Epigenetic Analysis through MSAP-NGS Coupled Technology: The Case Study of White Poplar Monoclonal Populations/Stands
Abstract
1. Introduction
2. Results
2.1. Technical Overview of MSAP-NGS Methodology
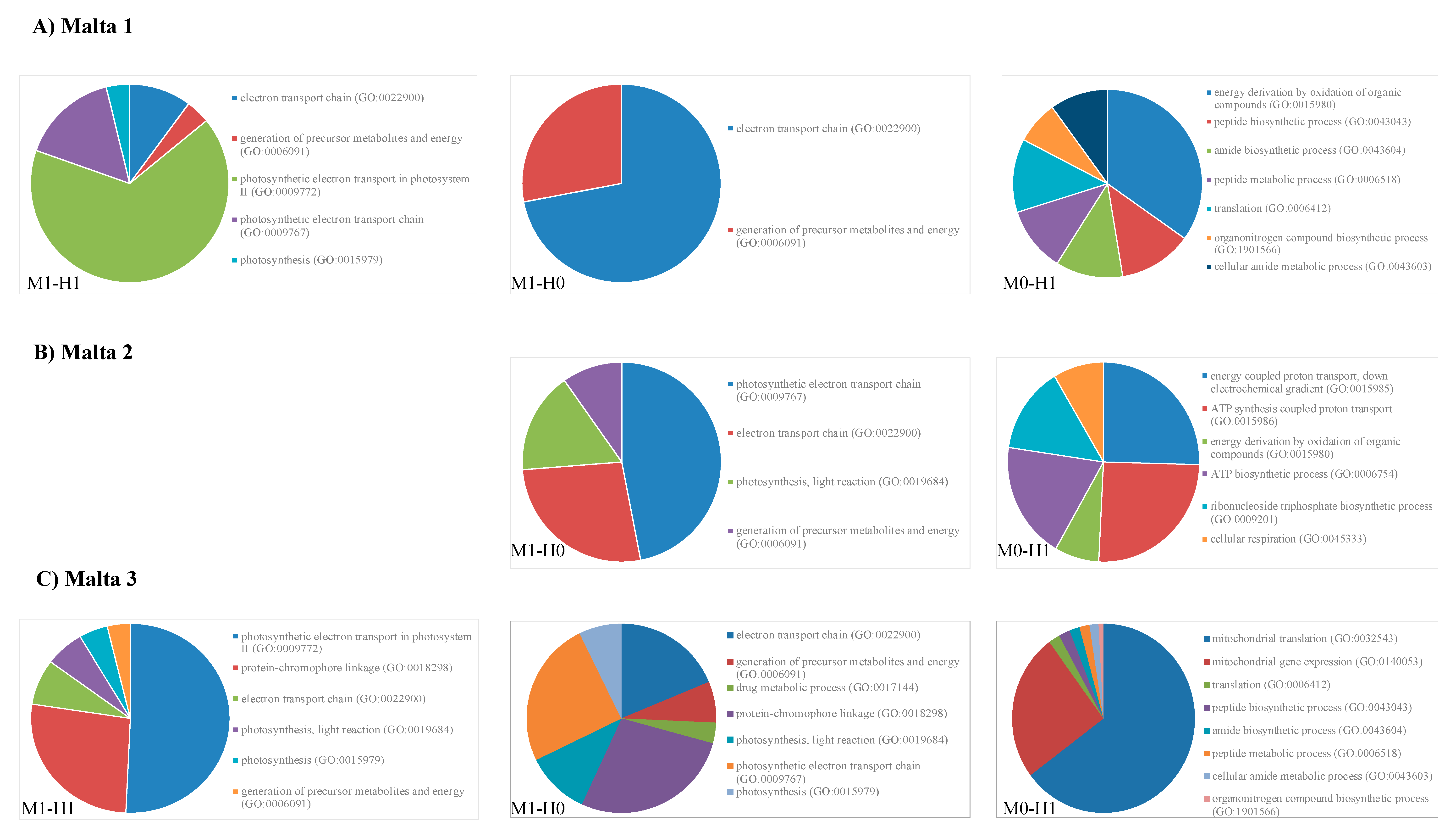
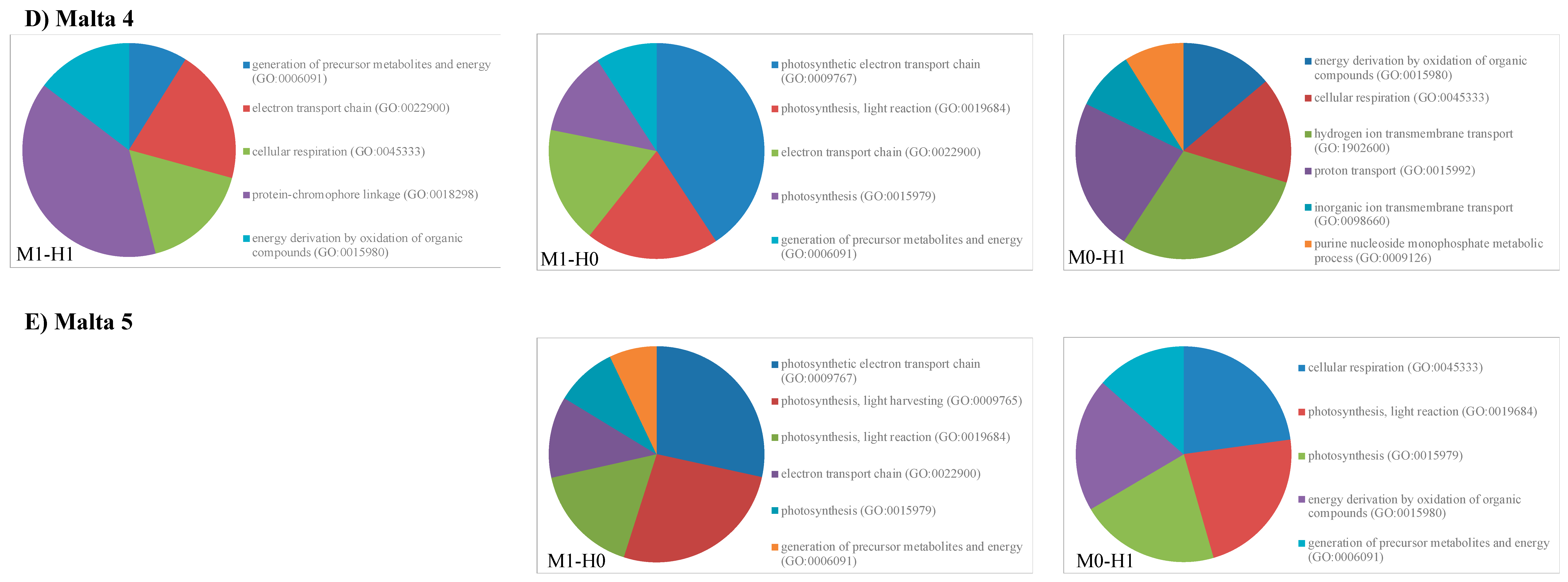
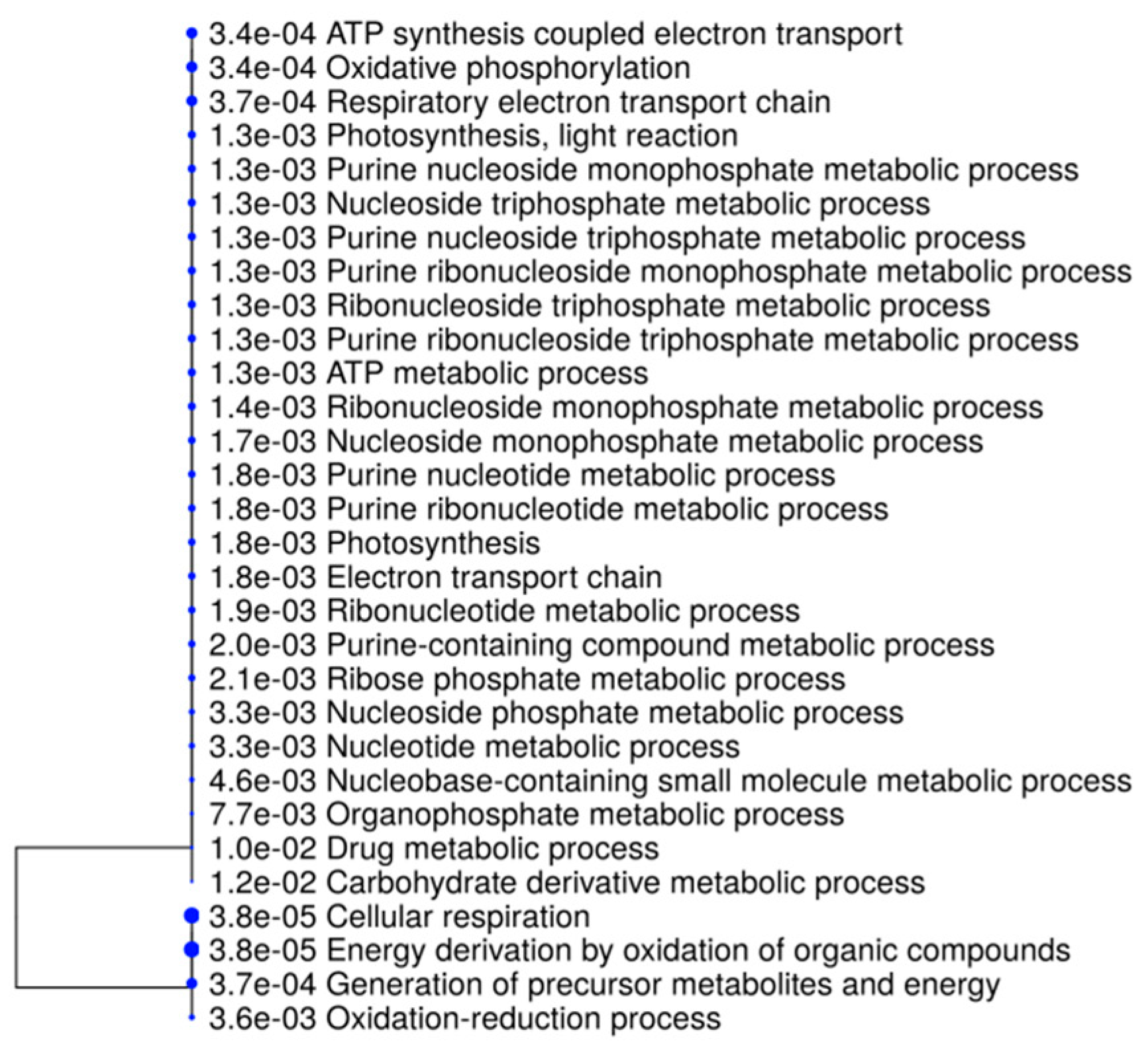
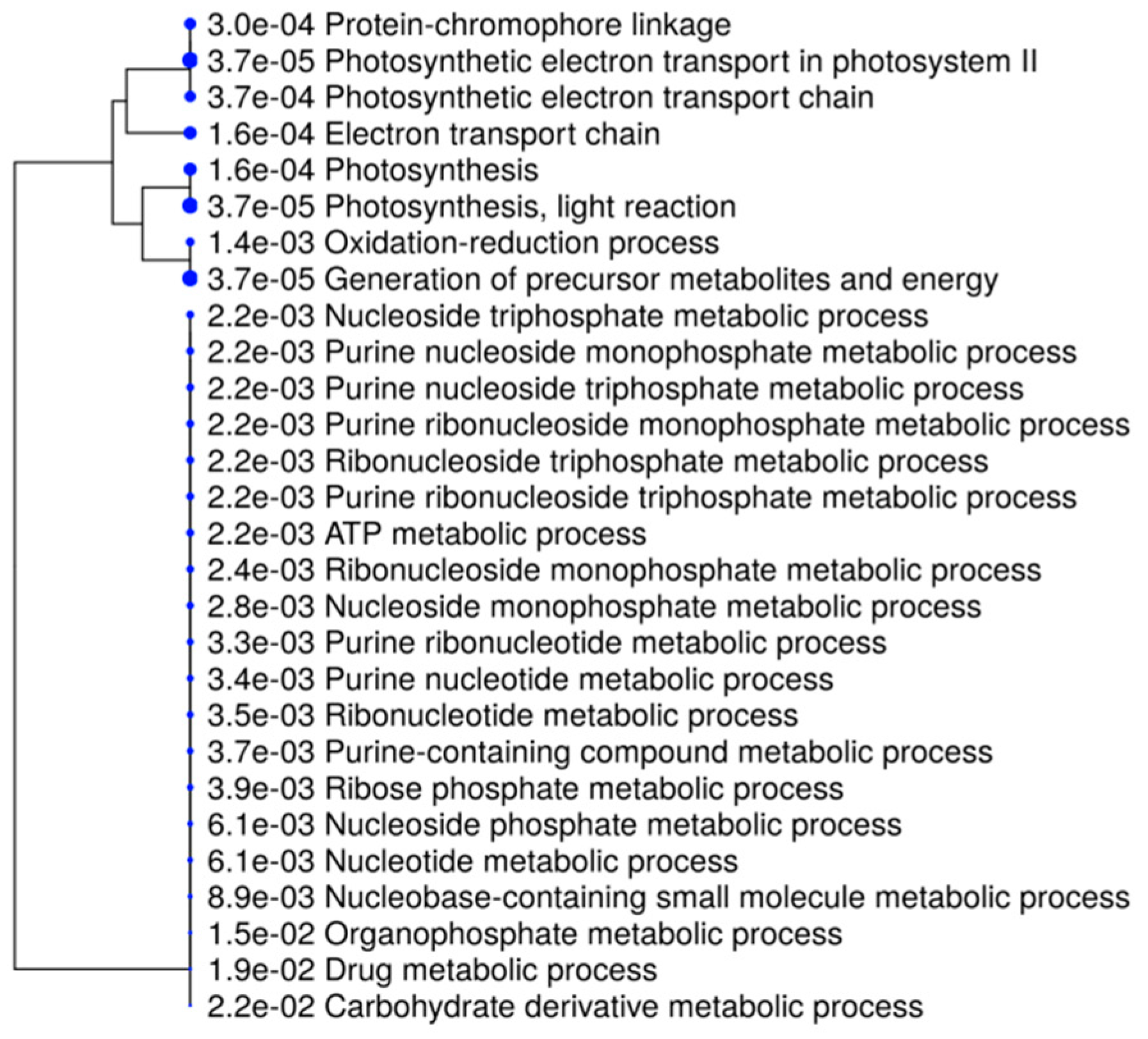
2.2. Gene Ontology Analyses
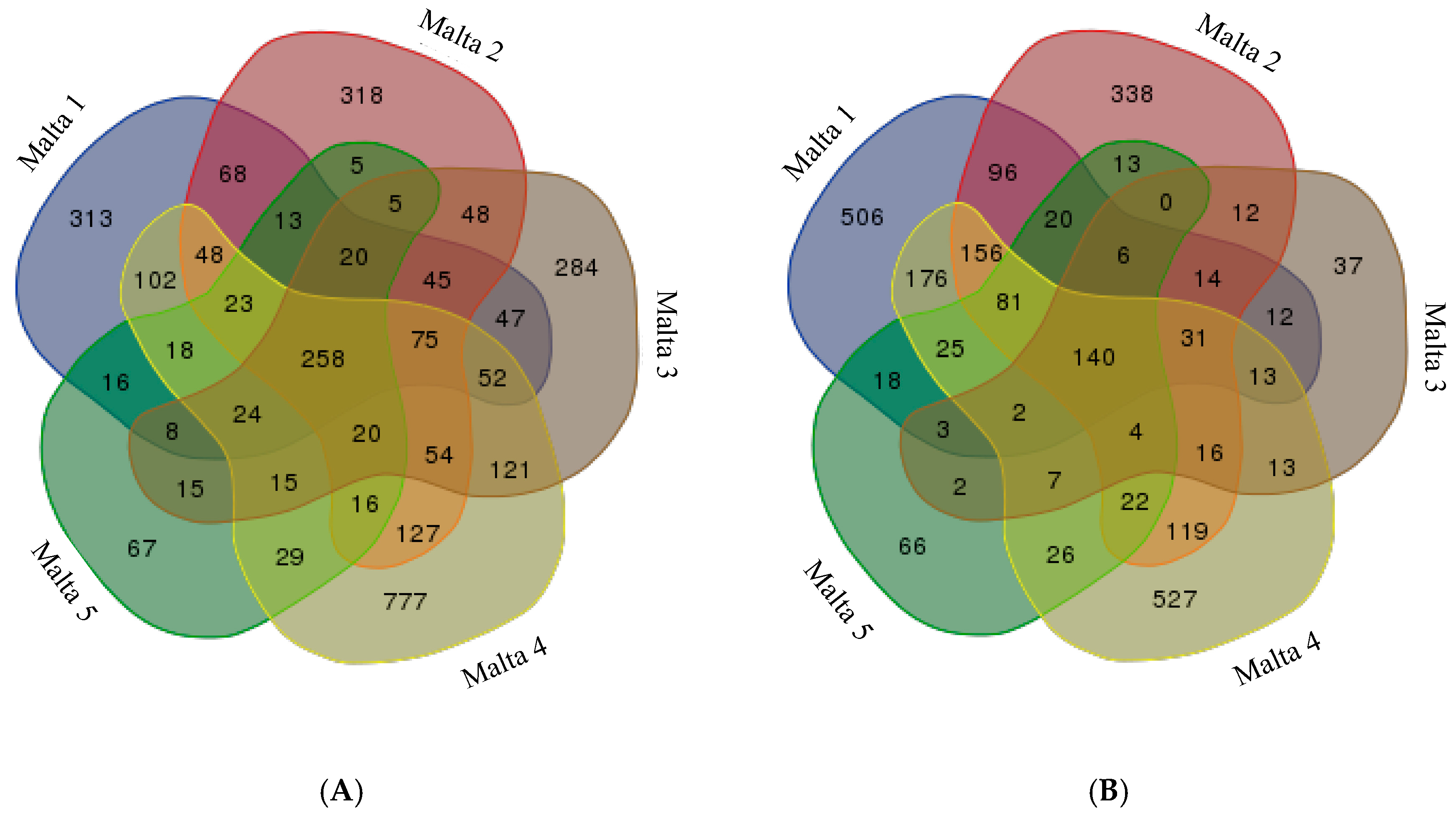
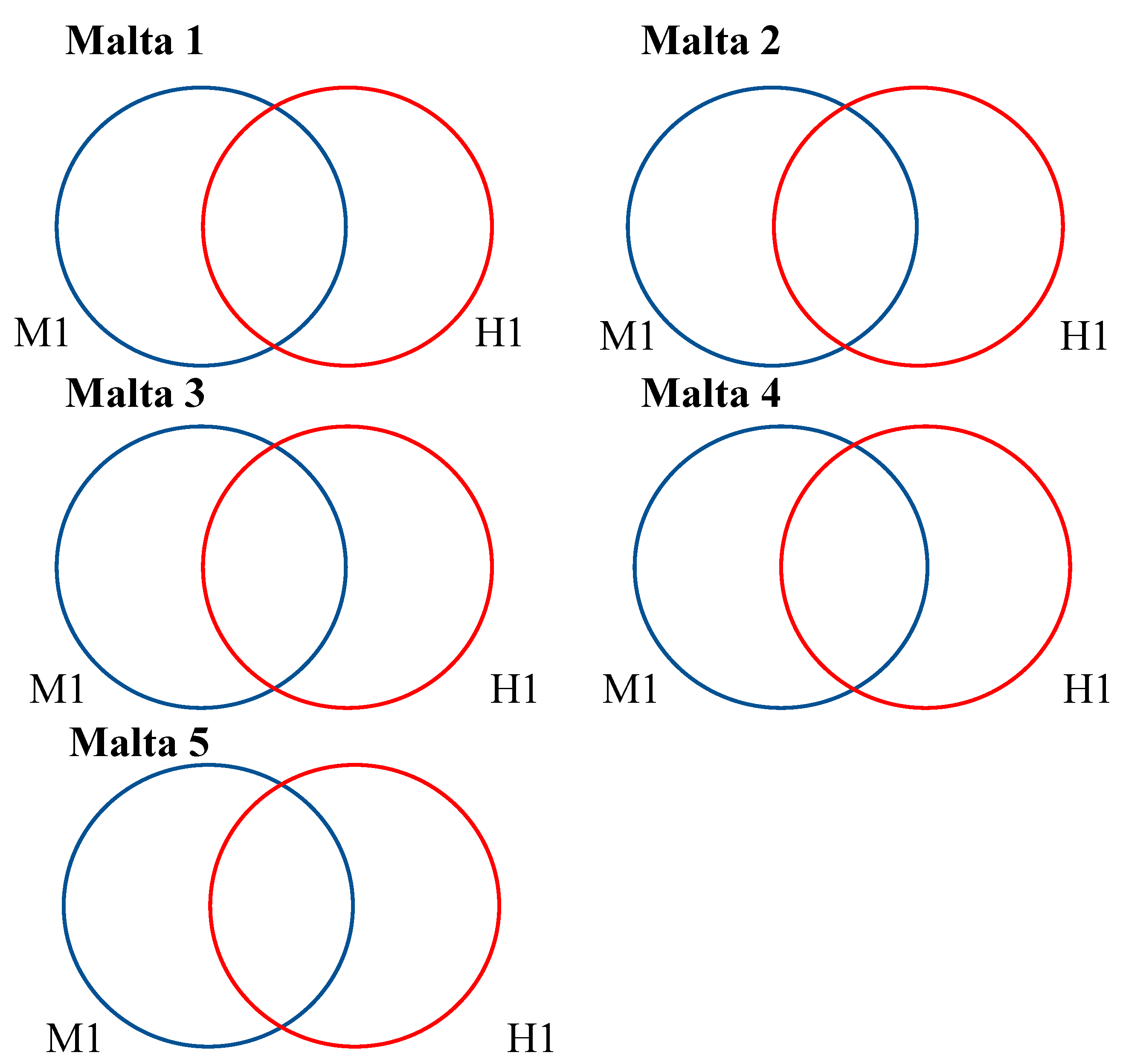
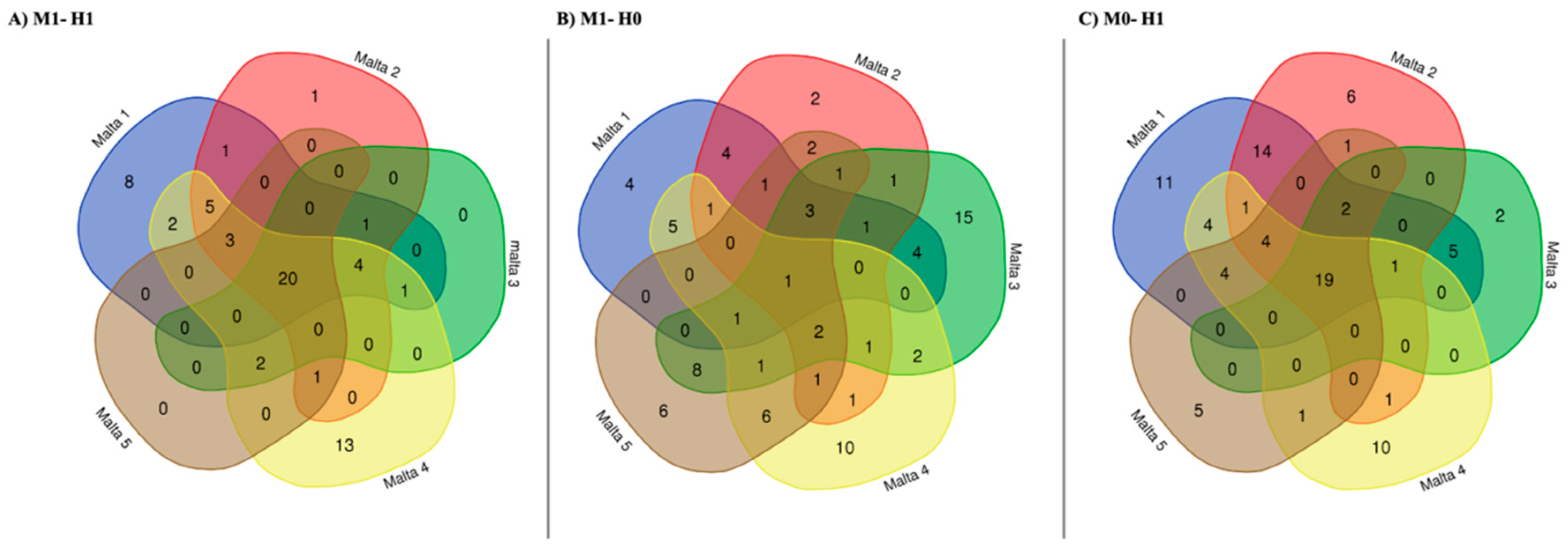
2.3. Venn Analysis of Differently Methylated Fragments
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Isolation and MSAP Analysis
4.3. Library Preparation for NGS Analysis of Amplicons
4.4. NGS Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lukens, L.N.; Zhan, S. The plant genome’s methylation status and response to stress: Implications for plant improvement. Curr. Opin. Plant Biol. 2007, 10, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Foret, S.; Kucharski, R.; Pellegrini, M.; Feng, S.; Jacobsen, S.E.; Robinson, G.E.; Maleszka, R. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honeybees. Proc. Natl. Acad. Sci. USA 2012, 109, 4968–4973. [Google Scholar] [CrossRef] [PubMed]
