Role of miRNA-mRNA Interaction in Neural Stem Cell Differentiation of Induced Pluripotent Stem Cells
Abstract
:1. Introduction
2. Results
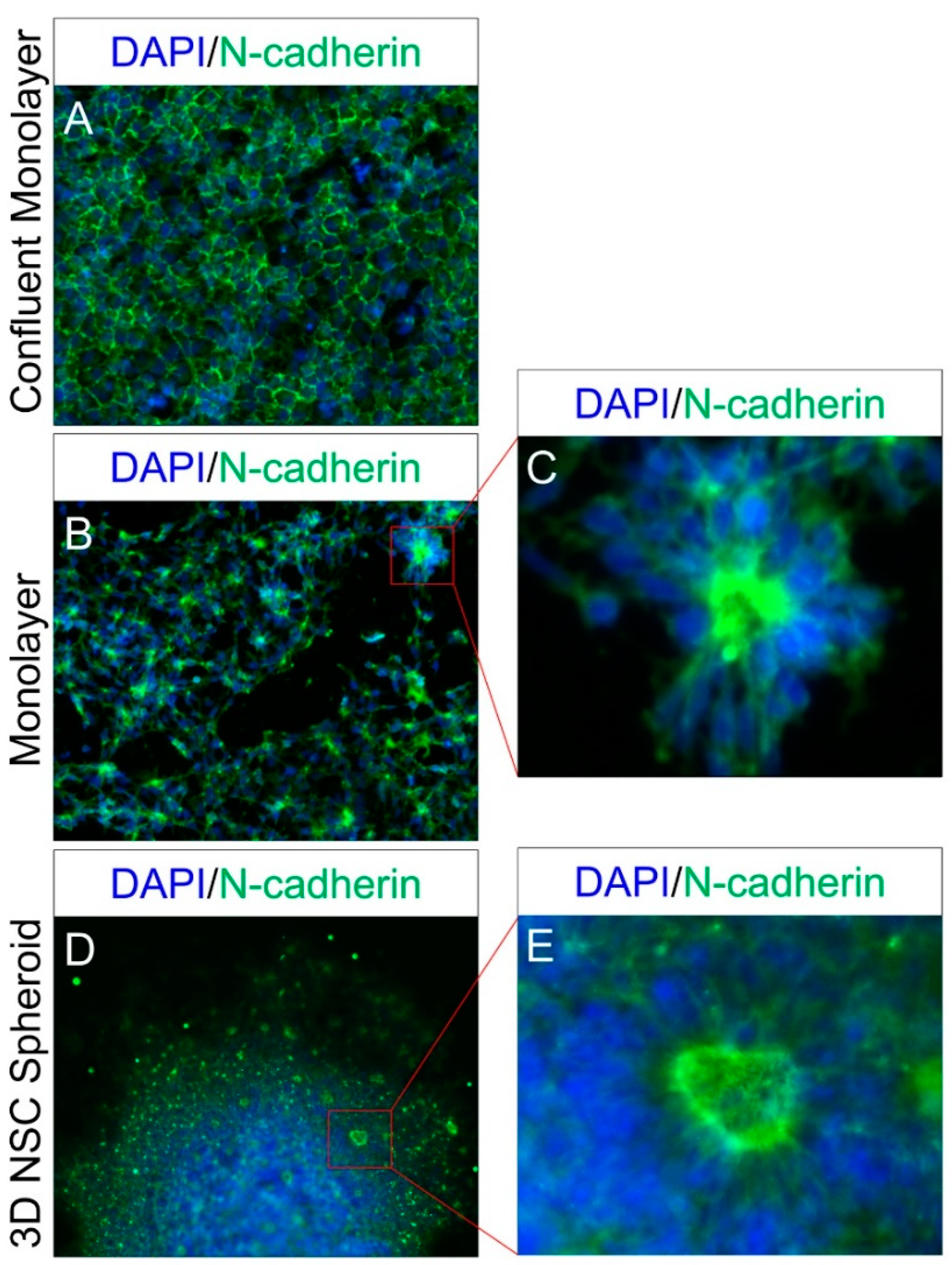
2.1. Differentiation and Lineage-Specific Functional Characteristics of Generated NSCs
2.2. Differentially Expressed (DE) Genes
2.3. In-Silico miRNA Target Prediction and Their Interaction with iPSC-NSC DE Transcriptome
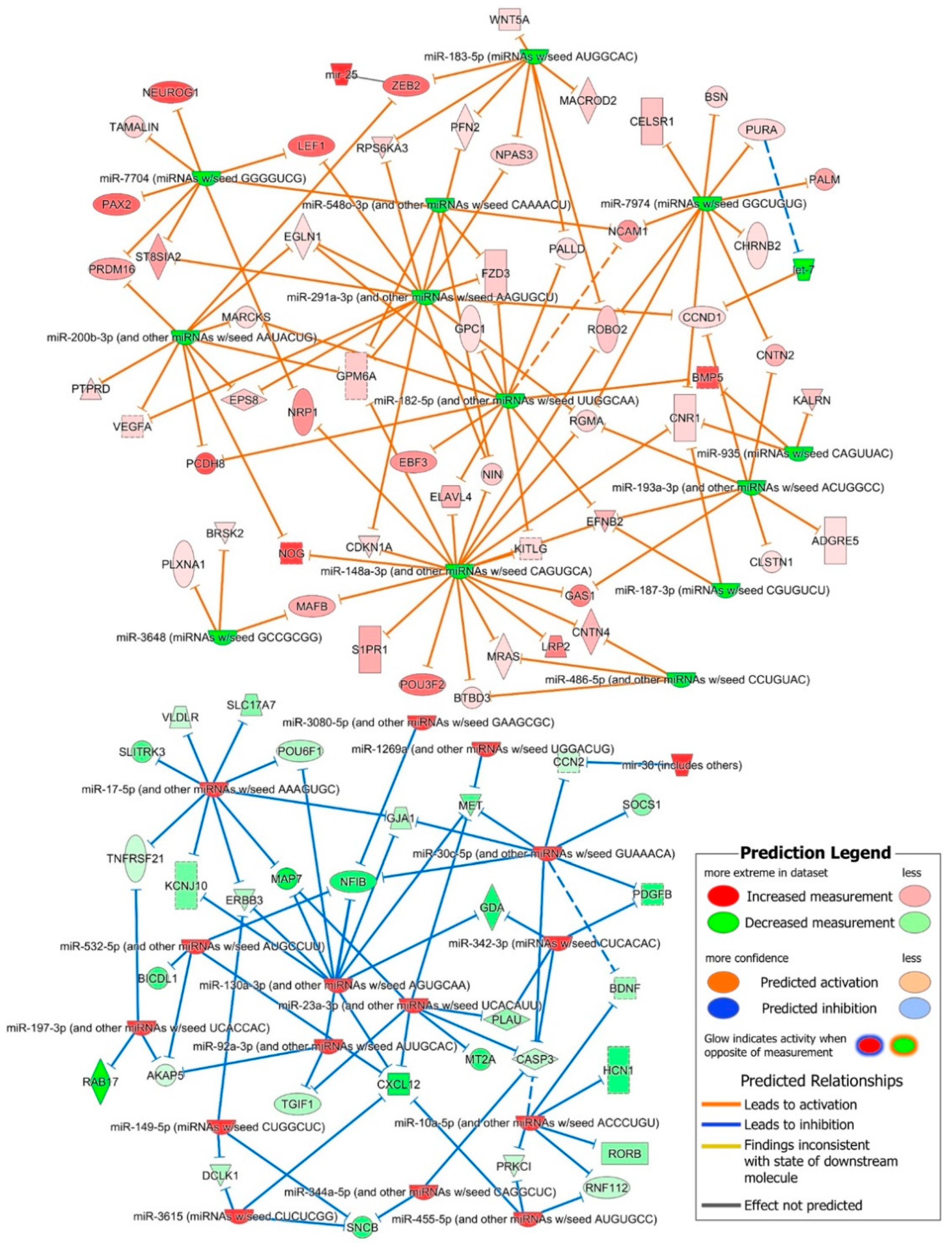
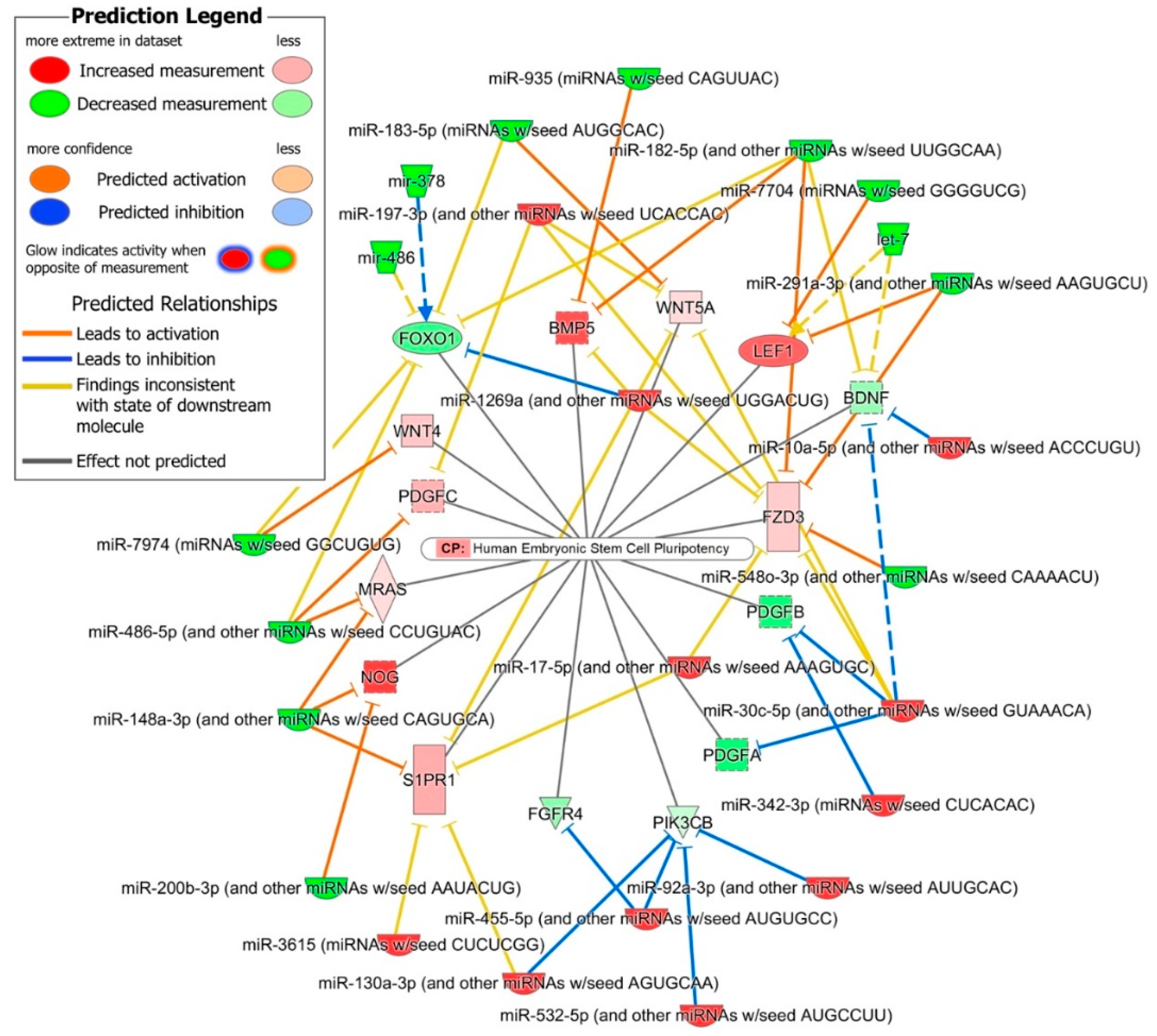
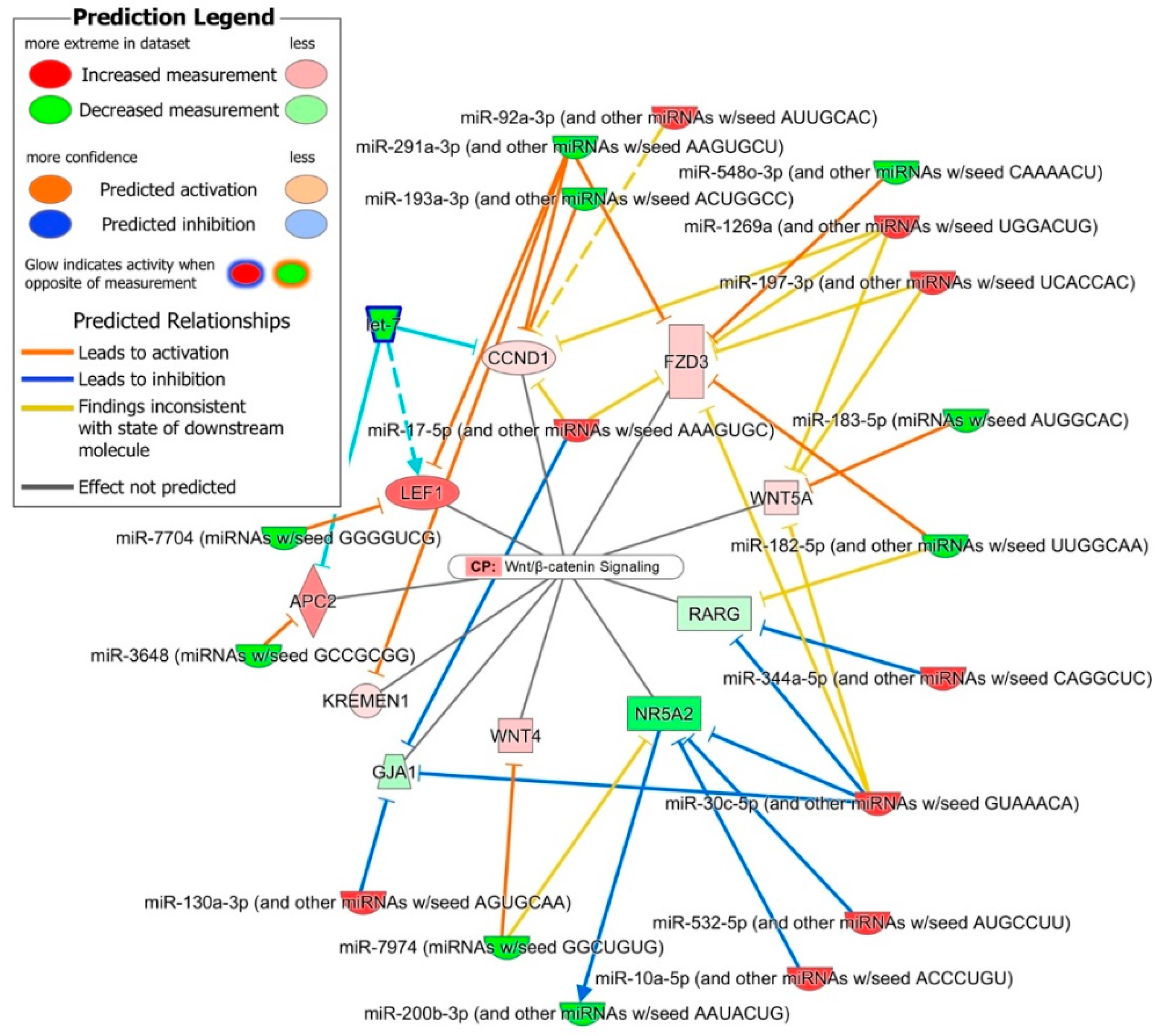
2.4. Role of DE miRNAs in the Regulation of Key iPSC and NSC Canonical Pathways
3. Discussion
4. Materials and Methods
4.1. NSC Differentiation
4.2. ICC-Based Characterization and Apical-Basal Polarity Assay of Generated NSCs
4.3. RNA Extraction and Sequencing
4.4. Differential Gene Expression and In-Silico miRNA Target Prediction Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Kloosterman, W.P.; Plasterk, R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell 2006, 11, 441–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivey, K.N.; Srivastava, D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol. Cell. Pharmacol. 2011, 3, 83–92. [Google Scholar] [PubMed]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Lall, S.; Grun, D.; Krek, A.; Chen, K.; Wang, Y.L.; Dewey, C.N.; Sood, P.; Colombo, T.; Bray, N.; Macmenamin, P.; et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006, 16, 460–471. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.P.; Fu, R.H.; Yu, H.H.; Li, K.W.; Tsai, C.H.; Shyu, W.C.; Lin, S.Z. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplant. 2009, 18, 1039–1045. [Google Scholar] [CrossRef]
