Plant Elongator—Protein Complex of Diverse Activities Regulates Growth, Development, and Immune Responses
Abstract
1. Introduction
2. In Plants Elongator is Located and Active in Nucleus and in Cytoplasm
3. Elongator in Nucleus
3.1. Plant Elongator Regulates Transcription
3.1.1. Processes, Mechanisms, and Pathways Regulated by Elongator via Control of Transcription
3.1.2. Transcript Levels of Individual Genes and Whole Transcriptomes are Altered in elo/elp Mutants and Plants Overexpressing Elongator Subunits
3.1.3. Elongator Regulates Elongation of Transcription
3.1.4. Elongator Promotes Biogenesis of miRNAs
3.1.5. Elongator Modifies DNA Methylation
3.2. Elongator Regulates Replication and Cell-Cycle Progression
4. Elongator in Cytoplasm
4.1. Processes and Mechanisms Regulated by Elongator via Activity in the Cytoplasm
4.2. Elongator Modifies Wobble U34 in tRNAs and Regulates Protein Translation
4.2.1. Conservation of Wobble U34 Modification between Yeast and Plants
4.2.2. The Role of the DRL1 Interactor of Elongator in tRNA Modification
4.2.3. The role of Elongator-Mediated tRNA Modification in Auxin Responses
4.2.4. Similar Role of Elongator and GRXS17
4.2.5. tRNA Wobble Uridine Modification Regulates Leaf Development
4.3. Elongator Acetylates alpha-Tubulin
5. Do Subunits of the Plant Elongator Complex Always Act Together?
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Otero, G.; Fellows, J.; Yang, L.; De Bizemont, T.; Dirac, A.M.G.; Gustafsson, C.M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 1999, 3, 109–118. [Google Scholar] [CrossRef]
- Krogan, N.J.; Greenblatt, J.F. Characterization of a Six-Subunit Holo-Elongator Complex Required for the Regulated Expression of a Group of Genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 8203–8212. [Google Scholar] [CrossRef] [PubMed]
- Winkler, S.G.; Petrakis, T.G.; Ethelberg, S.; Tokunaga, M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. RNA Polymerase II Elongator Holoenzyme Is Composed of Two Discrete Subcomplexes. J. Biol. Chem. 2001, 276, 32743–32749. [Google Scholar] [CrossRef] [PubMed]
- Glatt, S.; Létoquart, J.; Faux, C.; Taylor, N.M.I.; Séraphin, B.; Müller, C.W. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012, 19, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Dauden, M.I.; Kosinski, J.; Kolaj-Robin, O.; Desfosses, A.; Ori, A.; Faux, C.; Hoffmann, N.A.; Onuma, O.F.; Breunig, K.D.; Beck, M.; et al. Architecture of the yeast Elongator complex. EMBO Rep. 2017, 18, 264–279. [Google Scholar] [CrossRef]
- Setiaputra, D.T.; Cheng, D.T.; Lu, S.; Hansen, J.M.; Dalwadi, U.; Lam, C.H.; To, J.L.; Dong, M.; Yip, C.K. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017, 18, 280–291. [Google Scholar] [CrossRef]
- Winkler, G.S.; Kristjuhan, A.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 3517–3522. [Google Scholar] [CrossRef]
- Lin, T.Y.; Abbassi, N.E.H.; Zakrzewski, K.; Chramiec-Głąbik, A.; Jemioła-Rzemińska, M.; Różycki, J.; Glatt, S. The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Creppe, C.; Malinouskaya, L.; Volvert, M.L.; Gillard, M.; Close, P.; Malaise, O.; Laguesse, S.; Cornez, I.; Rahmouni, S.; Ormenese, S.; et al. Elongator Controls the Migration and Differentiation of Cortical Neurons through Acetylation of α-Tubulin. Cell 2009, 136, 551–564. [Google Scholar] [CrossRef]