- Labra, M.; Ghiani, A.; Citterio, S.; Sgorbati, S.; Sala, F.; Vannini, C.; Ruffini-Castiglione, M.; Bracale, M. Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol. 2002, 4, 694–699. [Google Scholar] [CrossRef]
- Yi, S.V. Insights into epigenome evolution from animal and plant methylomes. Genome Biol. Evol. 2017, 9, 3189–3201. [Google Scholar] [CrossRef]
- Putiri, E.L.; Robertson, K.D. Epigenetic mechanisms and genome stability. Clin. Epigenetics 2010, 2, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Ashikawa, I. Surveying CpG methylation at 5′-CCGG in the genomes of rice cultivars. Plant Mol. Biol. 2001, 45, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Latzel, V.; Allan, E.; Silveira, A.B.; Colot, V.; Fischer, M.; Bossdorf, O. Epigenetic diversity increases the productivity and stability of plant populations. Nat. Commun. 2013, 4, 2875. [Google Scholar] [CrossRef] [PubMed]
- Guarino, F.; Cicatelli, A.; Brundu, G.; Heinze, B.; Castiglione, S. Epigenetic diversity of clonal white poplar (Populus alba L.) populations: Could methylation support the success of vegetative reproduction strategy? PLoS ONE 2015, 10, e0131480. [Google Scholar] [CrossRef]