- Meza-Sosa, K.F.; Pedraza-Alva, G.; Perez-Martinez, L. microRNAs: Key triggers of neuronal cell fate. Front. Cell. Neurosci. 2014, 8, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Blangero, J.; Curran, J.E. Induced Pluripotent Stem Cells in Disease Modeling and Gene Identification. Methods Mol. Biol. 2018, 1706, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Abranches, E.; Silva, M.; Pradier, L.; Schulz, H.; Hummel, O.; Henrique, D.; Bekman, E. Neural differentiation of embryonic stem cells in vitro: A road map to neurogenesis in the embryo. PLoS ONE 2009, 4, e6286. [Google Scholar] [CrossRef] [PubMed]
- Denis, J.A.; Rochon-Beaucourt, C.; Champon, B.; Pietu, G. Global transcriptional profiling of neural and mesenchymal progenitors derived from human embryonic stem cells reveals alternative developmental signaling pathways. Stem Cells Dev. 2011, 20, 1395–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murry, C.E.; Keller, G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell 2008, 132, 661–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallier, L.; Pedersen, R.A. Human embryonic stem cells: An in vitro model to study mechanisms controlling pluripotency in early mammalian development. Stem Cells Rev. 2005, 1, 119–130. [Google Scholar] [CrossRef]
- Yan, Y.; Shin, S.; Jha, B.S.; Liu, Q.; Sheng, J.; Li, F.; Zhan, M.; Davis, J.; Bharti, K.; Zeng, X.; et al. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl. Med. 2013, 2, 862–870. [Google Scholar] [CrossRef]
