Lupinus albus γ-Conglutin, a Protein Structurally Related to GH12 Xyloglucan-Specific Endo-Glucanase Inhibitor Proteins (XEGIPs), Shows Inhibitory Activity against GH2 β-Mannosidase
Abstract
:1. Introduction
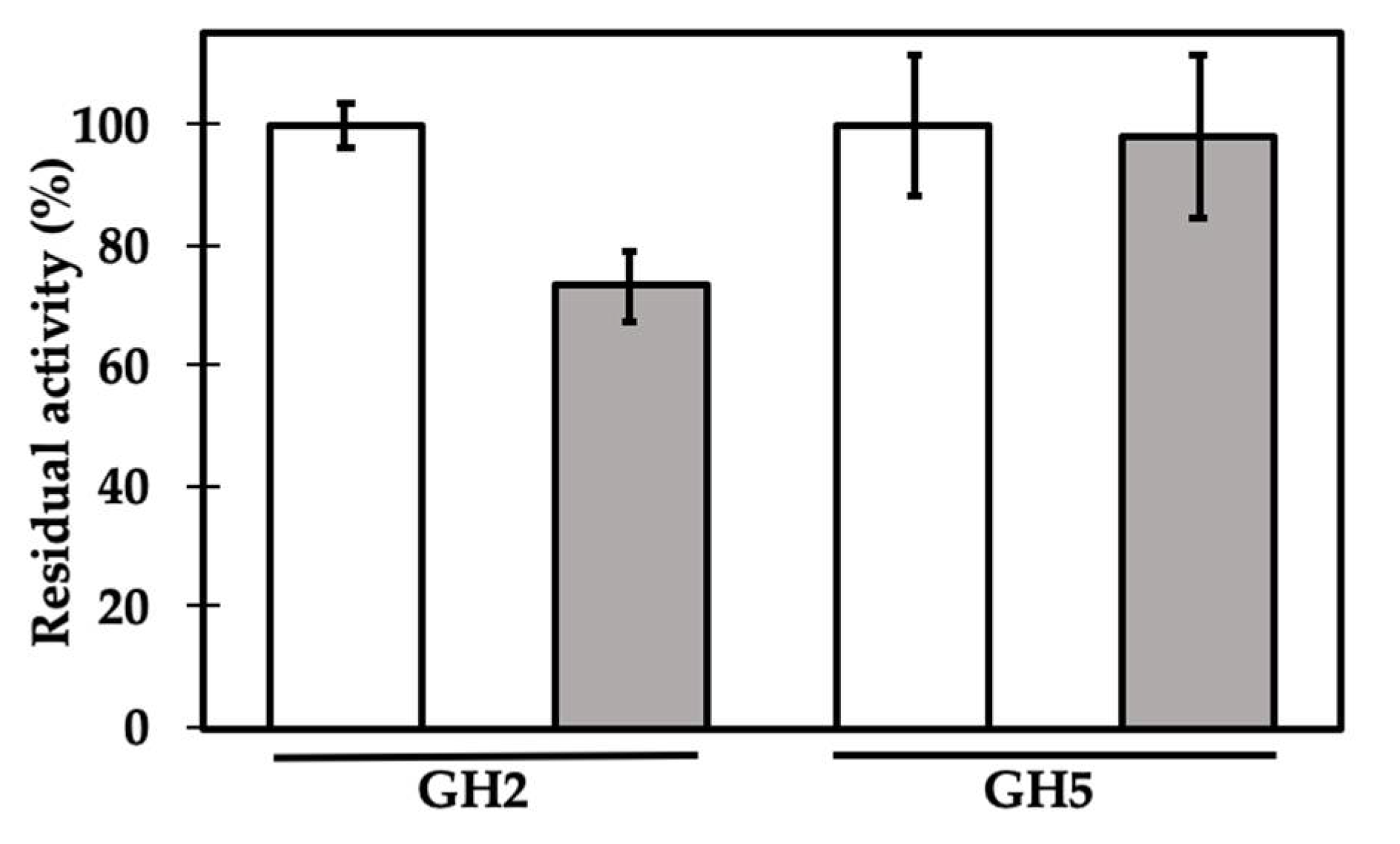
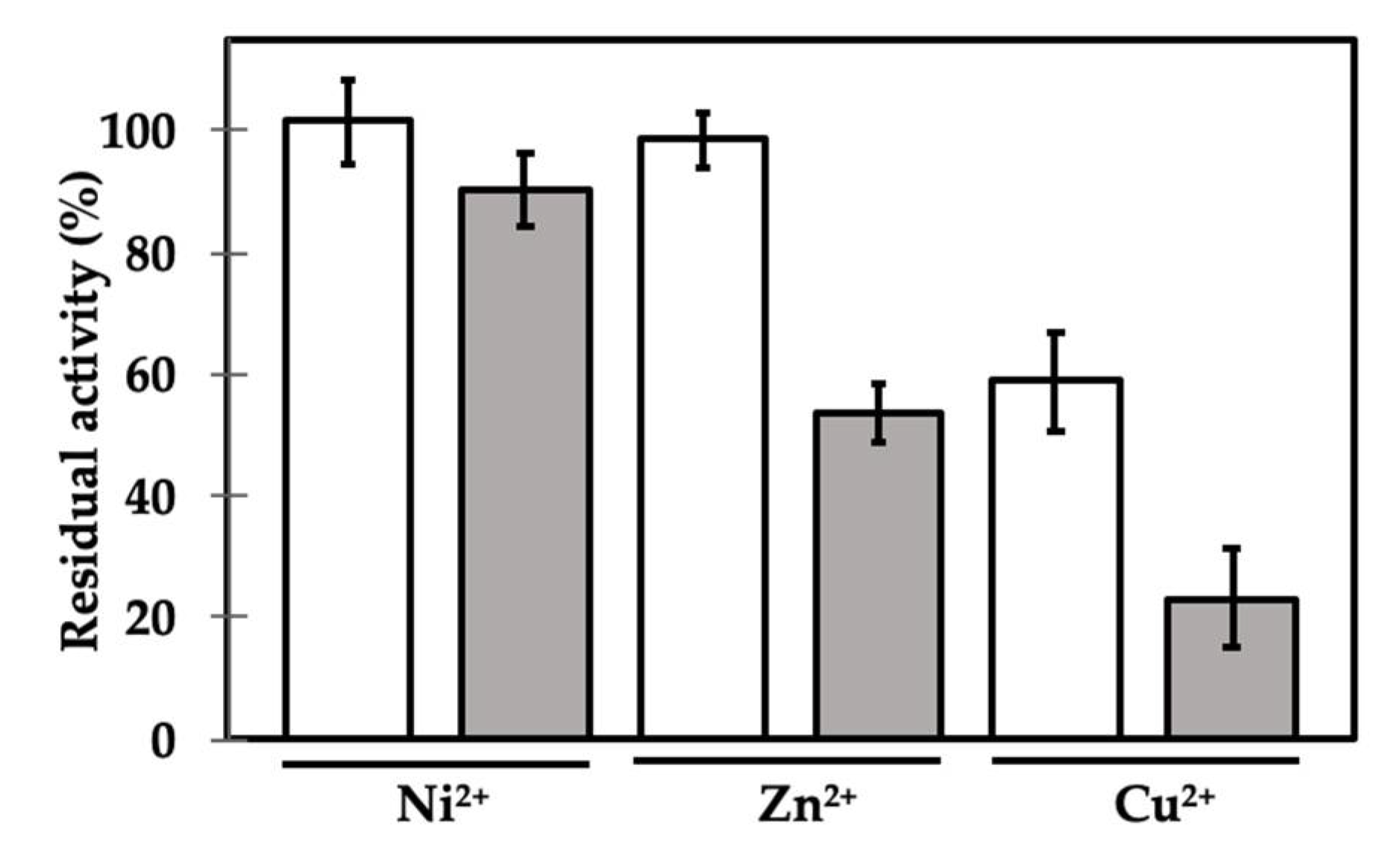
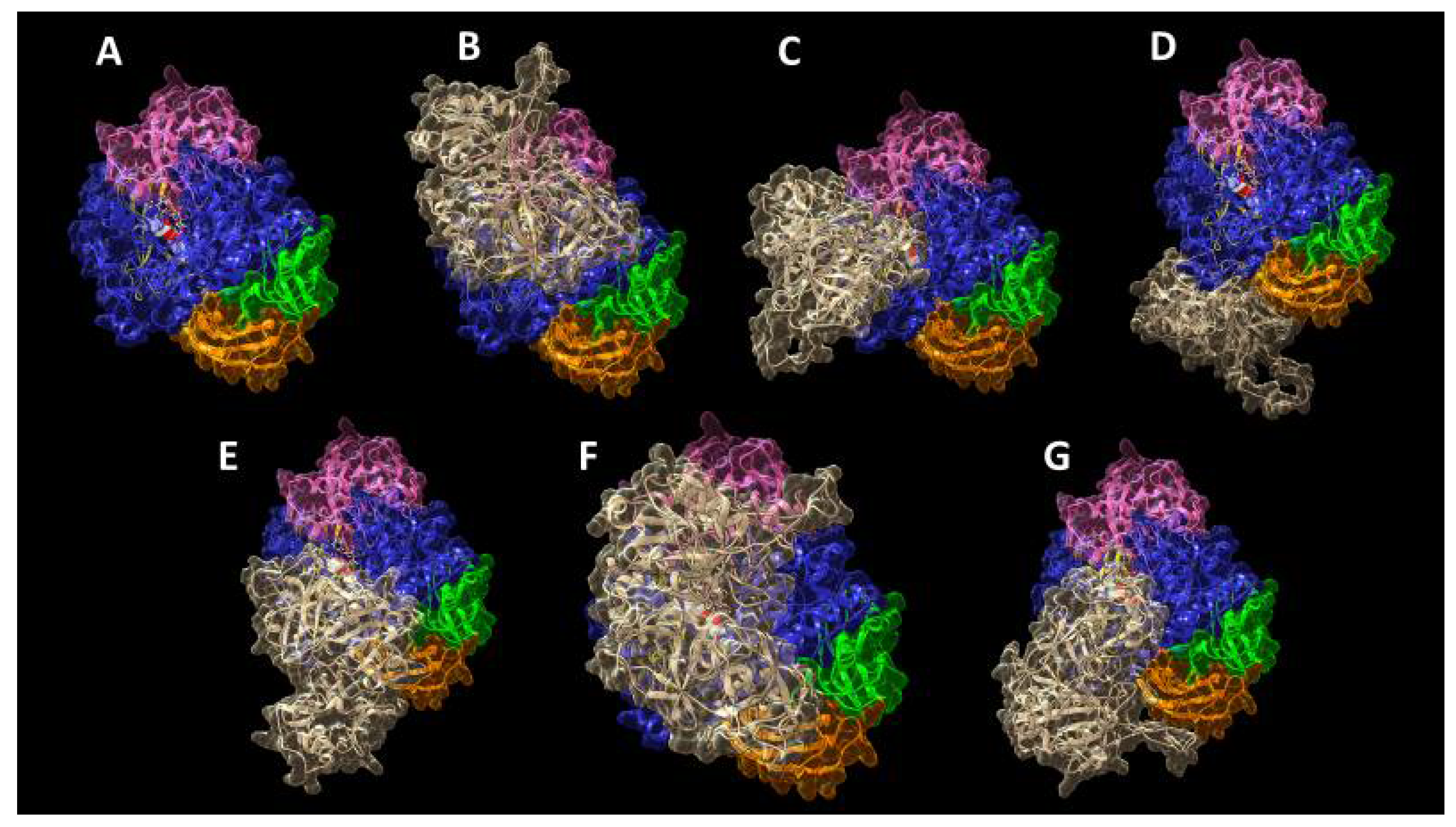
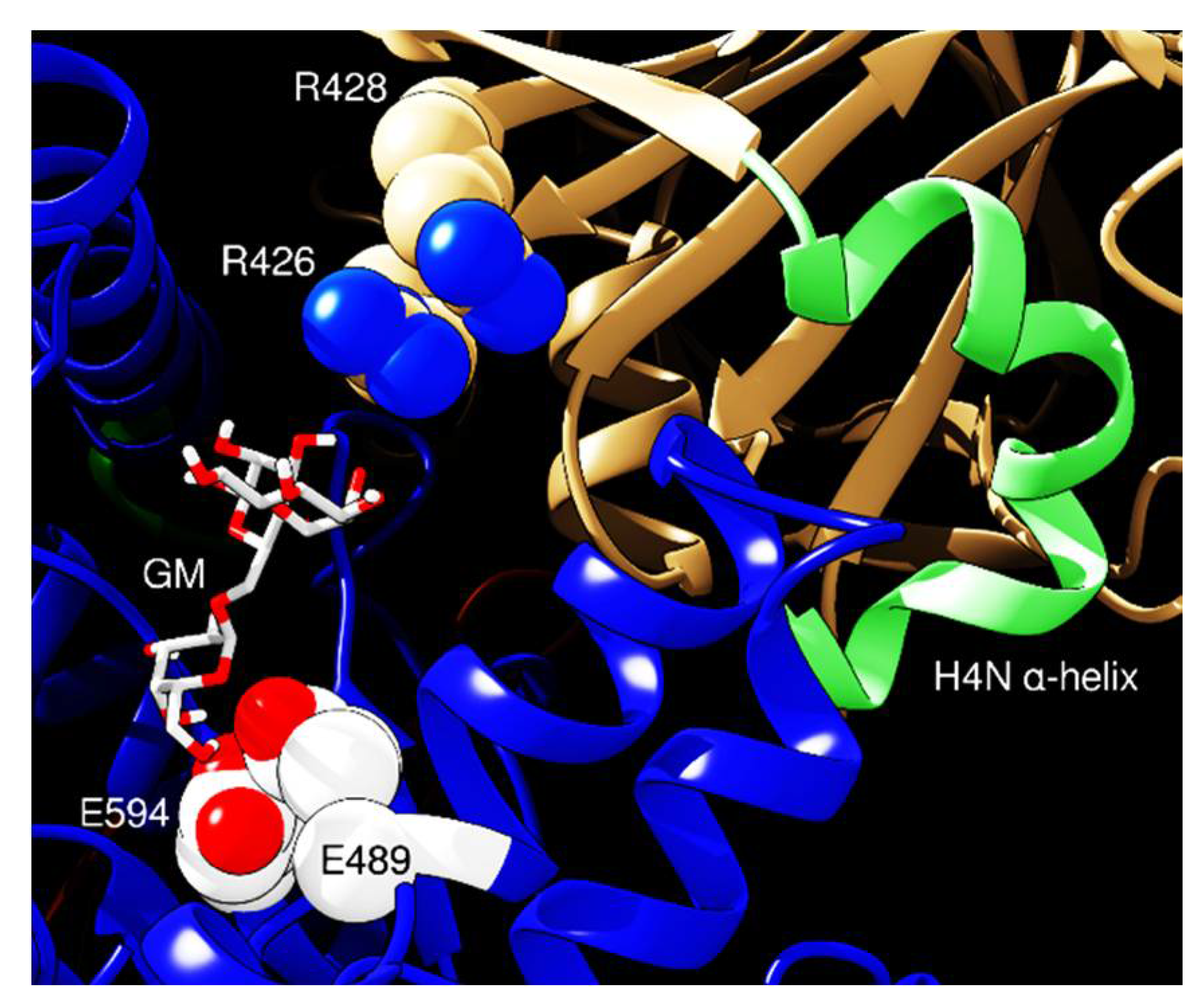
2. Results and Discussion
3. Materials and Methods
3.1. Reagents
3.2. γ-Conglutin Purification
3.3. Enzyme Activities
3.4. ICP-MS
3.5. Inhibition Data Analysis
3.6. Sequence Searches and In Silico Predictions
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GH | Glycoside hydrolase |
| GHIPs | Glycoside hydrolase inhibitor proteins |
| GM | β-galactomannan |
| GMQE | Global model quality estimation |
| IL1 | Inhibitory loop 1 |
| IL2 | Inhibitory loop 2 |
| pNP-β-d-mannopyranoside | para-nitrophenyl-β-d-mannopyranoside |
| QMEAN | Qualitative model energy analysis |
| RMSD | Root mean square deviation |
References
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.J.; Henrissat, B. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef] [Green Version]
- Henrissat, B.; Bairoch, A. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 1996, 316, 695–696. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, A.; Goyal, A.; Sharma, K. Insights into Structure and Reaction Mechanism of β-Mannanases. Curr. Protein Pept. Sci. 2017, 19, 34–47. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G.J. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef]
- Walton, J.D. Deconstructing the Cell Wall. Plant Physiol. 1994, 104, 1113–1118. [Google Scholar] [CrossRef]
- Doi, R.H.; Kosugi, A. Cellulosomes: Plant-cell-wall-degrading enzyme complexes. Nat. Rev. Genet. 2004, 2, 541–551. [Google Scholar] [CrossRef]
- Broxterman, S.E.; Schols, H.A. Interactions between pectin and cellulose in primary plant cell walls. Carbohydr. Polym. 2018, 192, 263–272. [Google Scholar] [CrossRef]
- Ebringerová, A. Structural Diversity and Application Potential of Hemicelluloses. Macromol. Symp. 2005, 232, 1–12. [Google Scholar] [CrossRef]
