PTBP1 Positively Regulates the Translation of Circadian Clock Gene, Period1
Abstract
:1. Introduction
2. Results
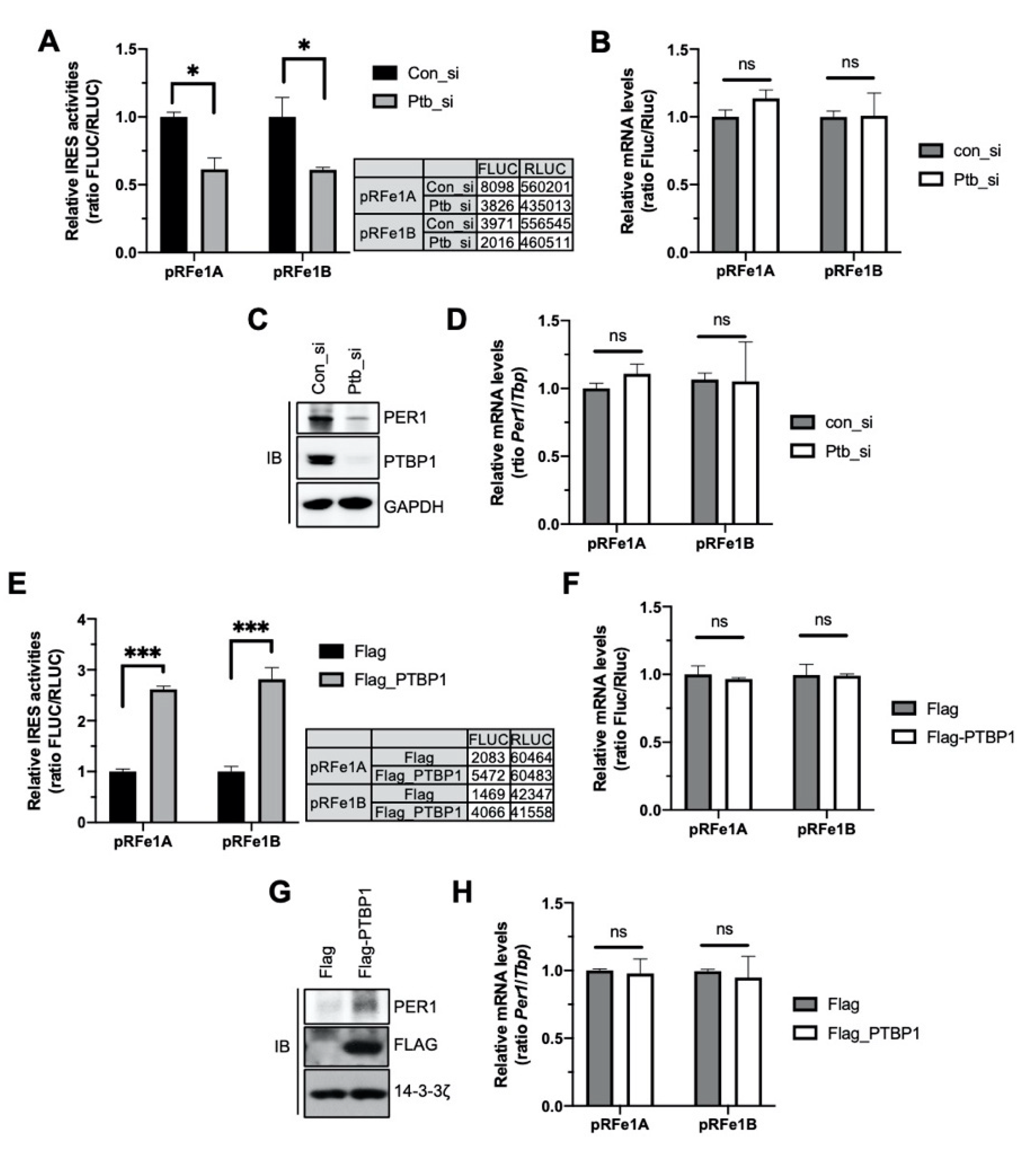
2.1. PTBP1 Activates the IRES Activity of Per1
2.2. PTBP1 Controls the IRES Activity of Per1 and Consequently the PER1 Levels
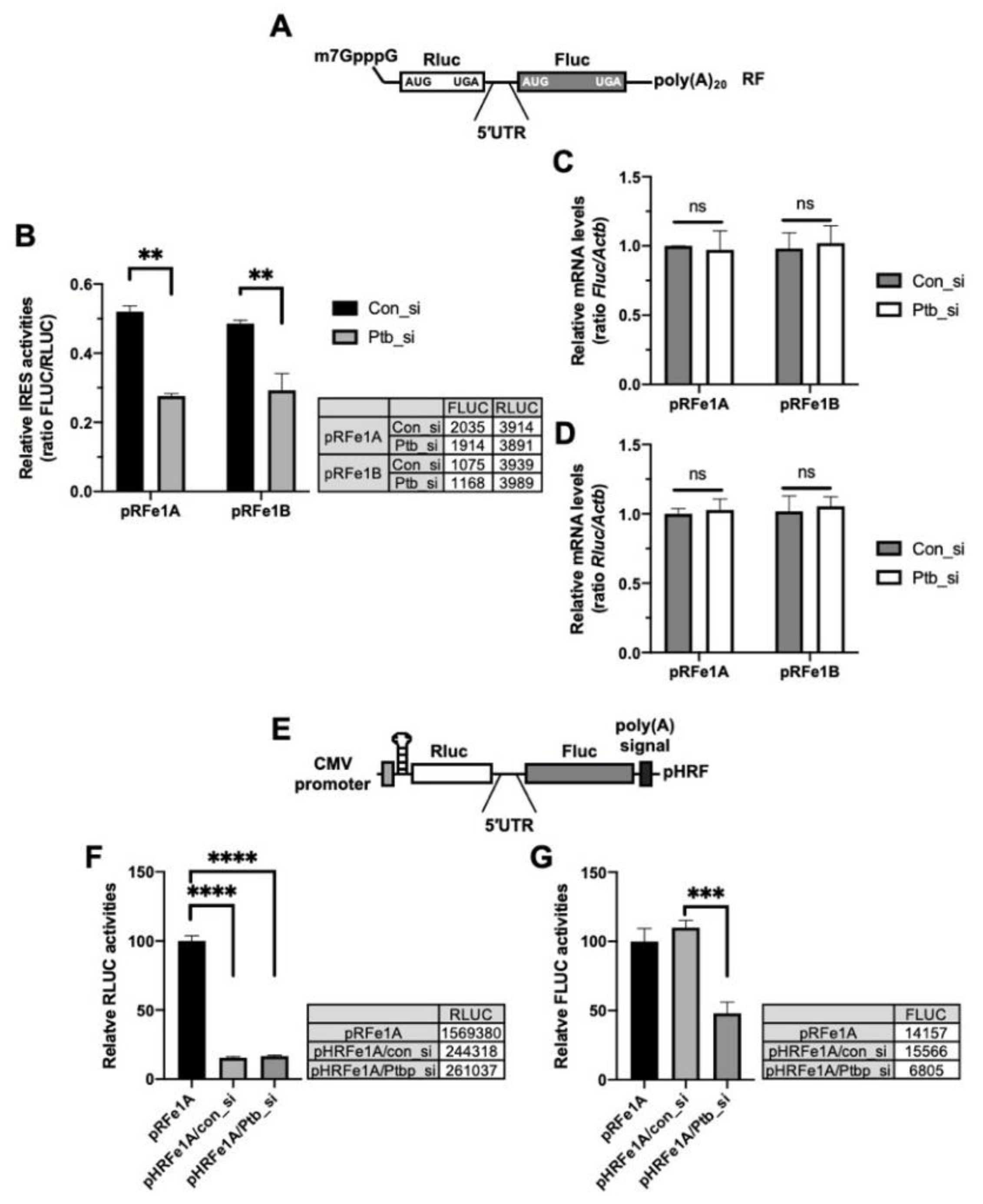
2.3. PTBP1 Specifically Modulates the IRES Activity of Per1
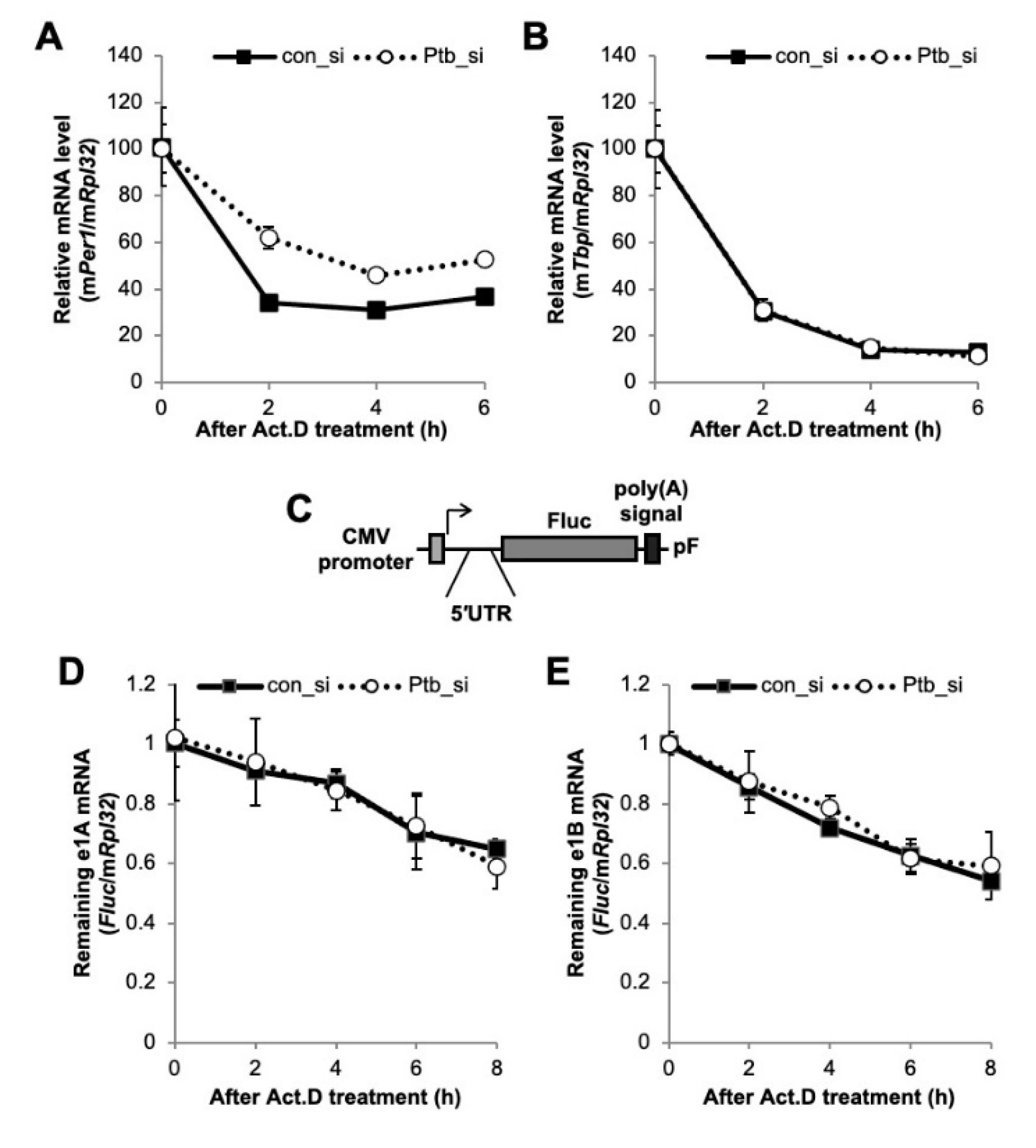
2.4. PTBP1 Does Not Affect the Expression of Per1 mRNA
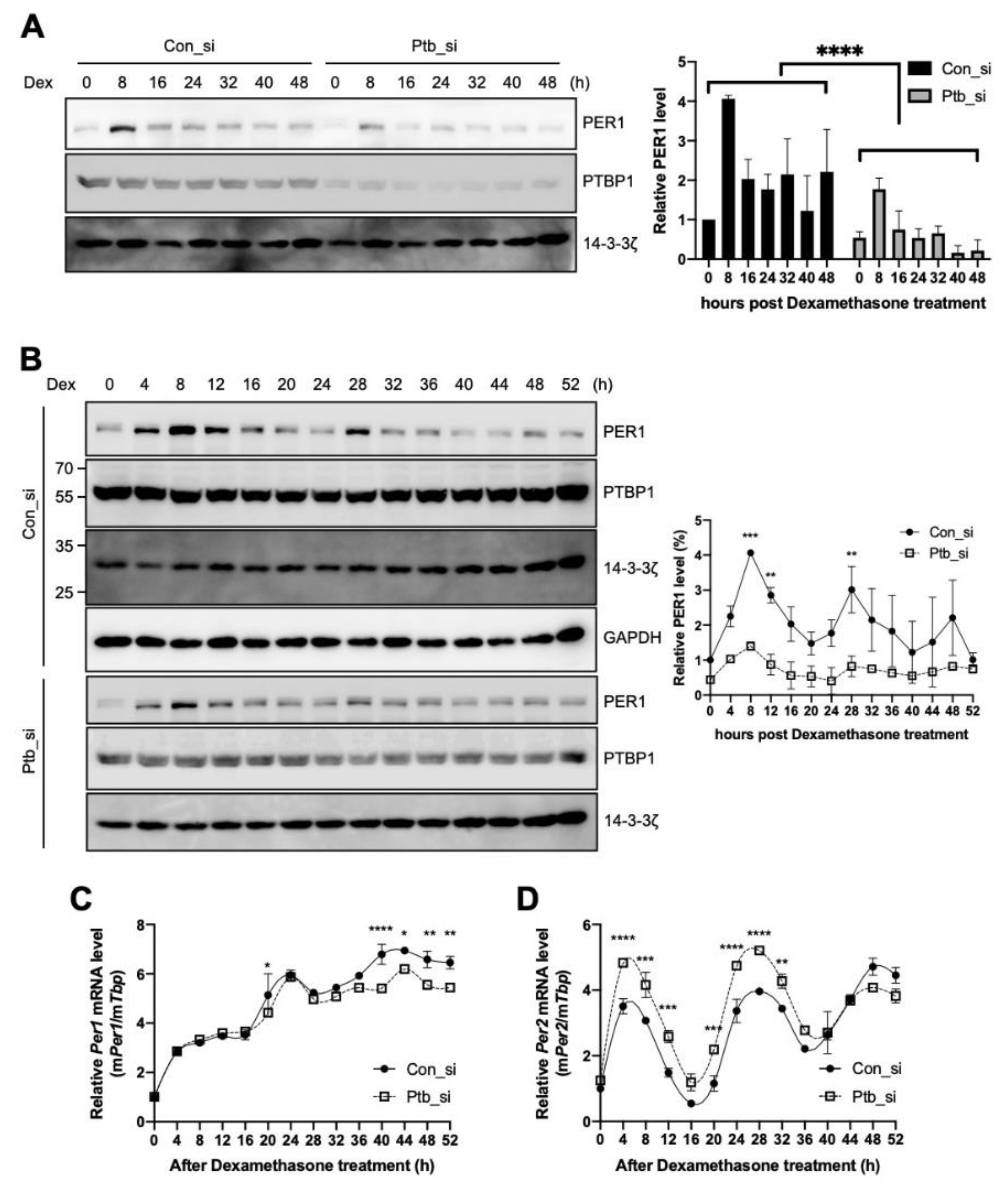
2.5. PTBP1 Affects the Circadian Expression of Per1
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs
4.2. Cell Culture and Drug Treatment
4.3. Transient Transfection and RNA Interference
4.4. In Vitro RNA Synthesis, In Vitro Binding
4.5. RNA Quantification
4.6. Immunoblot Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Per1 | Period1 |
| UTR | untranslated region |
| IRES | internal ribosomal entry site |
| PTBP1 | polypyrimidine tract-binding protein 1 |
| Cry1 | Cryptochrome 1 |
| ITAF | IRES trans-acting factors |
| Rluc | Renilla luciferase |
| Fluc | firefly luciferase |
| Act. D | Actinomycin D |
References
- Panda, S.; Hogenesch, J.B.; Kay, S.A. Circadian rhythms from flies to human. Nature 2002, 417, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Scheer, F.A.; Morris, C.J.; Shea, S.A. The internal circadian clock increases hunger and appetite in the evening independent of food intake and other behaviors. Obesity 2013, 21, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Refinetti, R. The circadian rhythm of body temperature. Front. Biosci. 2010, 15, 564–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maury, E.; Ramsey, K.M.; Bass, J. Circadian rhythms and metabolic syndrome: From experimental genetics to human disease. Circ. Res. 2010, 106, 447–462. [Google Scholar] [CrossRef] [Green Version]