- Tran, H.T.; Nimick, M.; Uhrig, R.G.; Templeton, G.; Morrice, N.; Gourlay, R.; Delong, A.; Moorhead, G.B.G. Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J. 2012, 71, 263–272. [Google Scholar] [CrossRef]
- Solinger, J.A.; Paolinelli, R.; Klöß, H.; Scorza, F.B.; Marchesi, S.; Sauder, U.; Mitsushima, D.; Capuani, F.; Stürzenbaum, S.R.; Cassata, G. The Caenorhabditis elegans elongator complex regulates neuronal α-tubulin acetylation. PLoS Genet. 2010, 6, e1000820. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Johansson, M.J.O.; Byström, A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. [Google Scholar] [CrossRef]
- Lin, Z.; Zhao, W.; Diao, W.; Xie, X.; Wang, Z.; Zhang, J.; Shen, Y.; Long, J. Crystal structure of elongator subcomplex Elp4-6. J. Biol. Chem. 2012, 287, 21501–21508. [Google Scholar] [CrossRef] [PubMed]
- Rahl, P.B.; Chen, C.Z.; Collins, R.N. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell 2005, 17, 841–853. [Google Scholar] [CrossRef]
- Li, Q.; Fazly, A.M.; Zhou, H.; Huang, S.; Zhang, Z.; Stillman, B. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009, 5, e1000684. [Google Scholar] [CrossRef] [PubMed]
- Frohloff, F.; Fichtner, L.; Jablonowski, D.; Breunig, K.D.; Schaffrath, R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001, 20, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Jablonowski, D.; Schaffrath, R. Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem. Soc. Trans. 2007, 35, 1533–1537. [Google Scholar] [CrossRef]
- Lu, J.; Huang, B.O.; Esberg, A.; Johansson, M.J.O.; Byström, A.S. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 2005, 11, 1648–1654. [Google Scholar] [CrossRef]
- Esberg, A.; Huang, B.; Johansson, M.J.O.; Byström, A.S. Elevated Levels of Two tRNA Species Bypass the Requirement for Elongator Complex in Transcription and Exocytosis. Mol. Cell 2006, 24, 139–148. [Google Scholar] [CrossRef]
- Hawkes, N.A.; Otero, G.; Sebastiaan Winkler, G.; Marshall, N.; Dahmus, M.E.; Krappmann, D.; Scheidereit, C.; Thomas, C.L.; Schiavo, G.; Erdjument-Bromage, H.; et al. Purification and characterization of the human elongator complex. J. Biol. Chem. 2002, 277, 3047–3052. [Google Scholar] [CrossRef]
- Close, P.; Gillard, M.; Ladang, A.; Jiang, Z.; Papuga, J.; Hawkes, N.; Nguyen, L.; Chapelle, J.P.; Bouillenne, F.; Svejstrup, J.; et al. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of elongator. J. Biol. Chem. 2012, 287, 32535–32545. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Yamagata, K.; Hong, K.; Wakayama, T.; Zhang, Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature 2010, 463, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tuck, S.; Byström, A.S. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009, 5, e1000561. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, H.; Fleury, D.; Bruno, L.; Robles, P.; De Veylder, L.; Traas, J.; Micol, J.L.; Van Montagu, M.; Inze, D.; Van Lijsebettens, M. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. USA 2005, 102, 7754–7759. [Google Scholar] [CrossRef]
- Nelissen, H.; De Groeve, S.; Fleury, D.; Neyt, P.; Bruno, L.; Bitonti, M.B.; Vandenbussche, F.; Van Der Straeten, D.; Yamaguchi, T.; Tsukaya, H.; et al. Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 2010, 107, 1678–1683. [Google Scholar] [CrossRef]