- McCleary, B.; Matheson, N. Galactomannan structure and β-mannanase and β-mannosidase activity in germinating legume seeds. Phytochemistry 1975, 14, 1187–1194. [Google Scholar] [CrossRef]
- Thambiraj, S.R.; Phillips, M.; Koyyalamudi, S.R.; Reddy, N. Yellow lupin (Lupinus luteus L.) polysaccharides: Antioxidant, immunomodulatory and prebiotic activities and their structural characterisation. Food Chem. 2018, 267, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant Cell Wall–Degrading Enzymes and Their Secretion in Plant-Pathogenic Fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Juge, N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 2006, 11, 359–367. [Google Scholar] [CrossRef]
- Duranti, M.; Consonni, A.; Magni, C.; Sessa, F.; Scarafoni, A. The major proteins of lupin seed: Characterisation and molecular properties for use as functional and nutraceutical ingredients. Trends Food Sci. Technol. 2008, 19, 624–633. [Google Scholar] [CrossRef]
- Magni, C.; Sessa, F.; Accardo, E.; Vanoni, M.; Morazzoni, P.; Scarafoni, A.; Duranti, M. Conglutin γ, a lupin seed protein, binds insulin in vitro and reduces plasma glucose levels of hyperglycemic rats. J. Nutr. Biochem. 2004, 15, 646–650. [Google Scholar] [CrossRef]
- Bertoglio, J.C.; Calvo, M.A.; Hancke, J.L.; Burgos, R.A.; Riva, A.; Morazzoni, P.; Ponzone, C.; Magni, C.; Duranti, M. Hypoglycemic effect of lupin seed gamma-conglutin in experimental animals and healthy human subjects. Fitoterapia 2011, 82, 933–938. [Google Scholar] [CrossRef]
- Guzmán, T.J.; Vargas-Guerrero, B.; García-López, P.M.; Gurrola-Díaz, C.M. Analysis of hepatic transcriptome modulation exerted by γ-conglutin from lupins in a streptozotocin-induced diabetes model. Gene 2020, 761, 145036. [Google Scholar] [CrossRef]
- Scarafoni, A.; Ronchi, A.; Duranti, M. γ-Conglutin, the Lupinus albus XEGIP-like protein, whose expression is elicited by chitosan, lacks of the typical inhibitory activity against GH12 endo-glucanases. Phytochemistry 2010, 71, 142–148. [Google Scholar] [CrossRef]