- Nakano, S.; Uchida, K.; Kigoshi, T.; Azukizawa, S.; Iwasaki, R.; Kaneko, M.; Morimoto, S. Circadian rhythm of blood pressure in normotensive NIDDM subjects. Its relationship to microvascular complications. Diabetes Care 1991, 14, 707–711. [Google Scholar] [CrossRef]
- Zelinski, E.L.; Deibel, S.H.; McDonald, R.J. The trouble with circadian clock dysfunction: Multiple deleterious effects on the brain and body. Neurosci. Biobehav. Rev. 2014, 40, 80–101. [Google Scholar] [CrossRef]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef] [Green Version]
- Storch, K.F.; Lipan, O.; Leykin, I.; Viswanathan, N.; Davis, F.C.; Wong, W.H.; Weitz, C.J. Extensive and divergent circadian gene expression in liver and heart. Nature 2002, 417, 78–83. [Google Scholar] [CrossRef]
- Akhtar, R.A.; Reddy, A.B.; Maywood, E.S.; Clayton, J.D.; King, V.M.; Smith, A.G.; Gant, T.W.; Hastings, M.H.; Kyriacou, C.P. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002, 12, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Delaunay, F.; Laudet, V. Circadian clock and microarrays: Mammalian genome gets rhythm. Trends Genet. 2002, 18, 595–597. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.D.; Woo, K.C.; Cho, S.; Ha, D.C.; Jang, S.K.; Kim, K.T. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007, 21, 797–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, S.; Shingle, D.L.; Green, C.B. Post-transcriptional control of circadian rhythms. J. Cell Sci. 2011, 124, 311–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Kim, S.H.; Kim, H.J.; Kim, W.; Lee, H.R.; Jung, Y.; Choi, J.H.; Hong, K.Y.; Jang, S.K.; Kim, K.T. AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 2014, 42, 3590–3606. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Woo, K.C.; Kim, D.Y.; Kim, T.D.; Shin, J.; Park, S.M.; Jang, S.K.; Kim, K.T. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol. Cell Biol. 2012, 32, 717–728. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, U.; Eichele, G. The mammalian circadian clock. Curr. Opin. Genet. Dev. 2003, 13, 271–277. [Google Scholar] [CrossRef]
- Yagita, K.; Tamanini, F.; van Der Horst, G.T.; Okamura, H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 2001, 292, 278–281. [Google Scholar] [CrossRef]
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Bushell, M.; Stoneley, M.; Kong, Y.W.; Hamilton, T.L.; Spriggs, K.A.; Dobbyn, H.C.; Qin, X.; Sarnow, P.; Willis, A.E. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell 2006, 23, 401–412. [Google Scholar] [CrossRef]
- Dobbyn, H.C.; Hill, K.; Hamilton, T.L.; Spriggs, K.A.; Pickering, B.M.; Coldwell, M.J.; de Moor, C.H.; Bushell, M.; Willis, A.E. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene 2008, 27, 1167–1174. [Google Scholar] [CrossRef] [Green Version]
- Schepens, B.; Tinton, S.A.; Bruynooghe, Y.; Parthoens, E.; Haegman, M.; Beyaert, R.; Cornelis, S. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007, 26, 158–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Kim, S.H.; Kim, D.Y.; Kim, S.; Kim, K.T. Internal ribosomal entry site-mediated translation is important for rhythmic PERIOD1 expression. PLoS ONE 2012, 7, e37936. [Google Scholar] [CrossRef] [PubMed]
