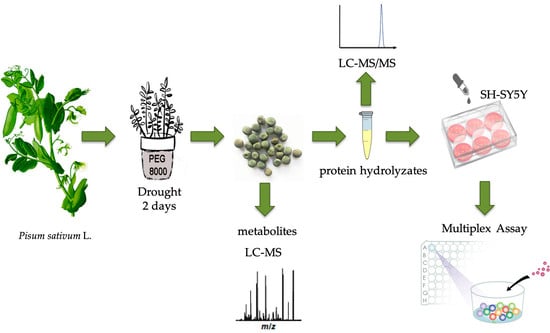
Does Protein Glycation Impact on the Drought-Related Changes in Metabolism and Nutritional Properties of Mature Pea (Pisum sativum L.) Seeds?
Abstract
1. Introduction
2. Results
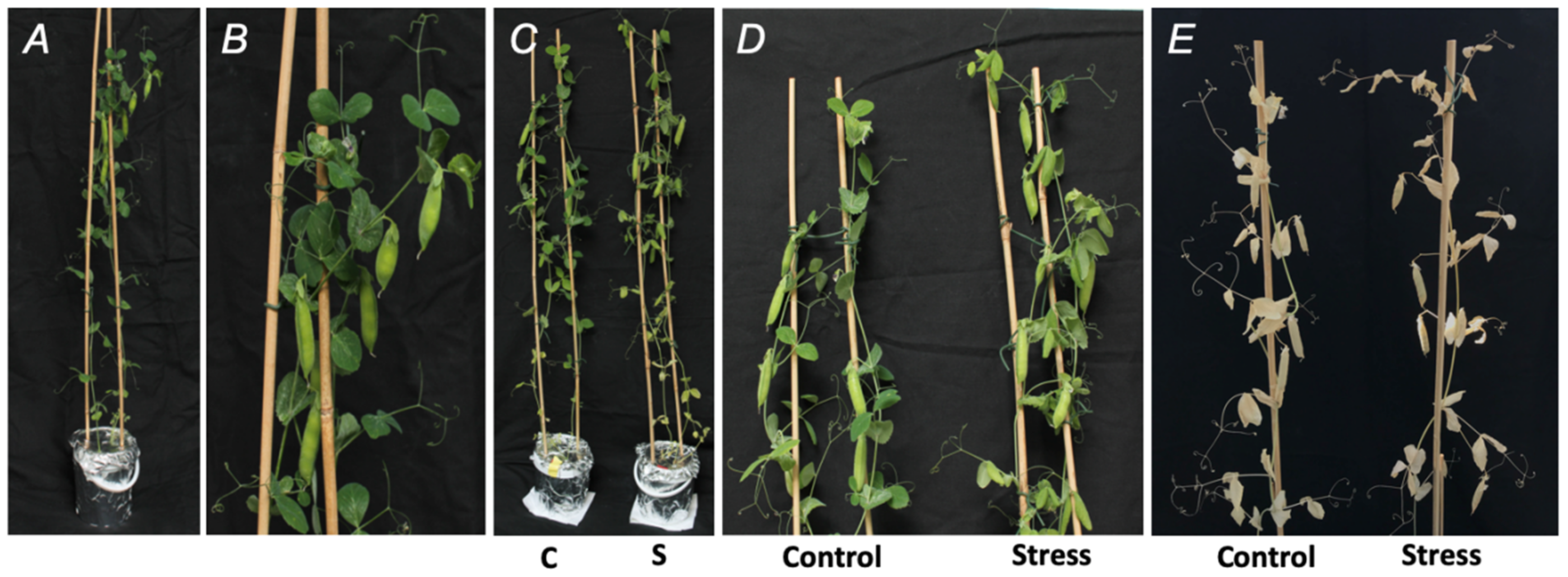
2.1. Establishment of Drought Stress and Characterization of Plant Stress Response
2.2. Assessment of Seed Quality
2.3. Metabolomics Analysis of Pea Seeds
2.4. Protein Extraction and Exhaustive Enzymatic Hydrolysis
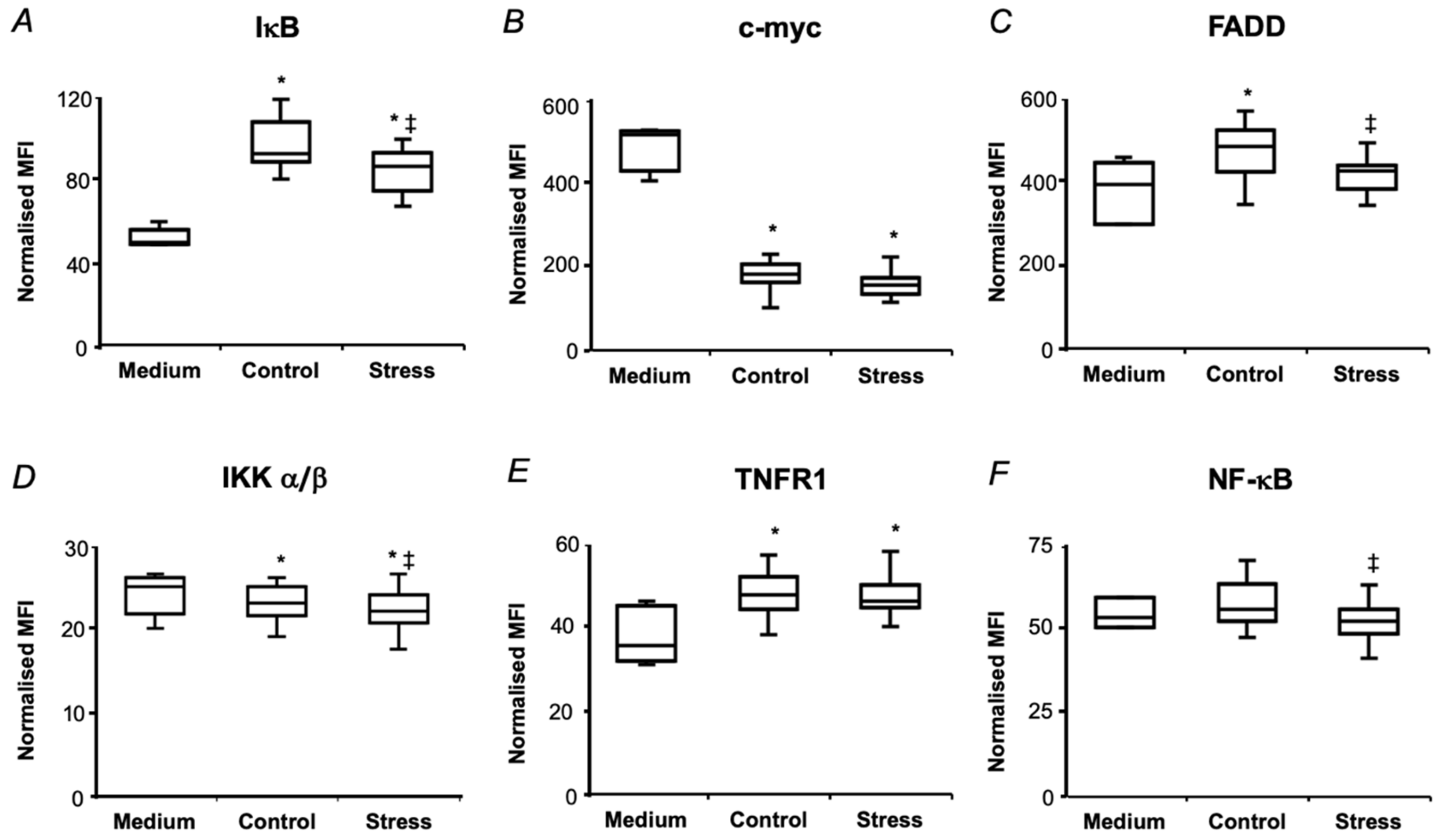
2.5. In Vitro Biological Effects of Pea Seed Proteins
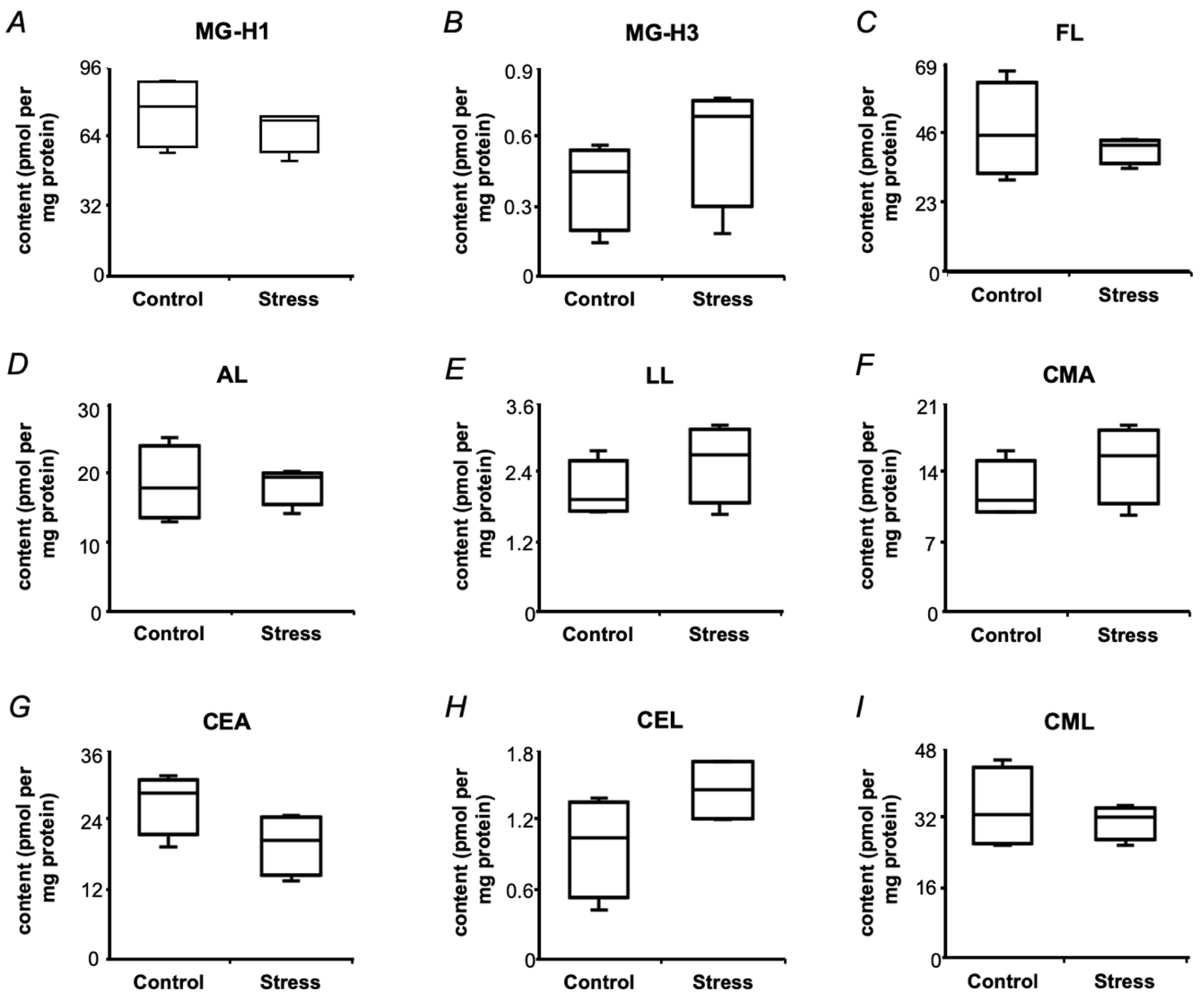
2.6. Protein-Bound Glycation Adducts in Pea Seeds
3. Discussion
3.1. Effect of Experimental Drought on Pea Seed Metabolome
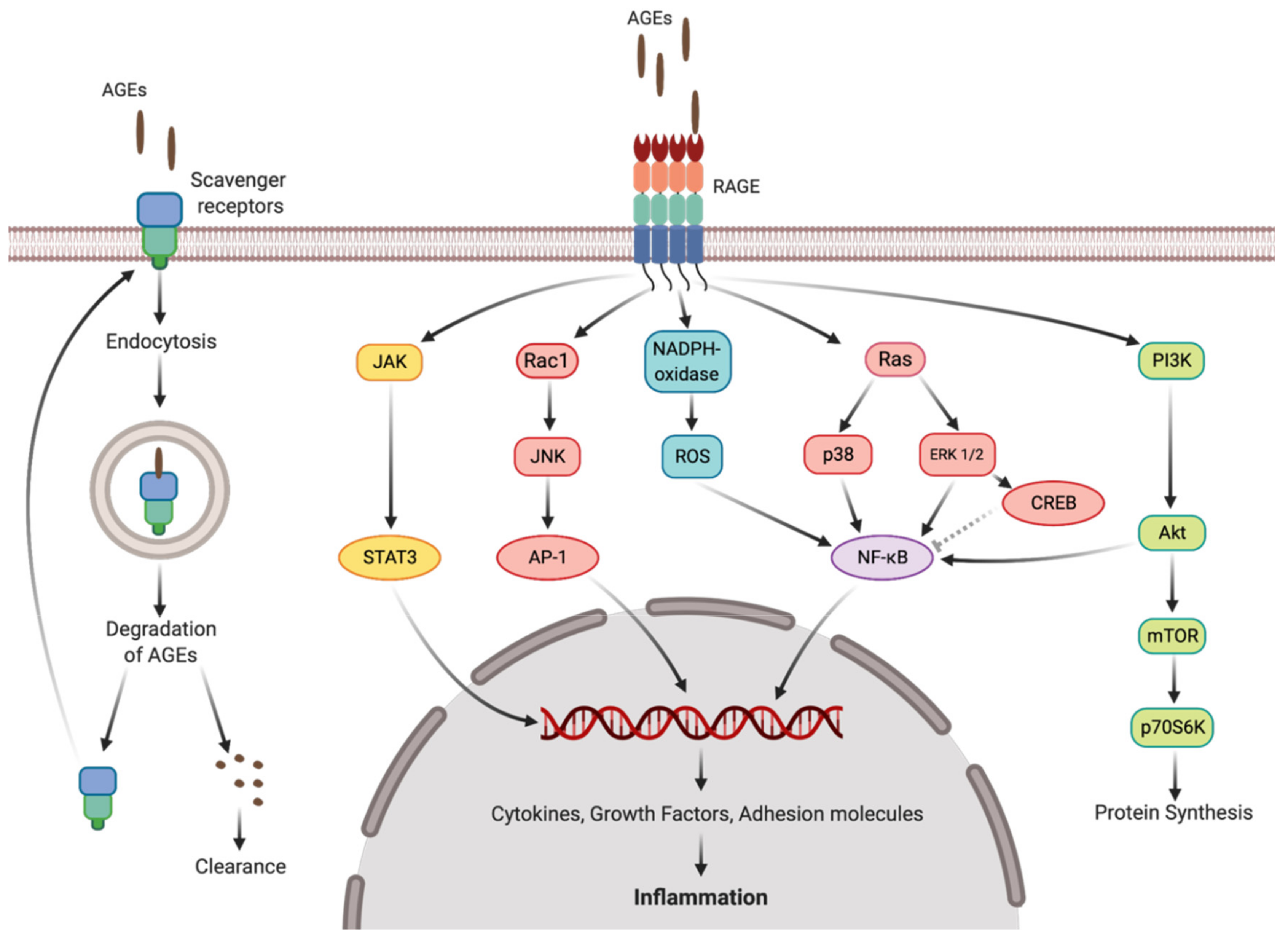
3.2. Short-Term Drought Modulates Activation of NF-kB in SH-SY5Y Human Neuroblastoma Cells by Seed Protein Hydrolyzates
4. Materials and Methods
4.1. Reagents, Plant Material and Rhizobial Culture
4.2. Plant Experiments
4.3. Determination of Water Potential
4.4. Physiological and Biochemical Assays
4.5. Assessment of Seed Quality
4.6. Metabolite Analysis
4.7. Protein Isolation
4.8. Exhaustive Enzymatic Hydrolysis
4.9. Quantitative Determination of Glycated Adducts
4.10. Cell Culture and Treatment with Protein Hydrolysates
4.11. Analysis of Signaling Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3-DG | 3-Deoxyglucosone |
| ABA | Abscisic acid |
| AGEs | Advanced glycation end products |
| Akt | RAC-alpha serine/threonine-protein kinase |
| AL | Nε-(acetyl)lysine |
| AP-1 | Activator protein 1 |
| AU | Arbitrary units |
| CEA | Nδ-(carboxyethyl)arginine |
| CEL | Nε-(carboxyethyl)lysine |
| CMA | Nδ-(carboxymethyl)arginine |
| CML | Nε-(carboxymethyl)lysine |
| CREB | cAMP response element-binding protein |
| DCM | Dichloromethane |
| DHA | Dehydroascorbic acid |
| DMEM:F12 | Dulbecco’s modified Eagle’s medium:Ham’s F12 medium |
| EDTA | Ethylenediaminetetraacetic acid |
| EI | Electron ionization |
| ERK | Extracellular signal-regulated kinases |
| ESI | Electrospray ionization |
| FADD | Fas-associated protein with death domain |
| FBS | Fetal bovine serum |
| FL | Nε-(formyl)lysine |
| FDR | False discovery rate |
| GC-MS | Gas chromatography-mass spectrometry |
| GMD | Golm Metabolome Database |
| GO | Glyoxal |
| HCA | Hierarchical cluster analysis |
| HMDB | Human Metabolome Database |
| IκB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor |
| IKK α/β | Inhibitor of nuclear factor kappa-B kinase subunits alpha/beta |
| IP-RP-UHPLC | Ion pair reversed phase-ultra-high-performance liquid chromatography |
| JAK | Janus kinase |
| JNK | c-Jun N-terminal kinases |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LL | Nε-(lactoyl)lysine |
| LRWC | Leaf relative water content |