- Scarafoni, A.; Consonni, A.; Pessina, S.; Balzaretti, S.; Capraro, J.; Galanti, E.; Duranti, M. Structural basis of the lack of endo-glucanase inhibitory activity of Lupinus albus γ-conglutin. Plant Physiol. Biochem. 2016, 99, 79–85. [Google Scholar] [CrossRef]
- Czubinski, J.; Barciszewski, J.; Gilski, M.; Szpotkowski, K.; Debski, J.; Lampart-Szczapa, E.; Jaskolski, M. Structure of gamma-conglutin: Insight into the quaternary structure of 7S basic globulins from legumes. Acta Crystallogr. D 2015, 71, 224–238. [Google Scholar] [CrossRef] [PubMed]
- Scarafoni, A.; Di Cataldo, A.; Vassilevskaia, T.D.; Bekman, E.P.; Rodrigues-Pousada, C.; Ceciliani, F.; Duranti, M. Cloning, sequencing and expression in the seeds and radicles of two Lupinus albus conglutin γ genes. Biochim. Biophys. Acta 2001, 1519, 147–151. [Google Scholar] [CrossRef]
- Schiarea, S.; Arnoldi, L.; Fanelli, R.; De Combarieu, E.; Chiabrando, C. In-Depth Glycoproteomic Characterization of γ-Conglutin by High-Resolution Accurate Mass Spectrometry. PLoS ONE 2013, 8, e73906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capraro, J.; Spotti, P.; Magni, C.; Scarafoni, A.; Duranti, M. Spectroscopic studies on the pH-dependent structural dynamics of γ-conglutin, the blood glucose-lowering protein of lupin seeds. Int. J. Biol. Macromol. 2010, 47, 502–507. [Google Scholar] [CrossRef]
- Duranti, M.; Faoro, F.; Harris, N. The Unusual Extracellular Localization of Conglutin γ in Germinating Lupinus albus Seeds Rules out its Role as a Storage Protein. J. Plant Physiol. 1994, 143, 711–716. [Google Scholar] [CrossRef]
- Duranti, M.; Scarafoni, A.; Di Cataldo, A.; Sessa, F. Interaction of metal ions with lupin seed conglutin γ. Phytochemistry 2001, 56, 529–533. [Google Scholar] [CrossRef]
- Scirè, A.; Baldassarre, M.; Tanfani, F.; Capraro, J.; Duranti, M.; Scarafoni, A. Interaction of γ-conglutin from Lupinus albus with model phospholipid membranes: Investigations on structure, thermal stability and oligomerization status. Biochim. Biophys. Acta 2018, 1866, 1242–1248. [Google Scholar] [CrossRef]