- Kumar, S.; Curran, J.E.; Espinosa, E.C.; Glahn, D.C.; Blangero, J. Highly efficient induced pluripotent stem cell reprogramming of cryopreserved lymphoblastoid cell lines. J. Biol. Methods 2020, 7, e124. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Curran, J.E.; Glahn, D.C.; Blangero, J. Utility of Lymphoblastoid cell lines for induced pluripotent stem cell generation. Stem Cells Int. 2016, 2016, 2349261. [Google Scholar] [CrossRef] [Green Version]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef] [Green Version]
- Punovuori, K.; Migueles, R.P.; Malaguti, M.; Blin, G.; Macleod, K.G.; Carragher, N.O.; Pieters, T.; van Roy, F.; Stemmler, M.P.; Lowell, S. N-cadherin stabilises neural identity by dampening anti-neural signals. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elkabetz, Y.; Panagiotakos, G.; Al Shamy, G.; Socci, N.D.; Tabar, V.; Studer, L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008, 22, 152–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, D.V.; Rubenstein, J.L.; Kriegstein, A.R. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron 2011, 70, 645–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Mariani, J.; Simonini, M.V.; Palejev, D.; Tomasini, L.; Coppola, G.; Szekely, A.M.; Horvath, T.L.; Vaccarino, F.M. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 12770–12775. [Google Scholar] [CrossRef] [Green Version]
- van de Leemput, J.; Boles, N.C.; Kiehl, T.R.; Corneo, B.; Lederman, P.; Menon, V.; Lee, C.; Martinez, R.A.; Levi, B.P.; Thompson, C.L.; et al. CORTECON: A temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron 2014, 83, 51–68. [Google Scholar] [CrossRef] [Green Version]
- Edri, R.; Yaffe, Y.; Ziller, M.J.; Mutukula, N.; Volkman, R.; David, E.; Jacob-Hirsch, J.; Malcov, H.; Levy, C.; Rechavi, G.; et al. Analysing human neural stem cell ontogeny by consecutive isolation of Notch active neural progenitors. Nat. Commun. 2015, 6, 6500. [Google Scholar] [CrossRef] [Green Version]
