Real-Time Visualization of Cellulase Activity by Microorganisms on Surface
Abstract
:1. Introduction
2. Results
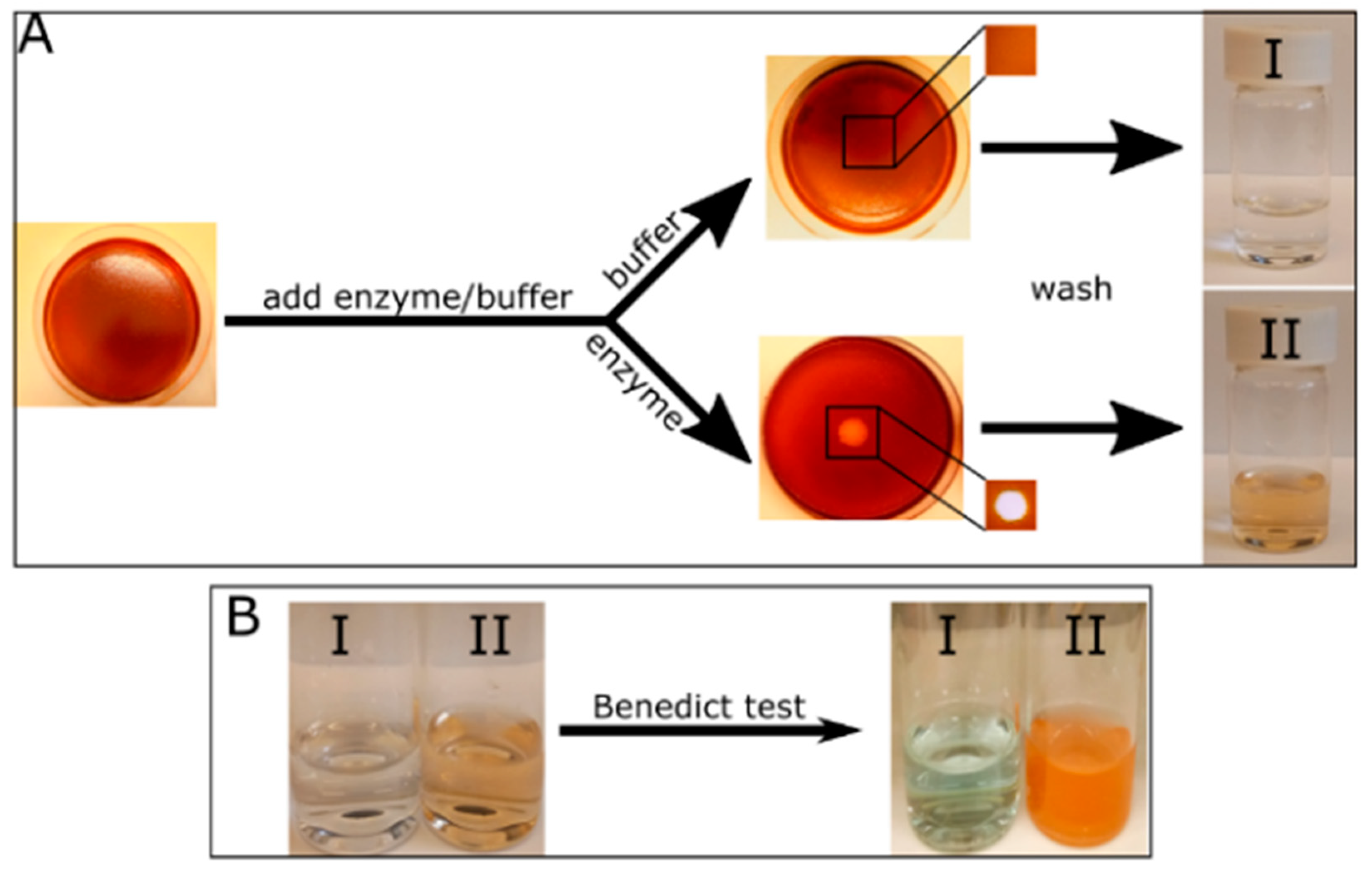
2.1. Enzyme Hydrolysis of Tagged CMC Films
2.2. Quantification of Products as a Function of Enzyme Concentration
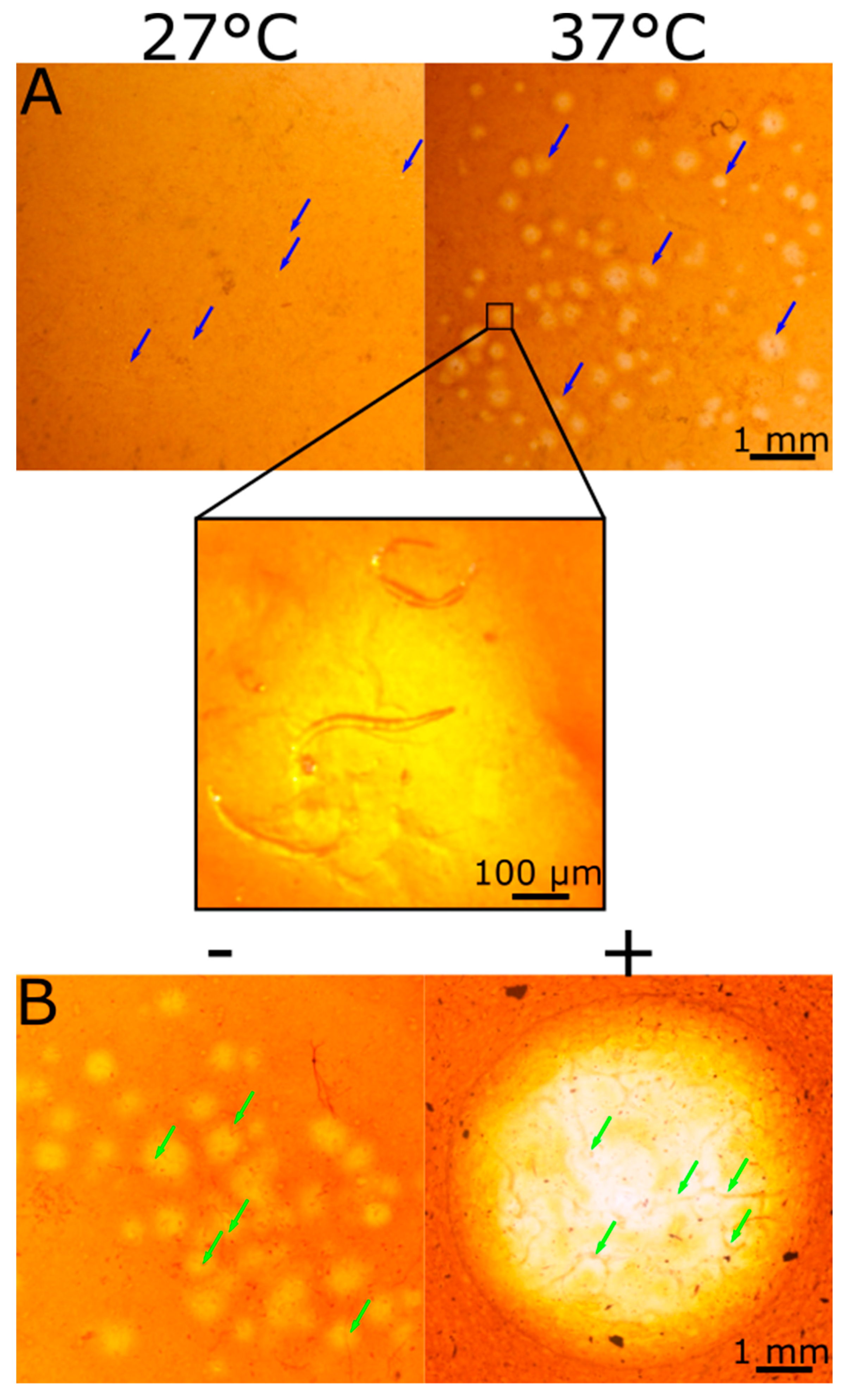
2.3. Change in Halo Size over Time
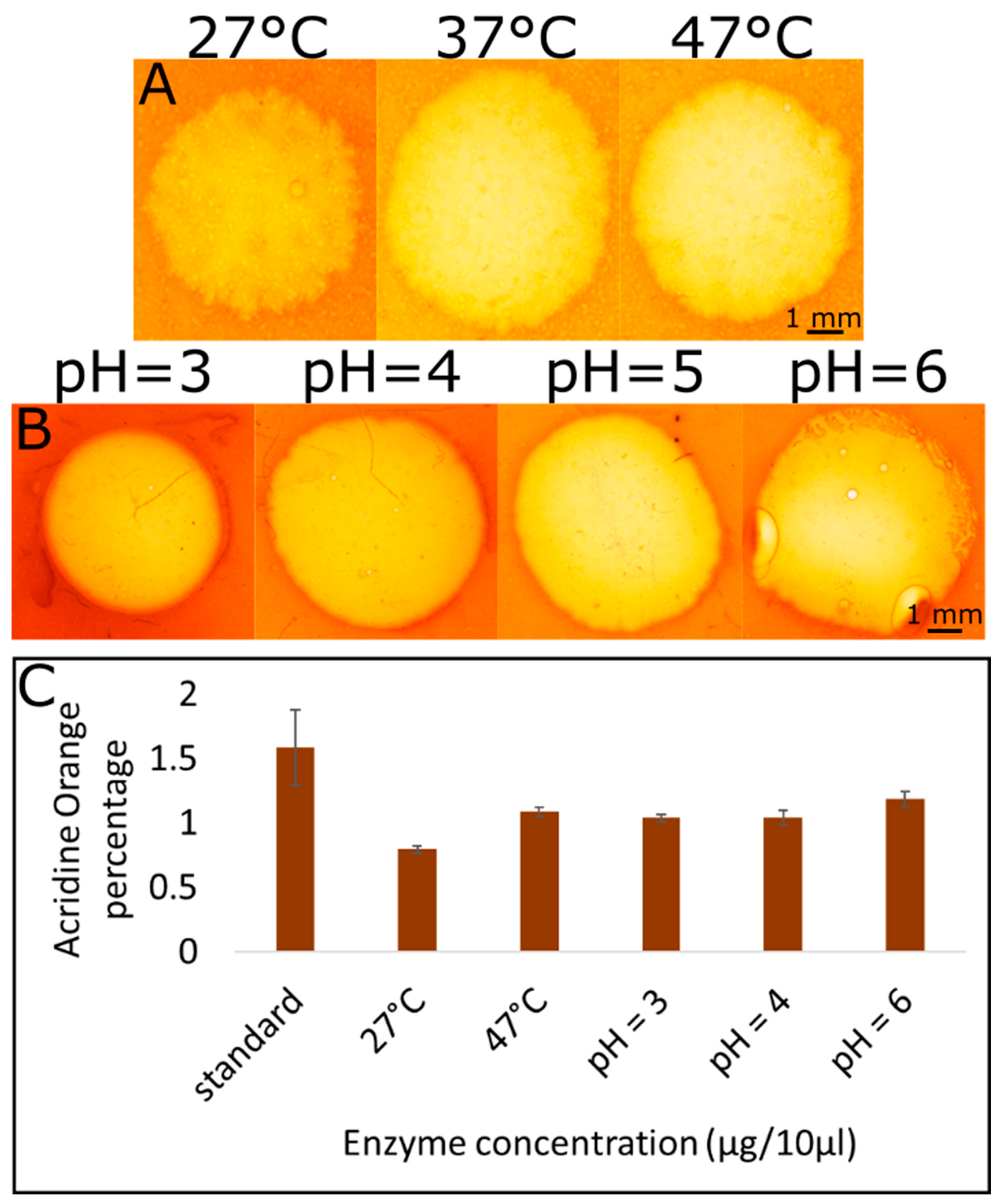
2.4. Hydrolysis Reaction under Different Temperature and pH Conditions