- Foley, R.C.; Jimenez-Lopez, J.C.; Kamphuis, L.G.; Hane, J.K.; Melser, S.; Singh, K.B. Analysis of conglutin seed storage proteins across lupin species using transcriptomic, protein and comparative genomic approaches. BMC Plant Biol. 2015, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pauly, M.; Andersen, L.N.; Kauppinen, S.; Kofod, L.V.; York, W.S.; Albersheim, P.; Darvill, A. A xyloglucan-specific endo--1,4-glucanase from Aspergillus aculeatus: Expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 1999, 9, 93–100. [Google Scholar] [CrossRef]
- Qin, Q.; Bergmann, C.W.; Rose, J.K.C.; Saladie, M.; Kolli, V.S.K.; Albersheim, P.; Darvill, A.G.; York, W.S. Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J. 2003, 34, 327–338. [Google Scholar] [CrossRef]
- Gebruers, K.; Brijs, K.; Courtin, C.M.; Fierens, K.; Goesaert, H.; Rabijns, A.; Raedschelders, G.; Robben, J.; Sansen, S.; Sørensen, J.F.; et al. Properties of TAXI-type endoxylanase inhibitors. Biochim. Biophys. Acta 2004, 1696, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Satoh, S.; Sturm, A.; Fujii, T.; Chrispeels, M. cDNA cloning of an extracellular dermal glycoprotein of carrot and its expression in response to wounding. Planta 1992, 188, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.S.; Harper, A.; Carter, C.; Ren, G.; Guirgis, A.; York, W.S.; Thornburg, R.W. Nectarin IV, a Potent Endoglucanase Inhibitor Secreted into the Nectar of Ornamental Tobacco Plants. Isolation, Cloning, and Characterization. Plant Physiol. 2005, 139, 1389–1400. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.W.; Ospina-Giraldo, M.; Deahl, K. Gene silencing indicates a role for potato endoglucanase inhibitor protein in germplasm resistance to late blight. Am. J. Potato Res. 2006, 83, 41–46. [Google Scholar] [CrossRef]
- Bai, S.; Dong, C.; Zhu, J.; Zhang, Y.; Dai, H. Identification of a xyloglucan-specific endo-(1-4)-beta-d-glucanase inhibitor protein from apple (Malus × domestica Borkh.) as a potential defense gene against Botryosphaeria dothidea. Plant Sci. 2015, 231, 11–19. [Google Scholar] [CrossRef]