- Lowell, S.; Benchoua, A.; Heavey, B.; Smith, A.G. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006, 4, e121. [Google Scholar] [CrossRef]
- Kulcenty, K.; Wroblewska, J.P.; Rucinski, M.; Kozlowska, E.; Jopek, K.; Suchorska, W.M. MicroRNA profiling during neural differentiation of induced pluripotent stem cells. Int. J. Mol. Sci. 2019, 20, 3651. [Google Scholar] [CrossRef] [Green Version]
- Maleki, F.; Ovens, K.; McQuillan, I.; Kusalik, A.J. Size matters: How sample size affects the reproducibility and specificity of gene set analysis. Hum. Genom. 2019, 13, 42. [Google Scholar] [CrossRef]
- Stretch, C.; Khan, S.; Asgarian, N.; Eisner, R.; Vaisipour, S.; Damaraju, S.; Graham, K.; Bathe, O.F.; Steed, H.; Greiner, R.; et al. Effects of sample size on differential gene expression, rank order and prediction accuracy of a gene signature. PLoS ONE 2013, 8, e65380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, M.; Kurokawa, D.; Nakao, H.; Ohmura, T.; Aizawa, S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. 2008, 28, 10415–10421. [Google Scholar] [CrossRef] [Green Version]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, J.W.; Lee, S.K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007, 21, 744–749. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mah, I.K.; Soloff, R.; Hedrick, S.M.; Mariani, F.V. Atypical PKC-iota controls stem cell expansion via regulation of the notch pathway. Stem Cell Rep. 2015, 5, 866–880. [Google Scholar] [CrossRef] [Green Version]
- Baines, K.J.; Hillier, D.M.; Haddad, F.L.; Rajakumar, N.; Schmid, S.; Renaud, S.J. Maternal immune activation alters fetal brain development and enhances proliferation of neural precursor cells in rats. Front. Immunol. 2020, 11, 1145. [Google Scholar] [CrossRef]