- Paraskevopoulou, C.; Fairhurst, S.A.; Lowe, D.J.; Brick, P.; Onesti, S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 2006, 59, 795–806. [Google Scholar] [CrossRef]
- Selvadurai, K.; Wang, P.; Seimetz, J.; Huang, R.H. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 2014, 10, 810–812. [Google Scholar] [CrossRef]
- Glatt, S.; Müller, C.W. Structural insights into Elongator function. Curr. Opin. Struct. Biol. 2013, 23, 235–242. [Google Scholar] [CrossRef]
- Frohloff, F.; Jablonowski, D.; Fichtner, L.; Schaffrath, R. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (γ-toxin target (TOT)) complex. J. Biol. Chem. 2003, 278, 956–961. [Google Scholar] [CrossRef]
- Singh, N.; Lorbeck, M.T.; Zervos, A.; Zimmerman, J.; Elefant, F. The histone acetyltransferase Elp3 plays in active role in the control of synaptic bouton expansion and sleep in Drosophila. J. Neurochem. 2010, 115, 493–504. [Google Scholar] [CrossRef]
- Walker, J.; Kwon, S.Y.; Badenhorst, P.; East, P.; McNeill, H.; Svejstrup, J.Q. Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics 2011, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Mehlgarten, C.; Jablonowski, D.; Wrackmeyer, U.; Tschitschmann, S.; Sondermann, D.; Jäger, G.; Gong, Z.; Byström, A.S.; Schaffrath, R.; Breunig, K.D. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010, 76, 1082–1094. [Google Scholar] [CrossRef] [PubMed]
- Slaugenhaupt, S.A.; Blumenfeld, A.; Gill, S.P.; Leyne, M.; Mull, J.; Cuajungco, M.P.; Liebert, C.B.; Chadwick, B.; Idelson, M.; Reznik, L.; et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001, 68, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.J.; Shen, L.; Jang, C.W.; Falnes, P.; Zhang, Y. Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression. PLoS Genet. 2013, 9, e1003516. [Google Scholar] [CrossRef]
- Kojic, M.; Gaik, M.; Kiska, B.; Salerno-Kochan, A.; Hunt, S.; Tedoldi, A.; Mureev, S.; Jones, A.; Whittle, B.; Genovesi, L.A.; et al. Elongator mutation in mice induces neurodegeneration and ataxia-like behavior. Nat. Commun. 2018, 9, 3195. [Google Scholar] [CrossRef]
- Miśkiewicz, K.; Jose, L.E.; Bento-Abreu, A.; Fislage, M.; Taes, I.; Kasprowicz, J.; Swerts, J.; Sigrist, S.; Versées, W.; Robberecht, W.; et al. ELP3 controls active zone morphology by acetylating the ELKS family member bruchpilot. Neuron 2011, 72, 776–788. [Google Scholar] [CrossRef]
- Falcone, A.; Nelissen, H.; Fleury, D.; Van Lijsebettens, M.; Bitonti, M.B. Cytological investigations of the Arabidopsis thaliana elo1 mutant give new insights into leaf lateral growth and elongator function. Ann. Bot. 2007, 100, 261–270. [Google Scholar] [CrossRef]
- Jia, Y.; Tian, H.; Li, H.; Yu, Q.; Wang, L.; Friml, J.; Ding, Z. The Arabidopsis thaliana elongator complex subunit 2 epigenetically affects root development. J. Exp. Bot. 2015, 66, 4631–4642. [Google Scholar] [CrossRef]
- Xu, D.; Huang, W.; Li, Y.; Wang, H.; Huang, H.; Cui, X. Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis. Plant J. 2012, 69, 792–808. [Google Scholar] [CrossRef]