| MAPK | Mitogen-activated protein kinase |
| MDA | Malondialdehyde |
| MES | 2-(N-morpholino)ethansulfonic acid |
| MFI | Median fluorescent intensity |
| MG-H1 | Methylglyoxal-derived hydroimidazolone 1, Nδ-(5-hydro-5-methyl-4-imidazolon-2-yl)ornithine |
| MG-H3 | Methyl lyoxal-derived hydroimidazolone 3, Nδ-(2-hydro-5-methyl-4-imidazolon-2-yl)ornithine |
| MGO | Methylglyoxal |
| MOA | Methoxyamine hydrochloride |
| MRM | Multiple reaction monitoring |
| MSTFA | N-methyl-N-(trimethylsilyl)trifluoroacetamide |
| mTOR | Mammalian target of rapamycin |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NIST | National Institute of Standards and Technology |
| p70S6K | Ribosomal protein S6 kinase |
| PBS | Phosphate buffered saline |
| PEG | Polyethylene glycol |
| PI3K | Phosphoinositide 3-kinases |
| PMSF | Phenylmethylsulfonyl fluoride |
| PLS-DA | Partial least squares discriminant analysis |
| PS II | Photosystem II |
| PVDF | Polyvinylidene fluoride |
| QqQ | Triple quadrupole |
| RAGE | Receptors for advanced glycation end-products |
| RSD | Relative standard deviation |
| ROS | Reactive oxygen species |
| SC | Stomatal conductivity |
| SDS | Sodium dodecyl sulfate |
| SDS-PAGE | Polyacrylamide gel electrophoresis in sodium dodecyl sulfate |
| SPE | Solid phase extraction |
| STAT | Signal transducer and activator of transcription |
| SWATH | Sequential window acquisition of all theoretical mass spectra |
| TBA | Thiobarbituric acid |
| TNFR1 | Tumor necrosis factor receptor 1 |
| XIC | Extracted ion chromatograms |
References
- Milkovska-Stamenova, S.; Schmidt, R.; Frolov, A.; Birkemeyer, C. GC-MS Method for the Quantitation of Carbohydrate Intermediates in Glycation Systems. J. Agric. Food Chem. 2015, 63, 5911–5919. [Google Scholar] [CrossRef] [PubMed]
- Hodge, J.E. The Amadori Rearrangement. Adv. Carbohydr. Chem. 1955, 10, 169–205. [Google Scholar] [PubMed]
- Heyns, K.; Noack, H. Die Umsetzung von D-Fructose mit L-Lysin und L-Arginin und deren Beziehung zu nichtenzymatischen Bräunungsreaktionen. Chem. Ber. 1961, 95, 720–727. [Google Scholar] [CrossRef]
- Kuzan, A.; Michel, O.; Gamian, A. Glycation of Matrix Proteins in the Artery Inhibits Migration of Smooth Muscle Cells from the Media to the Intima. Folia Biol. (Praha) 2017, 63, 105–114. [Google Scholar]
- Smith, B.A.; Mottishaw, C.R.; Hendricks, A.J.; Mitchell, J.; Becker, S.; Ropski, P.S.; Park, B.; Finkbeiner-Caufield, M.; Garay-Nontol, B.; Holman, R.W.; et al. Potential roles of inorganic phosphate on the progression of initially bound glucopyranose toward the nonenzymatic glycation of human hemoglobin: Mechanistic diversity and impacts on site selectivity. Cogent Biol. 2018, 4, 1425196. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, H.; Hanke, T.; Frolov, A.; Langrock, T.; Hoffmann, R.; Fischer, C.; Schwarzenbolz, U.; Henle, T.; Born, R.; Worch, H. Modification of collagen in vitro with respect to formation of Nε-carboxymethyllysine. Int. J. Biol. Macromol. 2009, 44, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, M.; Frolov, A.; Hoffmann, R. Fragmentation behavior of Amadori-peptides obtained by non-enzymatic glycosylation of lysine residues with ADP-ribose in tandem mass spectrometry. J. Mass Spectrom. 2010, 45, 664–669. [Google Scholar] [CrossRef]
- Wells-Knecht, K.J.; Zyzak, D.V.; Litchfield, J.E.; Thorpe, S.R.; Baynes, J.W. Mechanism of Autoxidative Glycosylation: Identification of Glyoxal and Arabinose as Intermediates in the Autoxidative Modification of Proteins by Glucose. Biochemistry 1995, 34, 3702–3709. [Google Scholar] [CrossRef]
- Soboleva, A.; Schmidt, R.; Vikhnina, M.; Grishina, T.; Frolov, A. Maillard proteomics: Opening new pages. Int. J. Mol. Sci. 2017, 18, 2677. [Google Scholar] [CrossRef]
- Münch, G.; Schinzel, R.; Loske, C.; Wong, A.; Durany, N.; Li, J.J.; Vlassara, H.; Smith, M.A.; Perry, G.; Riederer, P. Alzheimer’s disease-Synergistic effects of glucose deficit, oxidative stress and advanced glycation endproducts. J. Neural Transm. 1998, 105, 439–461. [Google Scholar] [CrossRef]
- Wolff, S.P.; Dean, R.T. Glucose autoxidation and protein modification. The potential role of “autoxidative glycosylation” in diabetes. Biochem. J. 1987, 245, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Semchyshyn, H.M. Fructation in vivo: Detrimental and protective effects of fructose. BioMed. Res. Int. 2013, 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Langborg, A.; Minhas, H.S. Formation of glyoxal, methylglyoxal and 8-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 1999, 344, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Dhar, A.; Desai, K.; Kazachmov, M.; Yu, P.; Wu, L. Methylglyoxal production in vascular smooth muscle cells from different metabolic precursors. Metabolism. 2008, 57, 1211–1220. [Google Scholar] [CrossRef]
- Kalapos, M.P. Methylglyoxal in living organisms: Chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 1999, 110, 145–175. [Google Scholar] [CrossRef]
- Nowotny, K.; Jung, T.; Höhn, A.; Weber, D.; Grune, T. Advanced glycation end products and oxidative stress in type 2 diabetes mellitus. Biomolecules 2015, 5, 194–222. [Google Scholar] [CrossRef]
- Boon Li Pun, P.; Murphy, M.P. Pathological Significance of Mitochondrial Glycation. Int. J. Cell Biol. 2012, 2012, 13. [Google Scholar]
- Nowotny, K.; Schröter, D.; Schreiner, M.; Grune, T. Dietary advanced glycation end products and their relevance for human health. Ageing Res. Rev. 2018, 47, 55–66. [Google Scholar] [CrossRef]