- Oishi, K.; Aramaki, M.; Nakajima, K. Mutually repressive interaction between Brn1/2 and Rorb contributes to the establishment of neocortical layer 2/3 and layer 4. Proc. Natl. Acad. Sci. USA 2016, 113, 3371–3376. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, F.F.; Ribeiro, F.F.; Rodrigues, R.S.; Sebastiao, A.M.; Xapelli, S. Brain-Derived Neurotrophic Factor (BDNF) Role in Cannabinoid-Mediated Neurogenesis. Front. Cell. Neurosci. 2018, 12, 441. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.Y.; Fan, X.R.; Shi, Z.J.; Fan, Z.C.; Luo, J.; Lin, N.; Liu, Y.C.; Wu, L.; Zeng, X.R.; Cao, J.M.; et al. Hyperpolarization-activated cyclic nucleotide-gated ion (HCN) channels regulate PC12 cell Differentiation toward sympathetic neuron. Front. Cell. Neurosci. 2019, 13, 415. [Google Scholar] [CrossRef] [Green Version]
- Bian, S.; Sun, T. Functions of noncoding RNAs in neural development and neurological diseases. Mol. Neurobiol. 2011, 44, 359–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Kim, S.; Song, Y.; Kim, W.; Ying, Q.L.; Jho, E.H. Dual Function of Wnt Signaling during Neuronal Differentiation of Mouse Embryonic Stem Cells. Stem Cells Int. 2015, 2015, 459301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, Y.; Sakane, F.; Hashimoto, K. N-cadherin-based adherens junction regulates the maintenance, proliferation, and differentiation of neural progenitor cells during development. Cell Adhes. Migr. 2015, 9, 183–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]


| Nervous System Development Functions | Enrichment p-Value | Activation z-Score | Enriched Interaction/mRNAs |
|---|---|---|---|