- Habrylo, O.; Forster, A.; Jeltsch, J.-M.; Phalip, V. The characterisation of xyloglucanase inhibitors from Humulus lupulus. Phytochemistry 2013, 90, 70–77. [Google Scholar] [CrossRef]
- Brutus, A.; Reca, I.B.; Herga, S.; Mattei, B.; Puigserver, A.; Chaix, J.-C.; Juge, N.; Bellincampi, D.; Giardina, T. A family 11 xylanase from the pathogen Botrytis cinerea is inhibited by plant endoxylanase inhibitors XIP-I and TAXI-I. Biochem. Biophys. Res. Commun. 2005, 337, 160–166. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Shimizu, T.; Hirano, H.; Sato, M.; Hashimoto, H. Structural Basis for Inhibition of Xyloglucan-specific Endo-β-1,4-glucanase (XEG) by XEG-Protein Inhibitor. J. Biol. Chem. 2012, 287, 18710–18716. [Google Scholar] [CrossRef] [Green Version]
- Yoshizawa, T.; Shimizu, T.; Yamabe, M.; Taichi, M.; Nishiuchi, Y.; Shichijo, N.; Unzai, S.; Hirano, H.; Sato, M.; Hashimoto, H. Crystal structure of basic 7S globulin, a xyloglucan-specific endo-b-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-b-glucanase. FEBS J. 2011, 278, 1944–1954. [Google Scholar] [CrossRef]
- Sandgren, M.; Ståhlberg, J.; Mitchinson, C. Structural and biochemical studies of GH family 12 cellulases: Improved thermal stability, and ligand complexes. Prog. Biophys. Mol. Biol. 2005, 89, 246–291. [Google Scholar] [CrossRef]
- Hylin, J.W.; Sawai, K. The enzymatic hydrolysis of Leucaena glauca galactomannan. Isolation of crystalline galactomannan depolymerase. J. Biol. Chem. 1964, 239, 990–992. [Google Scholar] [PubMed]
- Grishutin, S.G.; Gusakov, A.V.; Markov, A.V.; Ustinov, B.B.; Semenova, M.V.; Sinitsyn, A.P. Specific xyloglucanases as a new class of polysaccharide-degrading enzymes. Biochim. Biophys. Acta 2004, 1674, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Stoll, D.; Stålbrand, H.; Warren, R.A.J. Mannan-Degrading Enzymes from Cellulomonas fimi. Appl. Environ. Microbiol. 1999, 65, 2598–2605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaoi, K.; Nakai, T.; Kameda, Y.; Hiyoshi, A.; Mitsuishi, Y. Cloning and Characterization of Two Xyloglucanases from Paenibacillus sp. Strain KM21. Appl. Environ. Microbiol. 