- Yamagishi, S.; Nakamura, N.; Matsui, T. Glycation and cardiovascular disease in diabetes: A perspective on the concept of metabolic memory. J. Diabetes Investig. 2017, 9, 141–148. [Google Scholar] [CrossRef]
- Prasad, K. AGE–RAGE stress: A changing landscape in pathology and treatment of Alzheimer’s disease. Mol. Cell. Biochem. 2019, 459, 95–112. [Google Scholar] [CrossRef]
- Konig, A.; Miranda, H.V.; Outeiro, T.F. Alpha-synuclein glycation and the action of anti-diabetic agents in Parkinson’s disease. J. Parkinson’s Dis. 2018, 8, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Fournet, M.; Bonté, F.; Desmoulière, A. Glycation damage: A possible hub for major pathophysiological disorders and aging. Aging Dis. 2018, 9, 880–900. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, U.; Rabbani, N.; Mullineaux, P.M.; Thornalley, P.J. Quantitative Measurement of Specific Biomarkers for Protein Oxidation, Nitration and Glycation in Arabidopsis Leaves. Plant J. 2009, 59, 661–671. [Google Scholar] [CrossRef]
- Bilova, T.; Lukasheva, E.; Brauch, D.; Greifenhagen, U.; Paudel, G.; Tarakhovskaya, E.; Frolova, N.; Mittasch, J.; Balcke, G.U.; Tissier, A.; et al. A Snapshot of the Plant Glycated Proteome. Structural, Functional, and Mechanistic Aspects. J. Biol. Chem. 2016, 291, 7621–7636. [Google Scholar] [CrossRef] [PubMed]
- Bilova, T.; Greifenhagen, U.; Paudel, G.; Lukasheva, E.; Brauch, D.; Osmolovskaya, N.; Tarakhovskaya, E.; Balcke, G.U.; Tissier, A.; Vogt, T.; et al. Glycation of Plant Proteins under Environmental Stress—Methodological Approaches, Potential Mechanisms and Biological Role. In Abiotic and Biotic Stress in Plants-Recent Advances and Future Perspectives; IntechOpen: London, UK, 2016; pp. 295–316. [Google Scholar]
- Bilova, T.; Paudel, G.; Shilyaev, N.; Schmidt, R.; Brauch, D.; Milrud, S.; Smolikova, G.; Tissier, A.; Vogt, T.; Brandt, W.; et al. Global proteomic analysis of advanced glycation end products in the arabidopsis proteome provides evidence for age-related glycation hotspots. J. Biol. Chem. 2017, 292, 15758–15776. [Google Scholar] [CrossRef] [PubMed]
- Matamoros, M.A.; Kim, A.; Peñuelas, M.; Ihling, C.; Griesser, E.; Hoffmann, R.; Fedorova, M.; Frolov, A.; Becana, M. Protein carbonylation and glycation in legume nodules. Plant Physiol. 2018, 177, 1510–1528. [Google Scholar] [CrossRef]
- Chaplin, A.K.; Chernukhin, I.; Bechtold, U. Profiling of advanced glycation end products uncovers abiotic stress-specific target proteins in Arabidopsis. J. Exp. Bot. 2019, 70, 653–670. [Google Scholar] [CrossRef]
- Paudel, G.; Bilova, T.; Schmidt, R.; Greifenhagen, U.; Berger, R.; Tarakhovskaya, E.; Stöckhardt, S.; Balcke, G.U.; Humbeck, K.; Brandt, W.; et al. Osmotic stress is accompanied by protein glycation in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6283–6295. [Google Scholar] [CrossRef]