- Kamath, R.V.; Leary, D.J.; Huang, S. Nucleocytoplasmic shuttling of polypyrimidine tract-binding protein is uncoupled from RNA export. Mol. Biol. Cell 2001, 12, 3808–3820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Lee, J.A.; Kress, T.L.; Mowry, K.L.; Black, D.L. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 2003, 100, 8776–8781. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Blanco, M.A.; Jamison, S.F.; Sharp, P.A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989, 3, 1874–1886. [Google Scholar] [CrossRef]
- Kafasla, P.; Mickleburgh, I.; Llorian, M.; Coelho, M.; Gooding, C.; Cherny, D.; Joshi, A.; Kotik-Kogan, O.; Curry, S.; Eperon, I.C.; et al. Defining the roles and interactions of PTB. Biochem. Soc. Trans. 2012, 40, 815–820. [Google Scholar] [CrossRef] [Green Version]
- Lou, H.; Helfman, D.M.; Gagel, R.F.; Berget, S.M. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell Biol. 1999, 19, 78–85. [Google Scholar] [CrossRef] [Green Version]
- Knoch, K.P.; Bergert, H.; Borgonovo, B.; Saeger, H.D.; Altkruger, A.; Verkade, P.; Solimena, M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 2004, 6, 207–214. [Google Scholar] [CrossRef]
- Woo, K.C.; Kim, T.D.; Lee, K.H.; Kim, D.Y.; Kim, W.; Lee, K.Y.; Kim, K.T. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009, 37, 26–37. [Google Scholar] [CrossRef]
- Jang, S.K.; Wimmer, E. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990, 4, 1560–1572. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.A.; Brown, E.C.; Coldwell, M.J.; Jackson, R.J.; Willis, A.E. Protein factor requirements of the Apaf-1 internal ribosome entry segment: Roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell Biol. 2001, 21, 3364–3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, S.A.; Spriggs, K.A.; Coldwell, M.J.; Jackson, R.J.; Willis, A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 2003, 11, 757–771. [Google Scholar] [CrossRef]
