J. Boil. Chem. 2007, 282, 22460–22471. [Google Scholar] [CrossRef]
- Hillman, P.; Ickowicz, D.; Vizel, R.; Breitbart, H. Dissociation between AKAP3 and PKARII Promotes AKAP3 Degradation in Sperm Capacitation. PLoS ONE 2013, 8, e68873. [Google Scholar] [CrossRef]
- Zigo, M.; Manaskova-Postlerova, P.; Jonakova, V.; Kerns, K.C.; Sutovsky, P. Compartmentalization of the proteasome-interacting proteins during sperm capacitation. Sci. Rep. 2019, 9, 12583. [Google Scholar] [CrossRef]
- Yi, Y.-J.; Zimmerman, S.W.; Manandhar, G.; Odhiambo, J.F.; Kennedy, C.; Jonáková, V.; Maňásková, P.; Sutovsky, M.; Park, C.-S.; Šutovský, P. Ubiquitin-activating enzyme (UBA1) is required for sperm capacitation, acrosomal exocytosis and sperm-egg coat penetration during porcine fertilization. Int. J. Androl. 2011, 35, 196–210. [Google Scholar] [CrossRef]
- Zigo, M.; Jonakova, V.; Manaskova-Postlerova, P.; Kerns, K.; Sutovsky, P. Ubiquitin-proteasome system participates in the de-aggregation of spermadhesins and DQH protein during boar sperm capacitation. Reproduction 2019, 157, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Bernabò, N.; Barboni, B.; Maccarrone, M. The biological networks in studying cell signal transduction complexity: The examples of sperm capacitation and of endocannabinoid system. Comput. Struct. Biotechnol. J. 2014, 11, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Darszon, A.; Nishigaki, T.; Beltran, C.; Treviño, C.L. Calcium Channels in the Development, Maturation, and Function of Spermatozoa. Physiol. Rev. 2011, 91, 1305–1355. [Google Scholar] [CrossRef]
- Orta, G.; De La Vega-Beltran, J.L.; Hidalgo, D.M.; Santi, C.M.; Visconti, P.E.; Darszon, A. CatSper channels are regulated by protein kinase A. J. Boil. Chem. 2018, 293, 16830–16841. [Google Scholar] [CrossRef] [PubMed]
- Machado, S.A.; Sharif, M.; Wang, H.; Bovin, N.; Miller, D.J. Release of Porcine Sperm from Oviduct Cells is Stimulated by Progesterone and Requires CatSper. Sci. Rep. 2019, 9, 19546. [Google Scholar] [CrossRef] [PubMed]
- Gadella, B.M.; Tsai, P.-S.; Boerke, A.; Brewis, I.A. Sperm head membrane reorganisation during capacitation. Int. J. Dev. Boil. 2008, 52, 473–480. [Google Scholar] [CrossRef]
- Hickey, K.D.; Buhr, M.M. Lipid Bilayer Composition Affects Transmembrane Protein Orientation and Function. J. Lipids 2011, 2011, 1–9. [Google Scholar] [CrossRef]
- Bernabò, N.; Valbonetti, L.; Greco, L.; Capacchietti, G.; Sanchez, M.R.; Palestini, P.; Botto, L.; Mattioli, M.; Barboni, B. Aminopurvalanol A, a Potent, Selective, and Cell Permeable Inhibitor of Cyclins/Cdk Complexes, Causes the Reduction of in Vitro Fertilizing Ability of Boar Spermatozoa, by Negatively Affecting the Capacitation-Dependent Actin Polymerization. Front. Physiol. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Bernabò, N.; Ramal-Sanchez, M.; Valbonetti, L.; Machado-Simoes, J.; Ordinelli, A.; Capacchietti, G.; Taraschi, A.; Barboni, B.; Sanchez, R.; Simoes, M. Cyclin–CDK Complexes are Key Controllers of Capacitation-Dependent Actin Dynamics in Mammalian Spermatozoa. Int. J. Mol. Sci. 2019, 20, 4236. [Google Scholar] [CrossRef]
- Zimmerman, S.W.; Manandhar, G.; Yi, Y.-J.; Gupta, S.K.; Sutovsky, M.; Odhiambo, J.F.; Powell, M.D.; Miller, D.J.; Sutovsky, P. Sperm Proteasomes Degrade Sperm Receptor on the Egg Zona Pellucida during Mammalian Fertilization. PLoS ONE 2011, 6, e17256. [Google Scholar] [CrossRef]
- Gur, Y.; Breitbart, H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006, 20, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Ordinelli, A.; Bernabò, N.; Orsini, M.; Mattioli, M.; Barboni, B. Putative human sperm Interactome: A networks study. BMC Syst. Boil. 2018, 12, 52. [Google Scholar] [CrossRef] [PubMed]