- Guarino, F.; Cicatelli, A.; Brundu, G.; Improta, G.; Triassi, M.; Castiglione, S. The use of MSAP reveals epigenetic diversity of the invasive clonal populations of Arundo donax L. PLoS ONE 2019, 14, e0215096. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Raj, S.; Bräutigam, K.; Hamanishi, E.T.; Wilkins, O.; Thomas, B.R.; Schroeder, W.; Mansfield, S.D.; Plant, A.L.; Campbell, M.M. Clone history shapes Populus drought responses. Proc. Natl. Acad. Sci. USA 2011, 108, 12521–12526. [Google Scholar] [CrossRef] [PubMed]
- Reyna-López, G.E.; Simpson, J.; Ruiz-Herrera, J. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genet. 1997, 253, 703–710. [Google Scholar] [CrossRef]
- Baranek, M.; Cechova, J.; Kovacs, T.; Eichmeier, A.; Wang, S.; Raddova, J.; Necas, T.; Ye, X. Use of combined MSAP and NGS techniques to identify differentially methylated regions in somaclones: A case study of two stable somatic wheat mutants. PLoS ONE 2016, 11, e0165749. [Google Scholar] [CrossRef] [PubMed]
- Chwialkowska, K.; Korotko, U.; Kosinska, J.; Szarejko, I.; Kwasniewski, M. Methylation Sensitive Amplification Polymorphism Sequencing (MSAP-Seq)—A method for high-throughput analysis of differentially methylated CCGG sites in plants with large genomes. Front. Plant Sci. 2017, 8, 2056. [Google Scholar] [CrossRef] [PubMed]