2.5. CMC Film Hydrolysis by Root-Knot Nematode (RKN)
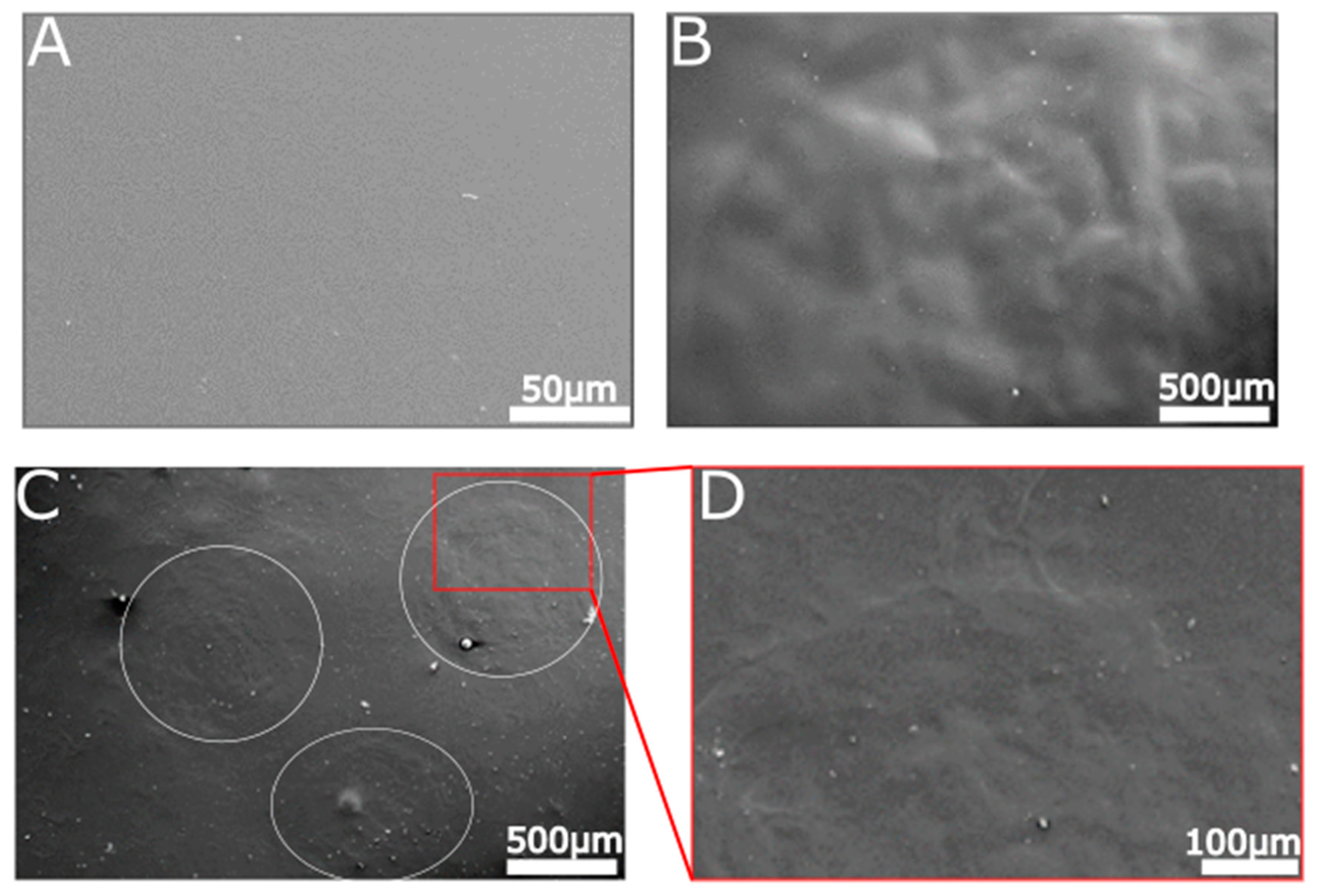
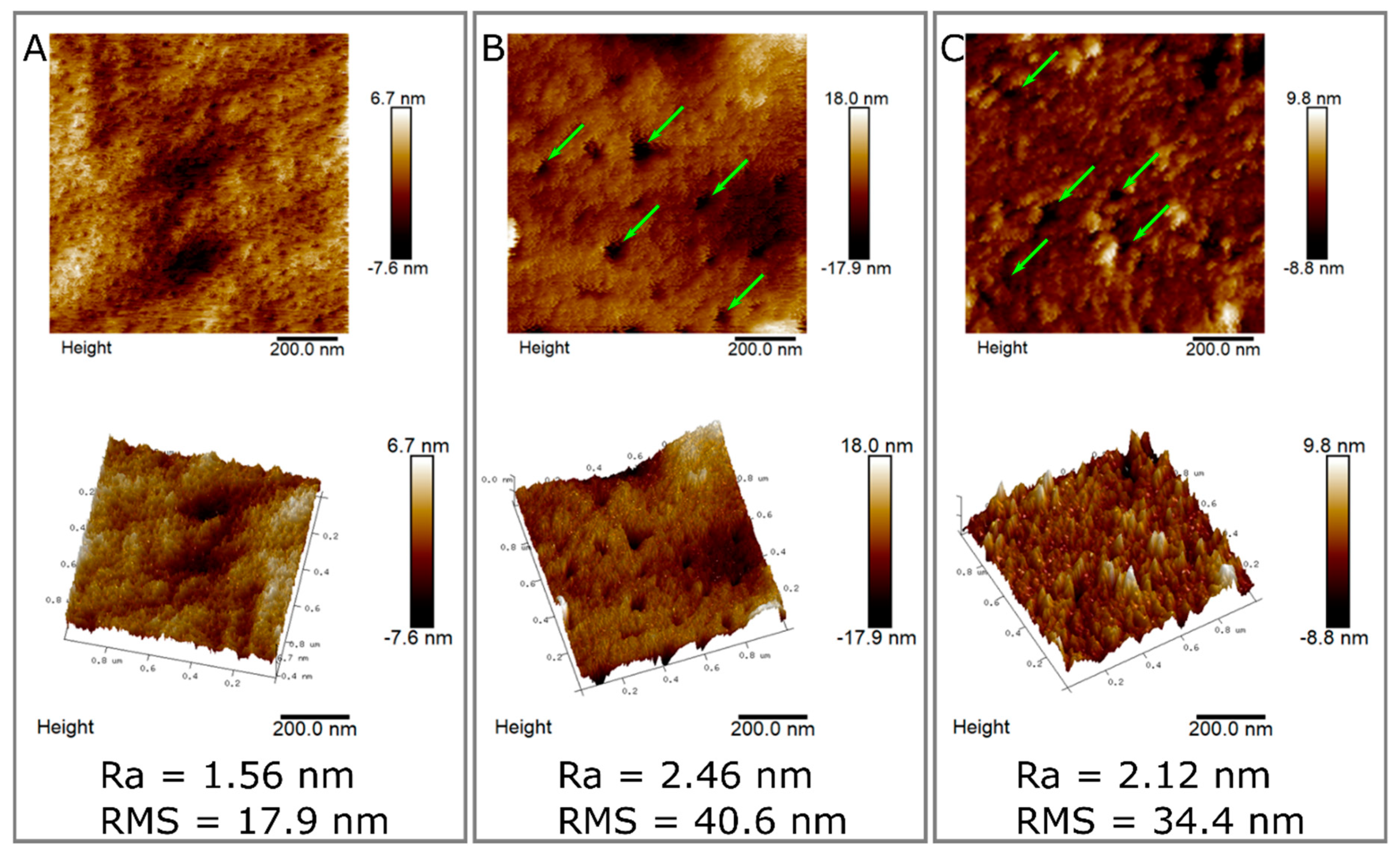
2.6. Surface Characterization of Hydrolyzed and Non-Hydrolyzed Films
3. Discussion
4. Materials and Methods
4.1. Preparation of Acridine Orange-CMC Films
4.2. Buffer and Enzyme Preparation
4.3. CMC Film Hydrolysis
- Time: Six CMC films were incubated at 37 °C and removed at different time points: after 5 min, 15 min, 30 min, 60 min, 90 min, and 120 min.
- Enzyme concentration: Seven different concentrations of cellulase enzyme were prepared in acetate buffer (1, 2, 5, 10, 15, 20, and 30 µg in 10 μL). Solutions with different enzyme concentrations were added to four different locations on the same film. The film was incubated at 37 °C for 2 h.
- Temperature: Three CMC films were used for the temperature-dependence experiment. The experiment was performed with 10 µg of enzyme and an incubation time of 2 h. The films were incubated at 3 different temperatures: 27 °C, 37 °C and 47 °C.
- pH: Four acetate buffer solutions at different pH values (3, 4, 5, 6) were prepared by titration with HCl or KOH. Four separate dyed CMC films were taken, and each one was washed with one of the buffers for 1 min. The experiment was performed with 10 µg of enzyme and incubation for 2 h at 37 °C.