- Song, Y.; Tzima, E.; Ochs, K.; Bassili, G.; Trusheim, H.; Linder, M.; Preissner, K.T.; Niepmann, M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 2005, 11, 1809–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilipenko, E.V.; Pestova, T.V.; Kolupaeva, V.G.; Khitrina, E.V.; Poperechnaya, A.N.; Agol, V.I.; Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000, 14, 2028–2045. [Google Scholar] [PubMed]
- Wang, Z.; Weaver, M.; Magnuson, N.S. Cryptic promoter activity in the DNA sequence corresponding to the pim-1 5′-UTR. Nucleic Acids Res. 2005, 33, 2248–2258. [Google Scholar] [CrossRef] [Green Version]
- Yap, K.; Lim, Z.Q.; Khandelia, P.; Friedman, B.; Makeyev, E.V. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012, 26, 1209–1223. [Google Scholar] [CrossRef] [Green Version]
- Wollerton, M.C.; Gooding, C.; Robinson, F.; Brown, E.C.; Jackson, R.J.; Smith, C.W. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 2001, 7, 819–832. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Guo, X.; Floros, J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am. J. Physiol. Lung Cell Mol. Physiol. 2005, 289, L497–L508. [Google Scholar] [CrossRef] [Green Version]
- Sobell, H.M. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA 1985, 82, 5328–5331. [Google Scholar] [CrossRef] [Green Version]
- Molcan, L. Time distributed data analysis by Cosinor.Online application. bioRxiv 2019, 805960. [Google Scholar] [CrossRef] [Green Version]