- Wang, Y.; An, C.; Zhang, X.; Yao, J.; Zhang, Y.; Sun, Y.; Yu, F.; Amador, D.M.; Mou, Z. The Arabidopsis Elongator Complex Subunit2 Epigenetically Regulates Plant Immune Responses. Plant Cell 2013, 25, 762–776. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, H.; Jablonowski, D.; Zhou, X.; Ren, X.; Hong, X.; Schaffrath, R.; Zhu, J.-K.; Gong, Z. Mutations in ABO1/ELO2, a Subunit of Holo-Elongator, Increase Abscisic Acid Sensitivity and Drought Tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 2006, 26, 6902–6912. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hua, D.; Chen, Z.; Zhou, Z.; Gong, Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 2009, 60, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Woloszynska, M.; Gagliardi, O.; Vandenbussche, F.; De Groeve, S.; Baez, L.A.; Neyt, P.; Le Gall, S.; Fung, J.; Mas, P.; Van Der Straeten, D.; et al. The Elongator complex regulates hypocotyl growth in darkness and during photomorphogenesis. J. Cell Sci. 2018, 131, jcs203927. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ding, Y.; Yao, J.; Zhang, Y.; Sun, Y.; Colee, J.; Mou, Z. Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 2015, 83, 1019–1033. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Cui, Y.; Li, Y.; Qi, Y. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 2015, 1, 15075. [Google Scholar] [CrossRef]
- Laubinger, S. MicroRNA transcription and processing: Elongator caught in the act. Nat. Plants 2015, 1, 15076. [Google Scholar] [CrossRef]
- Chen, P.; Jäger, G.; Zheng, B. Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 201. [Google Scholar] [CrossRef]
- Leitner, J.; Retzer, K.; Malenica, N.; Bartkeviciute, R.; Lucyshyn, D.; Jäger, G.; Korbei, B.; Byström, A.; Luschnig, C. Meta-regulation of Arabidopsis Auxin Responses Depends on tRNA Maturation. Cell Rep. 2015, 11, 516–526. [Google Scholar] [CrossRef]
- Iñigo, S.; Durand, A.N.; Ritter, A.; Le Gall, S.; Termathe, M.; Klassen, R.; Tohge, T.; De Coninck, B.; Van Leene, J.; De Clercq, R.; et al. Glutaredoxin GRXS17 associates with the cytosolic iron-sulfur cluster assembly pathway. Plant Physiol. 2016, 172, 858–873. [Google Scholar]
- Nakai, Y.; Horiguchi, G.; Iwabuchi, K.; Harada, A.; Nakai, M.; Hara-Nishimura, I.; Yano, T. tRNA Wobble Modification Affects Leaf Cell Development in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 2026–2039. [Google Scholar] [CrossRef]
- Fichtner, L.; Frohloff, F.; Jablonowski, D.; Stark, M.J.R.; Schaffrath, R. Protein interactions within Saccharomyces cerevisiae elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol. Microbiol. 2002, 45, 817–826. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, J.H.; Lane, W.S.; Reinberg, D. Human elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA 2002, 99, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Pokholok, D.K.; Hannett, N.M.; Young, R.A. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 2002, 9, 799–809. [Google Scholar] [CrossRef]
- Chen, C.; Huang, B.; Eliasson, M.; Rydén, P.; Byström, A.S. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011, 7, e1002258. [Google Scholar] [CrossRef]
- Durant, P.C.; Bajji, A.C.; Sundaram, M.; Kumar, R.K.; Davis, D.R. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: The effect of nucleosides s 2U, mcm5U, mcm5s2U, mnm 5s2U, t6A, and ms2t6A. Biochemistry 2005, 44, 8078–8089. [Google Scholar] [CrossRef]
- Begley, U.; Dyavaiah, M.; Patil, A.; Rooney, J.P.; DiRenzo, D.; Young, C.M.; Conklin, D.S.; Zitomer, R.S.; Begley, T.J. Trm9-Catalyzed tRNA Modifications Link Translation to the DNA Damage Response. Mol. Cell 2007, 28, 860–870. [Google Scholar] [CrossRef]