4.4. Benedict’s Test
4.5. Sumner’s Method
4.6. Acridine Orange Measurements
4.7. Root-Knot Nematode (RKN; Meloidogyne Javanica) Preparation
4.8. Root Extract Addition to CMC Film
4.9. Scanning Electron Microscopy (SEM) Visualization
4.10. Atomic Force Microscopy (AFM) Visualization
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CMC | Carboxymethyl Cellulose |
| PDMS | Polydimethyl Siloxane |
| PCWDE | Plant Cell Wall Degrading Enzymes |
| RKN | Root-Knot Nematode |
| SEM | Scanning Electron Microscopy |
| AFM | Atomic Force Microscopy |
References
- Genet, M.; Stokes, A.; Salin, F.; Mickovski, S.B.; Fourcaud, T.; Dumail, J.F.; Van Beek, R. The influence of cellulose content on tensile strength in tree roots. Plant Soil 2005, 278, 1–9. [Google Scholar] [CrossRef]
- Akkerman, M.; Franssen-Verheijen, M.A.W.; Immerzeel, P.; Den Hollander, L.; Schel, J.H.N.; Emons, A.M.C. Texture of cellulose microfibrils of root hair cell walls of Arabidopsis thaliana, Medicago truncatula, and Vicia sativa. J. Microsc. 2012, 247, 60–67. [Google Scholar] [CrossRef]
- Barthlott, W.; Mail, M.; Bhushan, B.; Koch, K. Plant surfaces: Structures and functions for biomimetic innovations. Nano-Micro Lett. 2017, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Sundarraj, A.A.; Ranganathan, T.V. A review on cellulose and its utilization from agro-industrial waste. Drug Inven Today 2018, 10, 89–94. [Google Scholar]
- Deng, J.; Xiong, T.; Wang, H.; Zheng, A.; Wang, Y. Effects of cellulose, hemicellulose, and lignin on the structure and morphology of porous carbons. ACS Sustain. Chem. Eng. 2016, 4, 3750–3756. [Google Scholar] [CrossRef]
- Zhang, J.; Choi, Y.S.; Yoo, C.G.; Kim, T.H.; Brown, R.C.; Shanks, B.H. Cellulose-Hemicellulose and cellulose-lignin interactions during fast pyrolysis. ACS Sustain. Chem. Eng. 2015, 3, 293–301. [Google Scholar] [CrossRef]
- Credou, J.; Berthelot, T. Cellulose: From biocompatible to bioactive material. J. Mater. Chem. B 2014, 2, 4767–4788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altaner, C.M.; Thomas, L.H.; Fernandes, A.N.; Jarvis, M.C. How cellulose stretches: Synergism between covalent and hydrogen bonding. Biomacromolecules 2014, 15, 791–798. [Google Scholar] [CrossRef] [Green Version]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Stahlberg, J.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Singh, I.K. Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018. [Google Scholar]
- Adewuyi, Y.G.; Deshmane, V.G. Intensification of enzymatic hydrolysis of cellulose using high-frequency ultrasound: An investigation of the effects of process parameters on glucose yield. Energy Fuels 2015, 29, 4998–5006. [Google Scholar] [CrossRef]
- Satari, B.; Karimi, K.; Kumar, R. Cellulose solvent-based pretreatment for enhanced second-generation biofuel production: A review. Sustain. Energy Fuels 2019, 3, 11–62. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Zyl, W.H.; Van Isak, S. Microbial cellulose utilization: Fundamentals and biotechnology microbial cellulose utilization: Fundamentals and biotechnology downloaded from http://mmbr.asm.org/ (accessed on 6 February 2013) by Indian Institute of Technology Madras. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, A.; Aggarwal, N.K.; Yadav, A. Isolation and screening of cellulose hydrolyzing bacteria from different ecological niches. J. Biosci. Bioeng. 2017, 5, 7–13. [Google Scholar] [CrossRef]
- Yousef, N.; Mawad, A.; Abeed, A. Enhancement the cellulase activity induced by endophytic bacteria using calcium nanoparticles. Curr. Microbiol. 2019, 76, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Samant, K.; Sahu, A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012, 2012, 578925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, F.; Roy, N. Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res. Notes 2018, 11, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Kumar, B.; Bhardwaj, N.; Alam, A.; Agrawal, K.; Prasad, H.; Verma, P. Production, purification and characterization of an acid/alkali and thermo tolerant cellulase from Schizophyllum commune NAIMCC-F-03379 and its application in hydrolysis of lignocellulosic wastes. AMB Express 2018, 8, 173–188. [Google Scholar] [CrossRef] [Green Version]
- He, D.; Bao, L.; Long, Y.; Wei, W.; Yao, S. A new study of the enzymatic hydrolysis of carboxymethyl cellulose with a bulk acoustic wave sensor. Talanta 2000, 50, 1267–1273. [Google Scholar] [CrossRef]