- Matoulkova, E.; Michalova, E.; Vojtesek, B.; Hrstka, R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012, 9, 563–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, S.; Matsumoto, K.; Hirose, M.; Shimada, M.; Nagano, M.; Shigeyoshi, Y.; Hoshino, S.; Ui-Tei, K.; Saigo, K.; Green, C.B.; et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. USA 2007, 104, 1859–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.; Allada, R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 2013, 16, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.G.; Mayer, S.A.; Tempst, P.; Nadal-Ginard, B. Characterization and molecular cloning of polypyrimidine tract-binding protein: A component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991, 5, 1237–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galban, S.; Kuwano, Y.; Pullmann, R., Jr.; Martindale, J.L.; Kim, H.H.; Lal, A.; Abdelmohsen, K.; Yang, X.; Dang, Y.; Liu, J.O.; et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell Biol. 2008, 28, 93–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hresko, R.C.; Mueckler, M. Identification of pp68 as the Tyrosine-phosphorylated Form of SYNCRIP/NSAP1. A cytoplasmic RNA-binding protein. J. Biol. Chem. 2002, 277, 25233–25238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passos, D.O.; Quaresma, A.J.; Kobarg, J. The methylation of the C-terminal region of hnRNPQ (NSAP1) is important for its nuclear localization. Biochem. Biophys. Res. Commun. 2006, 346, 517–525. [Google Scholar] [CrossRef]
- Cho, S.; Park, S.M.; Kim, T.D.; Kim, J.H.; Kim, K.T.; Jang, S.K. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell Biol. 2007, 27, 368–383. [Google Scholar] [CrossRef] [Green Version]
- Cote, C.A.; Gautreau, D.; Denegre, J.M.; Kress, T.L.; Terry, N.A.; Mowry, K.L. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell 1999, 4, 431–437. [Google Scholar] [CrossRef]