- Fussi, B.; Bonello, J.; Calleja, E.; Heinze, B. Combining the use of molecular techniques and archival documentary evidence to trace the origin of Populus alba in a Central Mediterranean archipelago. Eur. J. For. Res. 2012, 131, 347–354. [Google Scholar] [CrossRef]
- Brundu, G.; Lupi, R.; Zapelli, I.; Fossati, T.; Patrignani, G.; Camarda, I.; Sala, F.; Castiglione, S. The origin of clonal diversity and structure of Populus alba in Sardinia: Evidence from nuclear and plastid microsatellite markers. Ann. Bot. 2008, 102, 997–1006. [Google Scholar] [CrossRef]
- Santos-del-Blanco, L.; de-Lucas, A.I.; González-Martínez, S.C.; Sierra-de-Grado, R.; Hidalgo, E. Extensive clonal assemblies in Populus alba and Populus x canescens from the Iberian Peninsula. Tree Genet. Genomes 2013, 9, 499–510. [Google Scholar] [CrossRef]
- Dickman, D.I.; Isebrands, J.G.; Eckenwalder, J.E.; Richardson, J. Poplar culture in North America; NRC Research Press: Ottawa, ON, Canada, 2002; ISBN 978-0-660-18145-5. [Google Scholar]
- Barrett, S.C.H. Influences of clonality on plant sexual reproduction. Proc. Natl. Acad. Sci. USA 2015, 112, 8859–8866. [Google Scholar] [CrossRef]
- Greco, M.; Sáez, C.A.; Contreras, R.A.; Rodríguez-Rojas, F.; Bitonti, M.B.; Brown, M.T. Cadmium and/or copper excess induce interdependent metal accumulation, DNA methylation, induction of metal chelators and antioxidant defences in the seagrass Zostera marina. Chemosphere 2019, 224, 111–119. [Google Scholar] [CrossRef]