- Liang, Y.L.; Zhang, Z.; Wu, M.; Wu, Y.; Feng, J.X. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus terrae ME27-1. BioMed Res. Int. 2014, 2014, 512497. [Google Scholar] [CrossRef] [Green Version]
- Paljevac, M.; Primožič, M.; Habulin, M.; Novak, Z.; Knez, Ž. Hydrolysis of carboxymethyl cellulose catalyzed by cellulase immobilized on silica gels at low and high pressures. J. Supercrit. Fluids 2007, 43, 74–80. [Google Scholar] [CrossRef]
- Dashtban, M.; Maki, M.; Leung, K.T.; Mao, C.; Qin, W. Cellulase activities in biomass conversion: Measurement methods and comparison. Crit. Rev. Biotechnol. 2010, 30, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Dashtban, M.; Schraft, H.; Qin, W. Fungal bioconversion of lignocellulosic residues; Opportunities & perspectives. Int. J. Biol. Sci. 2009, 5, 578–595. [Google Scholar] [CrossRef] [PubMed]
- Hankin, L.; Anagnostakis, S.L. Solid media containing carboxymethylcellulose to detect Cx cellulase activity of micro organisms. J. Gen. Microbiol. 1977, 98, 109–115. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.P.; Hong, J.; Ye, X. Cellulase Assays. In Biofuels; Mielenz, J., Ed.; Humana Press: Totowa, NJ, USA, 2009. [Google Scholar]
- Yeoh, H.H.; Khew, E.; Lim, G. A simple method for screening cellulolytic fungi. Micologia 1985, 77, 161–162. [Google Scholar] [CrossRef]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- Johnsen, H.R.; Krause, K. Cellulase activity screening using pure carboxymethylcellulose: Application to soluble cellulolytic samples and to plant tissue prints. Int. J. Mol. Sci. 2014, 15, 830–838. [Google Scholar] [CrossRef] [Green Version]
- Meddeb-Mouelhi, F.; Moisan, J.K.; Beauregard, M. A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzym. Microb. Technol. 2014, 66, 16–19. [Google Scholar] [CrossRef]
- Iberkleid, I.; Sela, N.; Brown Miyara, S. Meloidogyne javanica fatty acid- and retinol-binding protein (Mj-FAR-1) regulates expression of lipid-, cell wall-, stress- and phenylpropanoid-related genes during nematode infection of tomato. BMC Genom. 2015, 16, 272. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Zhang, Y.; Qi, L.; Mei, X.; Liao, J.; Ding, X.; Deng, W.; Fan, L.; He, X.; Vivanco, J.M.; et al. Plant-Plant-Microbe mechanisms involved in soil-borne disease suppression on a maize and pepper intercropping system. PLoS ONE 2014, 9, e115052. [Google Scholar] [CrossRef] [Green Version]
- Cannesan, M.A.; Gangneux, C.; Lanoue, A.; Giron, D.; Laval, K.; Hawes, M.; Driouich, A.; Vicré-Gibouin, M. Association between border cell responses and localized root infection by pathogenic Aphanomyces euteiches. Ann. Bot. 2011, 108, 459–469. [Google Scholar] [CrossRef] [Green Version]
- Doan, H.K.; Leveau, J.H.J. Artificial surfaces in phyllosphere microbiology. Phytopathology 2015, 105, 1036–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Luo, Y.; Pearlstein, A.J.; Aplin, J.; Liu, Y.; Bauchan, G.R.; Payne, G.F.; Wang, Q.; Nou, X.; Millner, P.D. Fabrication of biomimetically patterned surfaces and their application to probing plant-bacteria interactions. ACS Appl. Mater. Interfaces 2014, 6, 12467–12478. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Ginzburg, N.; Sayas, T.; Saphier, S.; Bucki, P.; Miyara, S.B.; Caldwell, D.L.; Iyer-Pascuzzi, A.S.; Kleiman, M. A biomimetic platform for studying root-environment interaction. Plant Soil. 2020, 447, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Mutalik, V.; Manjeshwar, L.S.; Wali, A.; Sairam, M.; Sreedhar, B.; Raju, K.V.S.N.; Aminabhavi, T.M. Aqueous-Solution and solid-film properties of poly(vinyl alcohol), poly(vinyl pyrrolidone), gelatin, starch, and carboxymethylcellulose polymers. J. Appl. Polym. Sci. 2007, 106, 765–774. [Google Scholar] [CrossRef]
- Lauretti, F.; de Melo, F.L.; Benati, F.J.; de Mello Volotão, E.; Santos, N.; Linhares, R.E.C.; Nozawa, C. Use of acridine orange staining for the detection of rotavirus RNA in polyacrylamide gels. J. Virol. Methods 2003, 114, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, M.; Hu, X.; Wang, X.; Niu, J.; Ma, T. Adsorption of cationic dyes on a cellulose-based multicarboxyl adsorbent. J. Chem. Eng. Data 2013, 58, 413–421. [Google Scholar] [CrossRef]
- Byvaltsev, V.A.; Bardonova, L.A.; Polkin, R.A.; Ochkal, S.V.; Shepelev, V.V.; Aliyev, M.A.; Ptashnikov, D.A.; Potapov, A.A. Acridine orange: A review of novel applications for surgical cancer imaging and therapy. Front. Oncol. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Sreenath, H.K. Hydrolysis of carboxymethyl cellulose by cellulase. LWT-Food Sci. Technol. 1993, 26, 224–228. [Google Scholar] [CrossRef]