- Lee, H.; Chen, R.; Lee, Y.; Yoo, S.; Lee, C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2009, 106, 21359–21364. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Chen, S.; Li, S.; Cha, J.; Long, C.; Li, L.; He, Q.; Liu, Y. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 2007, 21, 3283–3295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- So, A.Y.; Bernal, T.U.; Pillsbury, M.L.; Yamamoto, K.R.; Feldman, B.J. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl. Acad. Sci. USA 2009, 106, 17582–17587. [Google Scholar] [CrossRef] [Green Version]
- Balsalobre, A.; Brown, S.A.; Marcacci, L.; Tronche, F.; Kellendonk, C.; Reichardt, H.M.; Schutz, G.; Schibler, U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000, 289, 2344–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bensaude, O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2011, 2, 103–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, W.; Shin, J.-C.; Lee, K.-H.; Kim, K.-T. PTBP1 Positively Regulates the Translation of Circadian Clock Gene, Period1. Int. J. Mol. Sci. 2020, 21, 6921. https://doi.org/10.3390/ijms21186921
Kim W, Shin J-C, Lee K-H, Kim K-T. PTBP1 Positively Regulates the Translation of Circadian Clock Gene, Period1. International Journal of Molecular Sciences. 2020; 21(18):6921. https://doi.org/10.3390/ijms21186921
Chicago/Turabian StyleKim, Wanil, Jae-Cheon Shin, Kyung-Ha Lee, and Kyong-Tai Kim. 2020. "PTBP1 Positively Regulates the Translation of Circadian Clock Gene, Period1" International Journal of Molecular Sciences 21, no. 18: 6921. https://doi.org/10.3390/ijms21186921
APA StyleKim, W., Shin, J.-C., Lee, K.-H., & Kim, K.-T. (2020). PTBP1 Positively Regulates the Translation of Circadian Clock Gene, Period1. International Journal of Molecular Sciences, 21(18), 6921. https://doi.org/10.3390/ijms21186921