- Bernabò, N.; Barboni, B.; Maccarrone, M. Systems Biology Analysis of the Endocannabinoid System Reveals a Scale-free Network with Distinct Roles for Anandamide and 2-Arachidonoylglycerol. OMICS A J. Integr. Boil. 2013, 17, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Bernabò, N.; Mattioli, M.; Barboni, B. Signal transduction in the activation of spermatozoa compared to other signalling pathways: A biological networks study. Int. J. Data Min. Bioinform. 2015, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Boil. 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed]
- Przulj, N.; A Wigle, D.; Jurisica, I. Functional topology in a network of protein interactions. Bioinformatics 2004, 20, 340–348. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, D447–D452. [Google Scholar] [CrossRef]
| Parameter | Value |
|---|---|
| Connected components | 1 |
| Number of nodes | 39 |
| Number of links | 47 |
| Clustering coefficient | 0.016 |
| Networks diameter | 8 |
| Characteristic path length | 3.121 |
| Averaged number of neighbors | 2.410 |
| In degree | |
| γ | −2.231 |
| r | 0.983 |
| R2 | 0.961 |
| Out degree | |
| γ | −1.150 |
| r | 0.988 |
| R2 | 0.934 |
| Most connected nodes | N° of links |
| 26S proteasome | 13 |
| PKA | 9 |
| Ca2+ | 7 |
| Parameter | Definition |
|---|---|
| Connected components | The number of networks in which any two vertices are connected to each other by links, and which are connected to no additional vertices in the network. |
| Number of nodes | The total number of molecules involved. |
| Number of edges | The total number of interactions found. |
| Clustering coefficient | Calculated as CI = 2nI/kI(kI − 1), where nI is the number of links connecting the kI neighbors of node I to each other. It is a measure of how the nodes tend to form clusters. |
| Network diameter | The longest of all the calculated shortest paths in a network. |
| Characteristic path length | The expected distance between two connected nodes. |
| Averaged number of neighbors | The mean number of connections of each node. |
| Node degree | The number of interactions of each node. |
| Node degree distribution | Represents the probability that a selected node has k links. |
| Exponent of node degree equation. | |
| R2 | Coefficient of determination of node degree vs. number of nodes, on logarithmized data. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taraschi, A.; Cimini, C.; Capacchietti, G.; Ramal-Sanchez, M.; Valbonetti, L.; Machado-Simoes, J.; Moussa, F.; Tagaram, I.; Mokh, S.; Al Iskandarani, M.; et al. Two-Player Game in a Complex Landscape: 26S Proteasome, PKA, and Intracellular Calcium Concentration Modulate Mammalian Sperm Capacitation by Creating an Integrated Dialogue—A Computational Analysis. Int. J. Mol. Sci. 2020, 21, 6256. https://doi.org/10.3390/ijms21176256
Taraschi A, Cimini C, Capacchietti G, Ramal-Sanchez M, Valbonetti L, Machado-Simoes J, Moussa F, Tagaram I, Mokh S, Al Iskandarani M, et al. Two-Player Game in a Complex Landscape: 26S Proteasome, PKA, and Intracellular Calcium Concentration Modulate Mammalian Sperm Capacitation by Creating an Integrated Dialogue—A Computational Analysis. International Journal of Molecular Sciences. 2020; 21(17):6256. https://doi.org/10.3390/ijms21176256
Chicago/Turabian StyleTaraschi, Angela, Costanza Cimini, Giulia Capacchietti, Marina Ramal-Sanchez, Luca Valbonetti, Juliana Machado-Simoes, Fadl Moussa, Israiel Tagaram, Samia Mokh, Mohamad Al Iskandarani, and et al. 2020. "Two-Player Game in a Complex Landscape: 26S Proteasome, PKA, and Intracellular Calcium Concentration Modulate Mammalian Sperm Capacitation by Creating an Integrated Dialogue—A Computational Analysis" International Journal of Molecular Sciences 21, no. 17: 6256. https://doi.org/10.3390/ijms21176256
APA StyleTaraschi, A., Cimini, C., Capacchietti, G., Ramal-Sanchez, M., Valbonetti, L., Machado-Simoes, J., Moussa, F., Tagaram, I., Mokh, S., Al Iskandarani, M., Colosimo, A., Barboni, B., & Bernabò, N. (2020). Two-Player Game in a Complex Landscape: 26S Proteasome, PKA, and Intracellular Calcium Concentration Modulate Mammalian Sperm Capacitation by Creating an Integrated Dialogue—A Computational Analysis. International Journal of Molecular Sciences, 21(17), 6256. https://doi.org/10.3390/ijms21176256