- Teather, R.M.; Wood, P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [CrossRef] [Green Version]
- Benedict, S.R. A reagent for the detection of reducing sugars. J. Biol. Chem. 1908, 5, 485–487. [Google Scholar]
- Ghose, T.K. Measurment of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Wood, T.M.; Bhat, K.M. Methods for measuring cellulase activities. Methods Enzymol. 1988, 160, 87–112. [Google Scholar]
- Shuangqi, T.; Zhenyu, W.; Ziluan, F.; Lili, Z.; Jichang, W. Determination methods of cellulase activity. Afr. J. Biotechnol. 2011, 10, 7122–7125. [Google Scholar] [CrossRef]
- Florencio, C.; Couri, S.; Farinas, C.S. Correlation between agar plate screening and solid-state fermentation for the prediction of cellulase production by trichoderma strains. Enzyme Res. 2012, 2012, 793708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carder, J.H. Detection and quantitation of cellulase by Congo red staining of substrates in a cup-plate diffusion assay. Anal. Biochem. 1986, 153, 75–79. [Google Scholar] [CrossRef]
- Guignard, R.; Pilet, P.-E. Viscosimetric determination of cellulase activity: Critical analyses. Plant Cell Physiol. 1976, 908, 899–908. [Google Scholar] [CrossRef] [Green Version]
- Levinson, H.S.; Reese, E.T. Enzymatic hydrolysis of soluble cellulose derivatives as measured by changes in viscosity. J. Gen. Physiol. 1950, 33, 601–628. [Google Scholar] [CrossRef]
- Gohel, H.R.; Contractor, C.N.; Ghosh, S.K.; Braganza, V.J. A comparative study of various staining techniques for determination of extra cellular cellulase activity on Carboxy Methyl Cellulose (CMC) agar plates. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 261–266. [Google Scholar]
- Nguyen, L.T.; Neo, K.R.S.; Yang, K.L. Continuous hydrolysis of carboxymethyl cellulose with cellulase aggregates trapped inside membranes. Enzyme Microb. Technol. 2015, 78, 34–39. [Google Scholar] [CrossRef]
- Diez, J.A.; Dusenbery, D.B. Preferred temperature of meloidogyne incognita. J. Nematol. 1989, 21, 99–104. [Google Scholar]
- Wang, K.H.; McSorley, R. Exposure time to lethal temperatures for Meloidogyne incognita suppression and its implication for soil solarization. J. Nematol. 2008, 40, 7–12. [Google Scholar] [PubMed]
- Smant, G.; Stokkermans, J.P.; Yan, Y.; De Boer, J.M.; Baum, T.J.; Wang, X.; Hussey, R.S.; Gommers, F.J.; Henrissat, B.; Davis, E.L.; et al. Endogenous cellulases in animals: Isolation of β-1, 4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc. Natl. Acad. Sci. USA 1998, 95, 4906–4911. [Google Scholar] [CrossRef] [Green Version]
- Mitreva-Dautova, M.; Roze, E.; Overmars, H.; De Graaff, L.; Schots, A.; Helder, J.; Goverse, A.; Bakker, J.; Smant, G. A Symbiont-Independent Endo-1,4-β-Xylanasefrom the Plant-Parasitic Nematode Meloidogyne incognita. Mol. Plant-Microbe Interact. 2006, 19, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Mitsumasu, K.; Seto, Y.; Yoshida, S. Apoplastic interactions between plants and plant root intruders. Front. Plant Sci. 2015, 6, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danchin, E.G.; Rosso, M.N.; Vieira, P.; de Almeida-Engler, J.; Coutinho, P.M.; Henrissat, B.; Abad, P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Natl. Acad. Sci. USA 2010, 107, 17651–17656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, R.N.; Moens, M. Introduction to Plant-Parasitic Nematodes; Modes of Parasitism. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Springer: Dordrecht, The Netherlands, 2011; pp. 3–20. [Google Scholar]
- Adegbite, A.A.; Adesiyan, S.O. Root extracts of plants to control root-knot nematode on edible soybean. J. Veg. Sci. 2006, 12, 5–12. [Google Scholar] [CrossRef]
- Iberkleid, I.; Vieira, P.; de Almeida Engler, J.; Firester, K.; Spiegel, Y.; Horowitz, S.B. Fatty acid-and retinol-binding protein, mj-far-1 induces tomato host susceptibility to root-knot nematodes. PLoS ONE 2013, 8, e64586. [Google Scholar] [CrossRef]
- Kennedy, M.W.; Harnett, W. Parasitic Nematodes: Molecular Biology, Biochemistry and Immunology; CABI: Wallingford, UK, 2013. [Google Scholar]
- Stetina, S.R.; McGawley, E.C.; Russin, J.S. Extraction of root-associated meloidogyne incognita and rotylenchulus reniformis. J. Nematol. 1997, 29, 209–215. [Google Scholar]