- Johansson, M.J.O.; Esberg, A.; Huang, B.; Björk, G.R.; Byström, A.S. Eukaryotic Wobble Uridine Modifications Promote a Functionally Redundant Decoding System. Mol. Cell. Biol. 2008, 28, 3301–3312. [Google Scholar] [CrossRef]
- Karlsborn, T.; Tükenmez, H.; Chen, C.; Byström, A.S. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem. Biophys. Res. Commun. 2014, 454, 441–445. [Google Scholar] [CrossRef]
- Yoshida, M.; Kataoka, N.; Miyauchi, K.; Ohe, K.; Iida, K.; Yoshida, S.; Nojima, T.; Okuno, Y.; Onogi, H.; Usui, T.; et al. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc. Natl. Acad. Sci. USA 2015, 112, 2764–2769. [Google Scholar] [CrossRef]
- Glatt, S.; Zabel, R.; Kolaj-Robin, O.; Onuma, O.F.; Baudin, F.; Graziadei, A.; Taverniti, V.; Lin, T.Y.; Baymann, F.; Séraphin, B.; et al. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 2016, 23, 794–802. [Google Scholar] [CrossRef]
- Antosz, W.; Pfab, A.; Ehrnsberger, H.F.; Holzinger, P.; Köllen, K.; Mortensen, S.A.; Bruckmann, A.; Schubert, T.; Längst, G.; Griesenbeck, J.; et al. The Composition of the Arabidopsis RNA Polymerase II Transcript Elongation Complex Reveals the Interplay between Elongation and mRNA Processing Factors. Plant Cell 2017, 29, 854–870. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Zhang, X.; Zhai, H.; Liu, J.; Wu, F.; Li, C.; Chen, Q. Elongator Is Required for Root Stem Cell Maintenance by Regulating SHORTROOT Transcription. Plant Physiol. 2019, 179, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Woloszynska, M.; Gagliardi, O.; Vandenbussche, F.; Van Lijsebettens, M. Elongator promotes germination and early post-germination growth. Plant Signal. Behav. 2018, 13, e1422465. [Google Scholar] [CrossRef] [PubMed]
- Himanen, K.; Woloszynska, M.; Boccardi, T.M.; De Groeve, S.; Nelissen, H.; Bruno, L.; Vuylsteke, M.; Van Lijsebettens, M. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 2012, 72, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Defraia, C.T.; Zhang, X.; Mou, Z. Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 2010, 64, 511–523. [Google Scholar] [CrossRef] [PubMed]
- DeFraia, C.T.; Wang, Y.; Yao, J.; Mou, Z. Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol. 2013, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Wang, C.; Mou, Z. The Arabidopsis Elongator complex is required for nonhost resistance against the bacterial pathogens Xanthomonas citri subsp. citri and Pseudomonas syringae pv. phaseolicola NPS3121. New Phytol. 2017, 214, 1245–1259. [Google Scholar] [CrossRef]
- An, C.; Ding, Y.; Zhang, X.; Wang, C.; Mou, Z. Elongator Plays a Positive Role in Exogenous NADInduced Defense Responses in Arabidopsis. Mol. Plant-Microbe Interact. 2016, 29, 396–404. [Google Scholar] [CrossRef]
- Silva, K.J.P.; Brunings, A.M.; Pereira, J.A.; Peres, N.A.; Folta, K.M.; Mou, Z. The Arabidopsis ELP3/ELO3 and ELP4/ELO1 genes enhance disease resistance in Fragaria vesca L. BMC Plant Biol. 2017, 17, 1–12. [Google Scholar] [CrossRef]
- Pereira, J.A.; Yu, F.; Zhang, Y.; Jones, J.B.; Mou, Z. The Arabidopsis Elongator Subunit ELP3 and ELP4 Confer Resistance to Bacterial Speck in Tomato. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Sanmartín, M.; Sauer, M.; Muñoz, A.; Zouhar, J.; Ordóñez, A.; Van De Ven, W.T.G.; Caro, E.; De La Paz Sánchez, M.; Raikhel, N.V.; Gutiérrez, C.; et al. A molecular switch for initiating cell differentiation in arabidopsis. Curr. Biol. 2011, 21, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Van Lijsebettens, M.; Durr, J.; Woloszynska, M.; Grasser, K.D. Elongator and SPT4/SPT5 complexes as proxy to study RNA polymerase II transcript elongation control of plant development. Proteomics 2014, 14, 2109–2114. [Google Scholar] [CrossRef] [PubMed]