2005, 71, 7670–7678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Huang, Y.; Zhang, Y.; Li, W.; Li, X.; Wang, F. High-level expression of a novel thermostable and mannose-tolerant β-mannosidase from Thermotoga thermarum DSM 5069 in Escherichia coli. BMC Biotechnol. 2013, 13, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; Hu, H.; Chen, H.; Wei, Q.; Yang, Z.; Huang, Q. Expression of a β-Mannosidase from Paenibacillus polymyxa A-8 in Escherichia coli and Characterization of the Recombinant Enzyme. PLoS ONE 2014, 9, e111622. [Google Scholar] [CrossRef] [PubMed]
- Pangsri, P.; Piwpankaew, Y.; Ingkakul, A.; Nitisinprasert, S.; Keawsompong, S. Characterization of mannanase from Bacillus circulans NT 6.7 and its application in mannooligosaccharides preparation as prebiotic. SpringerPlus 2015, 4, 771. [Google Scholar] [CrossRef] [Green Version]
- Adiguzel, A.; Nadaroglu, H.; Adiguzel, G. Purification and characterization of β-mannanase from Bacillus pumilus (M27) and its applications in some fruit juices. J. Food Sci. Technol. 2015, 52, 5292–5298. [Google Scholar] [CrossRef] [Green Version]
- Heinen, P.; Bauermeister, A.; Ribeiro, L.; Messias, J.; Almeida, P.; Moraes, L.A.B.; Vargas-Rechia, C.; De Oliveira, A.; Ward, R.; Filho, E.; et al. GH11 xylanase from Aspergillus tamarii Kita: Purification by one-step chromatography and xylooligosaccharides hydrolysis monitored in real-time by mass spectrometry. Int. J. Biol. Macromol. 2018, 108, 291–299. [Google Scholar] [CrossRef]
- Rawat, R.; Kumar, S.; Chadha, B.S.; Kumar, D.; Oberoi, H.S. An acido thermophilic functionally active novel GH12 family endoglucanase from Aspergillus niger HO: Purification, characterization and molecular interaction studies. Antonie Van Leeuwenhoek 2014, 107, 103–117. [Google Scholar] [CrossRef]