- Oota, M.; Tsai, A.Y.L.; Aoki, D.; Matsushita, Y.; Toyoda, S.; Fukushima, K.; Saeki, K.; Toda, K.; Perfus-Barbeoch, L.; Favery, B.; et al. Identification of naturally occurring polyamines as root-knot nematode attractants. Mol. Plant 2020, 13, 658–665. [Google Scholar] [CrossRef]
- Čepulyte, R.; Danquah, W.B.; Bruening, G.; Williamson, V.M. Potent attractant for root-knot nematodes in exudates from seedling root tips of two host species. Sci. Rep. 2018, 8, 10847. [Google Scholar] [CrossRef] [Green Version]
- Kepenekçi, I.; Erdoğuş, D.; Erdoğan, P. Effects of some plant extracts on root-knot nematodes in vitro and in vivo conditions. Turk. Entomoloji Derg. 2016, 40, 3–14. [Google Scholar] [CrossRef]
- Rady, M.M. The efficiency of some natural alternatives in root-knot nematode control. Adv. Plants Agric. Res. 2018, 8, 355–362. [Google Scholar] [CrossRef]
- Zhong, T.; Oporto, G.S.; Jaczynski, J.; Tesfai, A.T.; Armstrong, J. Antimicrobial properties of the hybrid copper nanoparticles-carboxymethyl cellulose. Wood Fiber Sci. 2013, 45, 215–222. [Google Scholar]
- Martelli, S.M.; Motta, C.; Caon, T.; Alberton, J.; Bellettini, I.C.; do Prado, A.C.P.; Barreto, P.L.M.; Soldi, V. Edible carboxymethyl cellulose films containing natural antioxidant and surfactants: Alpha-Tocopherol stability, in vitro release and film properties. LWT-Food Sci. Technol. 2017, 77, 21–29. [Google Scholar] [CrossRef]
- Fattahi, R.; Ghanbarzadeh, B.; Bahrami, A. Effect of emulsion condition of oil phase on microstructure and anti-fungal properties of emulsified films based on carboxymethyl cellulose. Int. J. Nutr. Sci. 2018, 3, 44–49. [Google Scholar]
- Biswas, A.; Furtado, R.F.; Bastos, M.D.S.R.; Benevides, S.D.; Oliveira, M.A.; Boddu, V.; Cheng, H.N. Preparation and characterization of carboxymethyl cellulose films with embedded essential oils. J. Mater. Sci. Res. 2018, 7, 16. [Google Scholar] [CrossRef]
- Saladino, M.L.; Markowska, M.; Carmone, C.; Cancemi, P.; Alduina, R.; Presentato, A.; Scaffaro, R.; Biały, D.; Hasiak, M.; Hreniak, D.; et al. Graphene oxide carboxymethylcellulose nanocomposite for dressing materials. Materials 2020, 13, 1980. [Google Scholar] [CrossRef] [PubMed]
- Nemazifard, M.; Kavoosi, G.; Marzban, Z.; Ezedi, N. Physical, mechanical, water binding, and antioxidant properties of cellulose dispersions and cellulose film incorporated with pomegranate seed extract. Int. J. Food Prop. 2017, 20, 1501–1514. [Google Scholar] [CrossRef]
- Quirk, A.; Lipkowski, J.; Vandenende, C.; Cockburn, D.; Clarke, A.J.; Dutcher, J.R.; Roscoe, S.G. Direct visualization of the enzymatic digestion of a single fiber of native cellulose in an aqueous environment by atomic force microscopy. Langmuir 2010, 26, 5007–5013. [Google Scholar] [CrossRef]
- Liu, H.; Fu, S.; Zhu, J.Y.; Li, H.; Zhan, H. Visualization of enzymatic hydrolysis of cellulose using AFM phase imaging. Enzyme Microb. Technol. 2009, 45, 274–281. [Google Scholar] [CrossRef]
- Martin, F.; Kohler, A.; Murat, C.; Veneault-Fourrey, C.; Hibbett, D.S. Unearthing the roots of ectomycorrhizal symbioses. Nat. Rev. Microbiol. 2016, 14, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Jayaprakash, A.; Rajeswary, T.R.; Annamalai, A.; Lakshmi, P.T.V. Genome-Wide annotation, comparison and functional genomics of carbohydrate-active enzymes in legumes infecting Fusarium oxysporum formae speciales. Mycology 2020, 11, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, H.C.; Rep, M. Adaptation to the host environment by plant-pathogenic fungi. Annu. Rev. Phytopatholoy 2017, 55, 427–450. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, P.; Sayas, T.; Bucki, P.; Brown-Miyara, S.; Kleiman, M. Real-Time Visualization of Cellulase Activity by Microorganisms on Surface. Int. J. Mol. Sci. 2020, 21, 6593. https://doi.org/10.3390/ijms21186593
Kumari P, Sayas T, Bucki P, Brown-Miyara S, Kleiman M. Real-Time Visualization of Cellulase Activity by Microorganisms on Surface. International Journal of Molecular Sciences. 2020; 21(18):6593. https://doi.org/10.3390/ijms21186593
Chicago/Turabian StyleKumari, Pallavi, Tali Sayas, Patricia Bucki, Sigal Brown-Miyara, and Maya Kleiman. 2020. "Real-Time Visualization of Cellulase Activity by Microorganisms on Surface" International Journal of Molecular Sciences 21, no. 18: 6593. https://doi.org/10.3390/ijms21186593
APA StyleKumari, P., Sayas, T., Bucki, P., Brown-Miyara, S., & Kleiman, M. (2020). Real-Time Visualization of Cellulase Activity by Microorganisms on Surface. International Journal of Molecular Sciences, 21(18), 6593. https://doi.org/10.3390/ijms21186593