- Woloszynska, M.; Le Gall, S.; Van Lijsebettens, M. Plant Elongator-mediated transcriptional control in a chromatin and epigenetic context. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Li, Y.; Chen, G.; Ren, L.; Xie, Q.; Zhao, Z.; Hu, Z. Silencing SlELP2L, a tomato Elongator complex protein 2-like gene, inhibits leaf growth, accelerates leaf, sepal senescence, and produces dark-green fruit. Sci. Rep. 2015, 5, 7693. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Skylar, A.; Matsuwaka, S.; Wu, X. ELONGATA3 is required for shoot meristem cell cycle progression in Arabidopsis thaliana seedlings. Dev. Biol. 2013, 382, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Malenica, N.; Abas, L.; Benjamins, R.; Kitakura, S.; Sigmund, H.F.; Jun, K.S.; Hauser, M.T.; Friml, J.; Luschnig, C. MODULATOR of PIN genes control steady-state levels of Arabidopsis PIN proteins. Plant J. 2007, 51, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, C.; Zhang, Y.; Yan, X.; Jin, X.; Yao, X.; Chen, P.; Zheng, B. PtKTI12 genes influence wobble uridine modifications and drought stress tolerance in hybrid poplar. Tree Physiol. 2020. [Google Scholar] [CrossRef]
- Agris, P.F. The importance of being modified: An unrealized code to RNA structure and function. RNA 2015, 21, 552–554. [Google Scholar] [CrossRef]
- Väre, V.Y.P.; Eruysal, E.R.; Narendran, A.; Sarachan, K.L.; Agris, P.F. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules 2017, 7, 29. [Google Scholar] [CrossRef]
- Vendeix, F.A.P.; Murphy IV, F.V.; Cantara, W.A.; Leszczyńska, G.; Gustilo, E.M.; Sproat, B.; Malkiewicz, A.; Agris, P.F. Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through Keto-Enol tautomerism. J. Mol. Biol. 2012, 416, 467–485. [Google Scholar] [CrossRef]
- Blanchet, S.; Cornu, D.; Hatin, I.; Grosjean, H.; Bertin, P.; Namy, O. Deciphering the reading of the genetic code by near-cognate tRNA. Proc. Natl. Acad. Sci. USA 2018, 115, 3018–3023. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Vendeix, F.A.P.; Graham, W.D. tRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rozov, A.; Demeshkina, N.; Khusainov, I.; Westhof, E.; Yusupov, M.; Yusupova, G. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016, 7, 10457. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, N.; Rodnina, M.V. Thio-Modification of tRNA at the Wobble Position as Regulator of the Kinetics of Decoding and Translocation on the Ribosome. J. Am. Chem. Soc. 2017, 139, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Leiber, R.M.; John, F.; Verhertbruggen, Y.; Diet, A.; Knox, J.P.; Ringli, C. The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 2010, 22, 1898–1908. [Google Scholar] [CrossRef]
- Nakai, Y.; Harada, A.; Hashiguchi, Y.; Nakai, M.; Hayashi, H. Arabidopsis molybdopterin biosynthesis protein Cnx5 collaborates with the ubiquitin-like protein urm11 in the thio-modification of tRNA. J. Biol. Chem. 2012, 287, 30874–30884. [Google Scholar] [CrossRef]