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef]
- Frolov, A.; Bilova, T.; Paudel, G.; Berger, R.; Balcke, U.G.; Birkemeyer, C.; Wessjohann, L.A. Early responses of mature Arabidopsis thaliana plants to reduced water potential in the agar-based polyethylene glycol infusion drought model. J. Plant Physiol. 2016, 208, 70–83. [Google Scholar] [CrossRef]
- Kar, R.K. Plant responses to water stress: Role of reactive oxygen species. Plant Signal. Behav. 2011, 6, 1741–1745. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Gupta, P.; Khan, F.; Singh, S.K.; Mishra, T.; Kumar, A.; Dhawan, S.S.; Shirke, P.A. Modulations in primary and secondary metabolic pathways and adjustment in physiological behaviour of Withania somnifera under drought stress. Plant Sci. 2018, 272, 42–54. [Google Scholar] [CrossRef]
- Rohn, S. Possibilities and limitations in the analysis of covalent interactions between phenolic compounds and proteins. Food Res. Int. 2014, 65, 13–19. [Google Scholar] [CrossRef]
- Xia, Q.; Zhao, Y.; Lin, G.; Beland, F.A.; Cai, L.; Fu, P.P. Pyrrolizidine Alkaloid-Protein Adducts: Potential Non-invasive Biomarkers of Pyrrolizidine Alkaloid-Induced Liver Toxicity and Exposure. Chem. Res. Toxicol. 2016, 29, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, G.; Benezra, C.; Schlewer, G.; Stampf, J.L. Allergic contact dermatitis to α-methylene-γ-butyrolactones: Preparation of alantolactone-protein conjugates and induction of contact sensitivity in the guinea pig by an alantolactone-skin protein conjugate. Mol. Immunol. 1980, 17, 1045–1051. [Google Scholar] [CrossRef]
- Nikolova, G.; Karamalakova, Y.; Gadjeva, V. Reducing oxidative toxicity of L-dopa in combination with two different antioxidants: An essential oil isolated from Rosa Damascena Mill., and vitamin C. Toxicol. Rep. 2019, 6, 267–271. [Google Scholar] [CrossRef]
- Krajcovicoa-Kudlackova, M.; Šebeková, K.; Schinzel, R.; Klvanová, J. Advanced Glycation End Products and Nutrition. Physiol. Res. 2002, 51, 313–316. [Google Scholar]
- Babu, M.; Nagaraja, B.; Reddy, K.B.; Reddy, B. Prevalence and distribution of atherosclerosis in different age groups in Tirupati area AP-Autopsy based study. Pharma Innov. J. 2015, 5, 87–91. [Google Scholar]
- Hellwig, M.; Geissler, S.; Matthes, R.; Peto, A.; Silow, C.; Brandsch, M.; Henle, T. Transport of Free and Peptide-Bound Glycated Amino Acids: Synthesis, Transepithelial Flux at Caco-2 Cell Monolayers, and Interaction with Apical Membrane Transport Proteins. ChemBioChem 2011, 12, 1270–1279. [Google Scholar] [CrossRef]
- Hellwig, M.; Matthes, R.; Peto, A.; Löbner, J.; Henle, T. N-ε-fructosyllysine and N-ε-carboxymethyllysine, but not lysinoalanine, are available for absorption after simulated gastrointestinal digestion. Amino Acids 2014, 46, 289–299. [Google Scholar] [CrossRef]