- Perrone, A.; Martinelli, F. Plant stress biology in epigenomic era. Plant Sci. 2020, 294, 110376. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. A role for epigenetic regulation in the adaptation and stress responses of non-model plants. Front. Plant Sci. 2019, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.; Tanti, B. Understanding epigenetic modifications in response to abiotic stresses in plants. Biocatal. Agric. Biotechnol. 2020, 27, 101673. [Google Scholar] [CrossRef]
- Kurdyukov, S.; Bullock, M. DNA methylation analysis: Choosing the right method. Biology 2016, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Herceg, Z. Deciphering the epigenetic code: An overview of DNA methylation analysis methods. Antioxid. Redox Signal. 2013, 18, 1972–1986. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; He, B.; Yi, C. Compilation of modern technologies to map genome-wide cytosine modifications in DNA. Chembiochem 2019, 20, 1898–1905. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Li, M.; Lou, Q.; Xia, H.; Wang, P.; Li, T.; Liu, H.; Luo, L. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS ONE 2013, 8, e80253. [Google Scholar] [CrossRef] [PubMed]
- Baránek, M.; Raddová, J.; Krizan, B.; Pidra, M. Genetic changes in grapevine genomes after stress induced by in vitro cultivation, thermotherapy and virus infection, as revealed by AFLP. Genet. Mol. Biol. 2009, 32, 834–839. [Google Scholar] [CrossRef][Green Version]
- Pereira, W.J.; Pappas, M.D.C.R.; Grattapaglia, D.; Pappas, G.J., Jr. A cost-effective approach to DNA methylation detection by Methyl Sensitive DArT sequencing. PLoS ONE 2020, 15, e233800. [Google Scholar] [CrossRef]
- Vekemans, X.; Beauwens, T.; Lemaire, M.; Roldan-Ruiz, I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X.; et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Liu, Y.J.; Wang, X.R.; Zeng, Q.Y. De novo assembly of white poplar genome and genetic diversity of white poplar population in Irtysh River basin in China. Sci. China Life Sci. 2019, 62, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Sello, S.; Meneghesso, A.; Alboresi, A.; Baldan, B.; Morosinotto, T. Plant biodiversity and regulation of photosynthesis in the natural environment. Planta 2019, 249, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Galili, G. Metabolic engineering of the plant primary-secondary metabolism interface. Curr. Opin. Biotechnol. 2011, 22, 239–244. [Google Scholar] [CrossRef]