- Mehlgarten, C.; Prochaska, H.; Hammermeister, A.; Abdel-Fattah, W.; Wagner, M.; Krutyhołowa, R.; Jun, S.E.; Kim, G.T.; Glatt, S.; Breunig, K.D.; et al. Use of a yeast tRNase killer toxin to diagnose Kti12 motifs required for tRNA modification by Elongator. Toxins 2017, 9, 272. [Google Scholar] [CrossRef]
- Krutyhołowa, R.; Reinhardt-Tews, A.; Chramiec-Głąbik, A.; Breunig, K.D.; Glatt, S. Fungal Kti12 proteins display unusual linker regions and unique ATPase p-loops. Curr. Genet. 2020, 66, 823–833. [Google Scholar] [CrossRef]
- Butler, A.R.; White, J.H.; Folawiyo, Y.; Edlin, A.; Gardiner, D.; Stark, M.J. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 1994, 14, 6306–6316. [Google Scholar] [CrossRef]
- Nelissen, H.; Clarke, J.H.; De Block, M.; De Block, S.; Vanderhaeghen, R.; Zielinski, R.E.; Dyer, T.; Lust, S.; Inzé, D.; Van Lijsebettens, M. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 2003, 15, 639–654. [Google Scholar] [CrossRef]
- Jun, S.E.; Cho, K.H.; Hwang, J.Y.; Abdel-Fattah, W.; Hammermeister, A.; Schaffrath, R.; Bowman, J.L.; Kim, G.T. Comparative analysis of the conserved functions of Arabidopsis DRL1 and yeast KTI12. Mol. Cells 2014, 38, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, X.; Li, J.L.; Zhang, Y.; Mou, Z. The Elongator complex-associated protein DRL1 plays a positive role in immune responses against necrotrophic fungal pathogens in Arabidopsis. Mol. Plant Pathol. 2018, 19, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Krutyhołowa, R.; Hammermeister, A.; Zabel, R.; Abdel-Fattah, W.; Reinhardt-Tews, A.; Helm, M.; Stark, M.J.R.; Breunig, K.D.; Schaffrath, R.; Glatt, S. Kti12, a PSTK-like tRNA dependent ATPase essential for tRNA modification by Elongator. Nucleic Acids Res. 2019, 47, 4814–4830. [Google Scholar] [CrossRef] [PubMed]
- Machida, C.; Nakagawa, A.; Kojima, S.; Takahashi, H.; Machida, Y. The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 2015. [Google Scholar] [CrossRef]
- Kojima, S.; Iwasaki, M.; Takahashi, H.; Imai, T.; Matsumura, Y.; Fleury, D.; Van Lijsebettens, M.; Machida, Y.; Machida, C. ASYMMETRIC LEAVES2 and elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1259–1273. [Google Scholar] [CrossRef]
- Wang, W.; Vignani, R.; Scali, M.; Sensi, E.; Cresti, M. Post-translational modifications of α-tubulin in Zea mays L. are highly tissue specific. Planta 2004, 218, 460–465. [Google Scholar] [CrossRef]
- Gu, J.; Sun, D.; Zheng, Q.; Wang, X.; Yang, H.; Miao, J.; Jiang, J.; Wei, W. Human Elongator complex is involved in cell cycle and suppresses cell growth in 293T human embryonic kidney cells. Acta Biochim. Biophys. Sin. 2009, 41, 831–838. [Google Scholar] [CrossRef]
- Turner, E.L.; Malo, M.E.; Pisclevich, M.G.; Dash, M.D.; Davies, G.F.; Arnason, T.G.; Harkness, T.A.A. The Saccharomyces cerevisiae anaphase-promoting complex interacts with multiple histone-modifying enzymes to regulate cell cycle progression. Eukaryot. Cell 2010, 9, 1418–1431. [Google Scholar] [CrossRef]

| Epigenetic Regulation | Genes | Biological Process | Reference |
|---|---|---|---|
| Histone acetylation | IAA3 and LAX2 | Auxin signaling pathway | [25] |
| NPR1, PR2, PR5 EDS1, PAD4 | Plant defense | [40] | |
| WRKY33, ORA59, PDF1.2 | Plant defense and ethylene signaling | [44] | |
| PLT1, PLT2, SHR, SCR, PIN1 | Root development | [38,62] | |
| RBOHD and ICS1 | Plant defense | [67] | |
| LHY, HYH, HFR1 | Circadian rhythms and phytochrome signaling pathway | [43] | |