- Smuda, M.; Henning, C.; Raghavan, C.T.; Johar, K.; Vasavada, A.R.; Nagaraj, R.H.; Glomb, M.A. Comprehensive Analysis of Maillard Protein Modifications in Human Lenses: Effect of Age and Cataract. Biochemistry 2015, 54, 2500–2507. [Google Scholar] [CrossRef] [PubMed]
- Antonova, K.; Vikhnina, M.; Soboleva, A.; Mehmood, T.; Heymich, M.-L.; Leonova, T.; Bankin, M.; Lukasheva, E.; Gensberger-Reigl, S.; Medvedev, S.; et al. Analysis of Chemically Labile Glycation Adducts in Seed Proteins: Case Study of Methylglyoxal-Derived Hydroimidazolone 1 (MG-H1). Int. J. Mol. Sci. 2019, 20, 3659. [Google Scholar] [CrossRef] [PubMed]
- Mamontova, T.; Afonin, A.M.; Ihling, C.; Soboleva, A.; Lukasheva, E.; Sulima, A.S.; Shtark, O.Y.; Akhtemova, G.A.; Povydysh, M.N.; Sinz, A.; et al. Profiling of seed proteome in pea (pisum sativum l.) lines characterized with high and low responsivity to combined inoculation with nodule bacteria and arbuscular mycorrhizal fungi. Molecules 2019, 24, 1603. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.W.; Hedegaard, R.V.; Andersen, J.M.; Courten, B.D.; Bügel, S.; Nielsen, J.; Skibsted, L.H.; Dragsted, L.O. Advanced glycation endproducts in food and their effects on health. Food Chem. Toxicol. 2013, 60, 10–37. [Google Scholar] [CrossRef]
- Shu, L.; Lou, Q.; Ma, C.; Ding, W.; Zhou, J.; Wu, J.; Feng, F.; Lu, X.; Luo, L.; Xu, G.; et al. Genetic, proteomic and metabolic analysis of the regulation of energy storage in rice seedlings in response to drought. Proteomics 2011, 11, 4122–4138. [Google Scholar] [CrossRef]
- Kosová, K.; Urban, M.O.; Vítámvás, P.; Prášil, I.T. Drought stress response in common wheat, durum wheat, and barley: Transcriptomics, proteomics, metabolomics, physiology, and breeding for an enhanced drought tolerance. In Drought Stress Tolerance in Plants, Vol 2: Molecular and Genetic Perspectives; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 277–314. ISBN 9783319324234. [Google Scholar]
- Das, A.; Rushton, P.; Rohila, J. Metabolomic Profiling of Soybeans (Glycine max L.) Reveals the Importance of Sugar and Nitrogen Metabolism under Drought and Heat Stress. Plants 2017, 6, 21. [Google Scholar] [CrossRef]
- Hatzig, S.V.; Nuppenau, J.-N.; Snowdon, R.J.; Schießl, S.V. Drought stress has transgenerational effects on seeds and seedlings in winter oilseed rape (Brassica napus L.). BMC Plant Biol. 2018, 18, 297. [Google Scholar] [CrossRef]
- Nakagawa, A.C.S.; Itoyama, H.; Ariyoshi, Y.; Ario, N.; Tomita, Y.; Kondo, Y.; Iwaya-Inoue, M.; Ishibashi, Y. Drought stress during soybean seed filling affects storage compounds through regulation of lipid and protein metabolism. Acta Physiol. Plant. 2018, 40, 111. [Google Scholar] [CrossRef]