| Development of neurons | 8.06 × 10−15 | 2.074 | 70 |
| Formation of brain | 5.94 × 10−10 | 1.841 | 44 |
| Development of cerebral cortex | 1.63 × 10−6 | 1.969 | 17 |
| Formation of hippocampus | 2.43 × 10−6 | 1.941 | 13 |
| Neurogenesis of hippocampus | 2.78 × 10−5 | 2.219 | 6 |
| Neurogenesis of brain | 4.09 × 10−5 | 2.102 | 8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Curran, J.E.; DeLeon, E.; Leandro, A.C.; Howard, T.E.; Lehman, D.M.; Williams-Blangero, S.; Glahn, D.C.; Blangero, J. Role of miRNA-mRNA Interaction in Neural Stem Cell Differentiation of Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2020, 21, 6980. https://doi.org/10.3390/ijms21196980
Kumar S, Curran JE, DeLeon E, Leandro AC, Howard TE, Lehman DM, Williams-Blangero S, Glahn DC, Blangero J. Role of miRNA-mRNA Interaction in Neural Stem Cell Differentiation of Induced Pluripotent Stem Cells. International Journal of Molecular Sciences. 2020; 21(19):6980. https://doi.org/10.3390/ijms21196980
Chicago/Turabian StyleKumar, Satish, Joanne E. Curran, Erica DeLeon, Ana C. Leandro, Tom E. Howard, Donna M. Lehman, Sarah Williams-Blangero, David C. Glahn, and John Blangero. 2020. "Role of miRNA-mRNA Interaction in Neural Stem Cell Differentiation of Induced Pluripotent Stem Cells" International Journal of Molecular Sciences 21, no. 19: 6980. https://doi.org/10.3390/ijms21196980
APA StyleKumar, S., Curran, J. E., DeLeon, E., Leandro, A. C., Howard, T. E., Lehman, D. M., Williams-Blangero, S., Glahn, D. C., & Blangero, J. (2020). Role of miRNA-mRNA Interaction in Neural Stem Cell Differentiation of Induced Pluripotent Stem Cells. International Journal of Molecular Sciences, 21(19), 6980. https://doi.org/10.3390/ijms21196980