| DNA (de)methylation | NPR1 and PAD4 | Plant defense | [40] |
| CYCB1 | Cell cycle | [38] |
| Localization | Molecular Process | Biochemical Activity | The Role of Elongator | Reference |
|---|---|---|---|---|
| Nucleus | Transcription elongation | Histone acetylation | Elongator regulates auxin signaling. | [25] |
| Elongator regulates mitotic cell cycle and leaf patterning. | [39] | |||
| Elongator is involved in plant defense. | [40] | |||
| The role of Elongator in B. cinerea infection. | [44] | |||
| Elongator regulates root development. | [38,62] | |||
| Elongator is involved in plant defense. | [67] | |||
| Elongator regulates skoto- and photomorphogenesis. | [43] | |||
| - | IYO interacts with RNAPII and Elongator to promote elongation and initiate cell differentiation. | [71] | ||
| Elongator and TEFs associate with elongating RNAPII. | [61] | |||
| Elongator is involved in root stem cell maintenance through unknown mechanism. | [62] | |||
| Transcription | DNA (de)methylation | Elongator is involved in plant defense. | [40] | |
| Elongator regulates root development. | [38] | |||
| miRNA biogenesis | Elongator regulates miRNA transcription and processing | [45] | ||
| [46] | ||||
| Unknown | Elongator regulates MYBL2 expression which is involved in anthocyanin biosynthesis. | [42] | ||
| Elongator mediates the establishment of leaf polarity through unknown gene expression system. | [95] | |||
| Elongator acts as meristem cell cycle activator. | [75] | |||
| Elongator controls expression of germination-related genes. | [63] | |||
| Cytoplasm | Translation | tRNA wobble uridine modification | Elongator-mediated tRNA modification is conserved in plants. Elongator is involved in mcm5s2U34 and ncm5U34 modification. | [32] |
| Elongator and DRL1 are involved in ncm5U34 modification. | [47] | |||
| Elongator regulates auxin responses by controlling PIN protein level. | [48] | |||
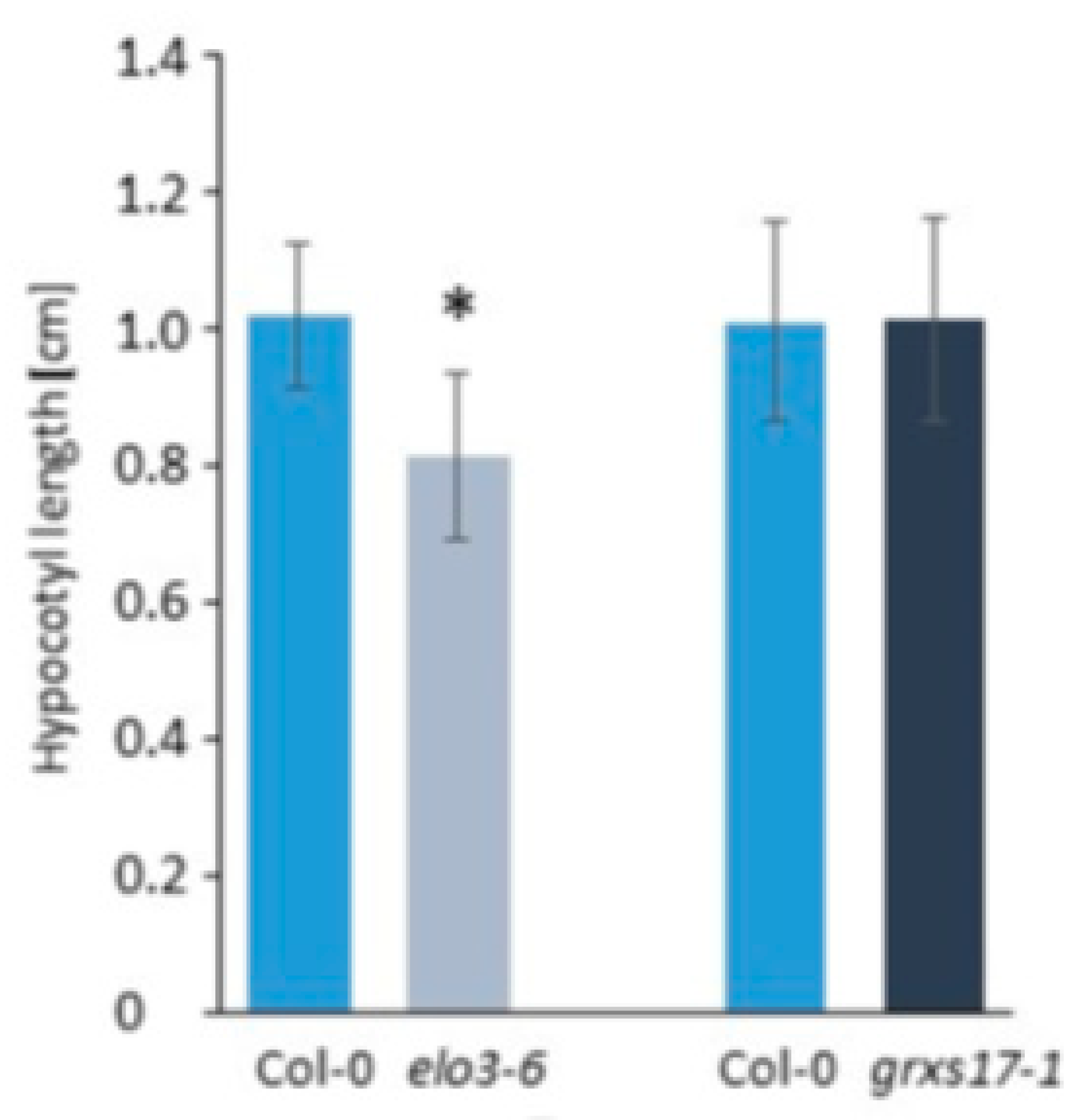
| Strong resemblance between elo3-6 and grxs17. GRXS17 is involved in tRNA modification. | [49] | |||
| Elongator-mediated tRNA modification regulates leaf morphogenesis. | [50] | |||
| Non-histone protein acetylation | Alpha tubulin acetylation | Elongator is involved in microtubules dynamics regulation. | [10] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarosz, M.; Van Lijsebettens, M.; Woloszynska, M. Plant Elongator—Protein Complex of Diverse Activities Regulates Growth, Development, and Immune Responses. Int. J. Mol. Sci. 2020, 21, 6912. https://doi.org/10.3390/ijms21186912
Jarosz M, Van Lijsebettens M, Woloszynska M. Plant Elongator—Protein Complex of Diverse Activities Regulates Growth, Development, and Immune Responses. International Journal of Molecular Sciences. 2020; 21(18):6912. https://doi.org/10.3390/ijms21186912
Chicago/Turabian StyleJarosz, Magdalena, Mieke Van Lijsebettens, and Magdalena Woloszynska. 2020. "Plant Elongator—Protein Complex of Diverse Activities Regulates Growth, Development, and Immune Responses" International Journal of Molecular Sciences 21, no. 18: 6912. https://doi.org/10.3390/ijms21186912
APA StyleJarosz, M., Van Lijsebettens, M., & Woloszynska, M. (2020). Plant Elongator—Protein Complex of Diverse Activities Regulates Growth, Development, and Immune Responses. International Journal of Molecular Sciences, 21(18), 6912. https://doi.org/10.3390/ijms21186912