- Refsgaard, H.H.F.; Tsai, L.; Stadtman, E.R. Modifications of proteins by polyunsaturated fatty acid peroxidation products. Proc. Natl. Acad. Sci. USA 2000, 97, 611–616. [Google Scholar] [CrossRef]
- Vistoli, G.; De Maddis, D.; Cipak, A.; Zarkovic, N.; Carini, M.; Aldini, G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): An overview of their mechanisms of formation. Free Radic. Res. 2013, 47, 3–27. [Google Scholar] [CrossRef]
- Pham, L.B.; Wang, B.; Zisu, B.; Adhikari, B. Covalent modification of flaxseed protein isolate by phenolic compounds and the structure and functional properties of the adducts. Food Chem. 2019, 293, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Sun, C.; Yang, W.; Yuan, F.; Gao, Y. Structural characterization and functional evaluation of lactoferrin-polyphenol conjugates formed by free-radical graft copolymerization. RSC Adv. 2015, 5, 15641–15651. [Google Scholar] [CrossRef]
- Tang, C.B.; Zhang, W.G.; Zou, Y.F.; Xing, L.J.; Zheng, H.B.; Xu, X.L.; Zhou, G.H. Influence of RosA-protein adducts formation on myofibrillar protein gelation properties under oxidative stress. Food Hydrocoll. 2017, 67, 197–205. [Google Scholar] [CrossRef]
- Liu, W.; Cai, A.; Carley, R.; Rocchio, R.; Petrovas, Z.M.; Chartier, C.A.; Meng, X.; Su, J.; Cho, B.P.; Dain, J.A.; et al. Bioactive anthraquinones found in plant foods interact with human serum albumin and inhibit the formation of advanced glycation endproducts. J. Food Bioact. 2018, 4, 130–138. [Google Scholar] [CrossRef]
- Mann, R.K.; Beachy, P.A. Cholesterol modification of proteins. Biochim. Biophys. Acta 2000, 1529, 188–202. [Google Scholar] [CrossRef]
- Ott, C.; Jacobs, K.; Haucke, E.; Navarrete Santos, A.; Grune, T.; Simm, A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014, 2, 411–429. [Google Scholar] [CrossRef]
- Rojas, A.; Delgado-Lopez, F.; Gonzalez, I.; Perez-Castro, R.; Romero, J.; Rojas, I. The receptor for advanced glycation end-products: A complex signaling scenario for a promiscuous receptor. Cell. Signal. 2013, 25, 609–614. [Google Scholar] [CrossRef]
- Kellow, N.J.; Coughlan, M.T. Effect of diet-derived advanced glycation end products on inflammation. Nutr. Rev. 2015, 73, 737–759. [Google Scholar] [CrossRef]
- Wang, X.; Yu, S.; Wang, C.; Wang, Y. Advanced glycation end products induce oxidative stress and mitochondrial dysfunction in SH-SY5Y cells. Vitr. Cell. Dev. Biol. Anim. 2015, 51, 204–209. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Yu, H.-T.; Pu, X.-P.; Du, G.-H. Myricitrin Alleviates Methylglyoxal-Induced Mitochondrial Dysfunction and AGEs / RAGE / NF- κ B Pathway Activation in SH-SY5Y Cells. J. Mol. Neurosci. 2014, 53, 562–570. [Google Scholar] [CrossRef]