- Oh, C.H.; Park, C.S.; Lee, Y.G.; Song, Y.; Bae, H.-J. Characterization of acidic endoglucanase Cel12A from Gloeophyllum trabeum and its synergistic effects on hydrogen peroxide–acetic acid (HPAC)-pretreated lignocellulose. J. Wood Sci. 2019, 65, 24. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Microbial xylanases: Engineering, production and industrial applications. Biotechnol. Adv. 2012, 30, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Cer, R.Z.; Mudunuri, U.; Stephens, R.; Lebeda, F.J. IC50-to-Ki: A web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009, 37, W441–W445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansen, S.; De Ranter, C.; Gebruers, K.; Brijs, K.; Courtin, C.M.; Delcour, J.A.; Rabijns, A. Structural Basis for Inhibition of Aspergillus niger Xylanase by Triticum aestivum Xylanase Inhibitor-I. J. Biol. Chem. 2004, 279, 36022–36028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nascimento, A.S.; Muniz, J.R.C.; Aparicio, R.; Golubev, A.M.; Polikarpov, I.; Nascimento, A.S.; Muniz, J.R.C.; Aparício, R. Insights into the structure and function of fungal β-mannosidases from glycoside hydrolase family 2 based on multiple crystal structures of the Trichoderma harzianum enzyme. FEBS J. 2014, 281, 4165–4178. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; Beer, T.A.P.D.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.W.; Kim, N.H.; Lee, Y.K.; Hwang, B.K. The Pepper Extracellular Xyloglucan-Specific Endo-β-1,4-Glucanase Inhibitor Protein Gene, CaXEGIP1, Is Required for Plant Cell Death and Defense Responses. Plant Physiol. 2012, 161, 384–396. [Google Scholar] [CrossRef] [Green Version]
- Duranti, M.; Sessa, F.; Scarafoni, A.; Bellini, T.; Dallocchio, F. Thermal Stabilities of Lupin Seed Conglutin γ Protomers and Tetramers. J. Agric. Food Chem. 2000, 48, 1118–1123. [Google Scholar] [CrossRef]
- Lever, M. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 1972, 47, 273–279. [Google Scholar] [CrossRef]
- Mermet, J.M.; Poussel, E. ICP Emission Spectrometers: 1995 Analytical Figures of Merit. Appl. Spectrosc. 1995, 49, 12A–18A. [Google Scholar] [CrossRef]
- IC50-toKi Converter. Available online: https://bioinfo-abcc.ncifcrf.gov/IC50_Ki_Converter/index.php (accessed on 3 October 2020).