- Moradi, N.; Rezaei, A.; Bahramnejad, B.; Goodwin, P.H. Methylation-sensitive amplified polymorphism analysis of resistant and susceptible interactions of cucumber with Podosphaera xanthii. Physiol. Mol. Plant Pathol. 2019, 106, 64–73. [Google Scholar] [CrossRef]
- Guarino, F.; Miranda, A.; Castiglione, S.; Cicatelli, A. Arsenic phytovolatilization and epigenetic modifications in Arundo donax L. assisted by a PGPR consortium. Chemosphere 2020, 251, 126310. [Google Scholar] [CrossRef]
- Vanden Broeck, A.; Cox, K.; Brys, R.; Castiglione, S.; Cicatelli, A.; Guarino, F.; Heinze, B.; Steenackers, M.; Vander Mijnsbrugge, K. Variability in DNA methylation and generational plasticity in the Lombardy poplar, a single genotype worldwide distributed since the eighteenth century. Front. Plant Sci. 2018, 9, 1635. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.Y.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]


| HpaII | MspI | Methylation Status |
|---|---|---|
| 1 | 1 | No methylation |
| 1 | 0 | Hemi-methylated CHG-sites (hemi-methylation of inner and outer cytosine) |
| 0 | 1 | Double-strand methylation of inner cytosine or hemi-methylation of inner cytosine |