- Brauer, A.B.E.; Domingo, G.J.; Cooke, R.M.; Matthews, S.J.; Leatherbarrow, R.J. A ConservedcisPeptide Bond Is Necessary for the Activity of Bowman-Birk Inhibitor Protein. Biochemistry 2002, 41, 10608–10615. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-García, B.; Pons, C.; Fernandez-Recio, J. pyDockWEB: A web server for rigid-body protein–protein docking using electrostatics and desolvation scoring. Bioinformatics 2013, 29, 1698–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Tuncbag, N.; Gursoy, A.; Nussinov, R.; Keskin, O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat. Protoc. 2011, 6, 1341–1354. [Google Scholar] [CrossRef]
- Baspinar, A.; Cukuroglu, E.; Nussinov, R.; Keskin, O.; Gursoy, A. PRISM: A web server and repository for prediction of protein–protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014, 42, W285–W289. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Ramírez-Aportela, E.; López-Blanco, J.R.; Chacon, P. FRODOCK 2.0: Fast protein–protein docking server. Bioinformatics 2016, 32, 2386–2388. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- MIB: Metal Ion-Binding Site Prediction and Docking Server. Available online: http://bioinfo.cmu.edu.tw/MIB/ (accessed on 3 October 2020).
- Lu, C.-H.; Lin, Y.-F.; Lin, J.-J.; Yu, C.-S. Prediction of Metal Ion–Binding Sites in Proteins Using the Fragment Transformation Method. PLoS ONE 2012, 7, e39252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-F.; Cheng, C.-W.; Shih, C.-S.; Hwang, J.-K.; Yu, C.-S.; Lu, C.-H. MIB: Metal Ion-Binding Site Prediction and Docking Server. J. Chem. Inf. Model. 2016, 56, 2287–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohne-Lang, A.; Von Der Lieth, C.-W. GlyProt: In silico glycosylation of proteins. Nucleic Acids Res. 2005, 33, W214–W219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GlyProt—In Silico Glycosylation of Proteins. Available online: http://www.glycosciences.de/modeling/glyprot/php/main.php (accessed on 3 October 2020).
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.L. Peptide promiscuity: An evolutionary concept for plant defense. FEBS Lett. 2011, 585, 995–1000. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.B.; Melo, T.; Teixeira, A. Catabolism of the Seed Storage Proteins from Lupinus albus: Fate of Globulins during Germination and Seedling Growth. Funct. Plant Biol. 1995, 22, 373–381. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benedetti, S.D.; Galanti, E.; Capraro, J.; Magni, C.; Scarafoni, A. Lupinus albus γ-Conglutin, a Protein Structurally Related to GH12 Xyloglucan-Specific Endo-Glucanase Inhibitor Proteins (XEGIPs), Shows Inhibitory Activity against GH2 β-Mannosidase. Int. J. Mol. Sci. 2020, 21, 7305. https://doi.org/10.3390/ijms21197305
Benedetti SD, Galanti E, Capraro J, Magni C, Scarafoni A. Lupinus albus γ-Conglutin, a Protein Structurally Related to GH12 Xyloglucan-Specific Endo-Glucanase Inhibitor Proteins (XEGIPs), Shows Inhibitory Activity against GH2 β-Mannosidase. International Journal of Molecular Sciences. 2020; 21(19):7305. https://doi.org/10.3390/ijms21197305
Chicago/Turabian StyleBenedetti, Stefano De, Elisabetta Galanti, Jessica Capraro, Chiara Magni, and Alessio Scarafoni. 2020. "Lupinus albus γ-Conglutin, a Protein Structurally Related to GH12 Xyloglucan-Specific Endo-Glucanase Inhibitor Proteins (XEGIPs), Shows Inhibitory Activity against GH2 β-Mannosidase" International Journal of Molecular Sciences 21, no. 19: 7305. https://doi.org/10.3390/ijms21197305
APA StyleBenedetti, S. D., Galanti, E., Capraro, J., Magni, C., & Scarafoni, A. (2020). Lupinus albus γ-Conglutin, a Protein Structurally Related to GH12 Xyloglucan-Specific Endo-Glucanase Inhibitor Proteins (XEGIPs), Shows Inhibitory Activity against GH2 β-Mannosidase. International Journal of Molecular Sciences, 21(19), 7305. https://doi.org/10.3390/ijms21197305