| 0 | 0 | Un-informative state caused either by different types of methylation or due to restriction site polymorphism |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guarino, F.; Heinze, B.; Castiglione, S.; Cicatelli, A. Epigenetic Analysis through MSAP-NGS Coupled Technology: The Case Study of White Poplar Monoclonal Populations/Stands. Int. J. Mol. Sci. 2020, 21, 7393. https://doi.org/10.3390/ijms21197393
Guarino F, Heinze B, Castiglione S, Cicatelli A. Epigenetic Analysis through MSAP-NGS Coupled Technology: The Case Study of White Poplar Monoclonal Populations/Stands. International Journal of Molecular Sciences. 2020; 21(19):7393. https://doi.org/10.3390/ijms21197393
Chicago/Turabian StyleGuarino, Francesco, Berthold Heinze, Stefano Castiglione, and Angela Cicatelli. 2020. "Epigenetic Analysis through MSAP-NGS Coupled Technology: The Case Study of White Poplar Monoclonal Populations/Stands" International Journal of Molecular Sciences 21, no. 19: 7393. https://doi.org/10.3390/ijms21197393
APA StyleGuarino, F., Heinze, B., Castiglione, S., & Cicatelli, A. (2020). Epigenetic Analysis through MSAP-NGS Coupled Technology: The Case Study of White Poplar Monoclonal Populations/Stands. International Journal of Molecular Sciences, 21(19), 7393. https://doi.org/10.3390/ijms21197393