- Yin, Q.Q.; Dong, C.F.; Dong, S.Q.; Dong, X.L.; Hong, Y.; Hou, X.Y.; Luo, D.Z.; Pei, J.J.; Liu, X.P. AGEs induce cell death via oxidative and endoplasmic reticulum stresses in both human SH-SY5Y neuroblastoma cells and rat cortical neurons. Cell. Mol. Neurobiol. 2012, 32, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Palanissami, G.; Paul, S.F.D. RAGE and Its Ligands: Molecular Interplay Between Glycation, Inflammation, and Hallmarks of Cancer—A Review. Horm. Cancer 2018, 9, 295–325. [Google Scholar] [CrossRef] [PubMed]
- Buitimea-Cantúa, N.E.; Gutiérrez-Uribe, J.A.; Serna-Saldívar, S.O. Phenolic-Protein Interactions: Effects on Food Properties and Health Benefits. J. Med. Food 2018, 21, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Wen, A.Y.; Sakamoto, K.M.; Miller, L.S. The Role of the Transcription Factor CREB in Immune Function. J. Immunol. 2010, 185, 6413–6419. [Google Scholar] [CrossRef] [PubMed]
- Greifenhagen, U.; Frolov, A.; Hoffmann, R. Oxidative degradation of N ε-fructosylamine-substituted peptides in heated aqueous systems. Amino Acids 2015, 47, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Frolov, A.; Schmidt, R.; Spiller, S.; Greifenhagen, U.; Hoffmann, R. Arginine-derived advanced glycation end products generated in peptide-glucose mixtures during boiling. J. Agric. Food Chem. 2014, 62, 3626–3635. [Google Scholar] [CrossRef]
- Henle, T. Dietary advanced glycation end products - A risk to human health? a call for an interdisciplinary debate. Mol. Nutr. Food Res. 2007, 51, 1075–1078. [Google Scholar] [CrossRef]
- Johansen, J.T.; Breddam, K.; Ottesen, M. Isolation of carboxypeptidase Y by affinity chromatography. Carlsberg Res. Commun. 1976, 41, 1–14. [Google Scholar] [CrossRef]
- Money, N.P. Osmotic Pressure of Aqueous Polyethylene Glycols. Plant Physiol. 1989, 91, 766–769. [Google Scholar] [CrossRef]
- Morison, J.I.L. Increasing atmospheric CO2 and stomata. New Phytol. 2001, 149, 154–156. [Google Scholar] [CrossRef]
- Silva, M.D.A.; Jifon, J.L.; Da Silva, J.A.G.; Sharma, V. Use of physiological parameters as fast tools to screen for drought tolerance in sugarcane. Braz. J. Plant Physiol. 2007, 19, 193–201. [Google Scholar] [CrossRef]
- Balcke, G.U.; Handrick, V.; Bergau, N.; Fichtner, M.; Henning, A.; Stellmach, H.; Tissier, A.; Hause, B.; Frolov, A. An UPLC-MS / MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 2012, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hampton, J.M.; Tekrony, D.M. Handbook of Vigour Test. Methods, 3rd ed.; ISTA: Zurich, Switzerland; pp. 1–117.
- Chantseva, V.; Bilova, T.; Smolikova, G.; Frolov, A.; Medvedev, S. 3D-clinorotation induces specific alterations in metabolite profiles of germinating Brassica napus L. seeds. Biol. Commun. 2019, 64, 55–74. [Google Scholar] [CrossRef]
- Balcke, G.U.; Bennewitz, S.; Bergau, N.; Athmer, B.; Henning, A.; Majovsky, P.; Jiménez-Gómez, J.M.; Hoehenwarter, W.; Tissier, A. Multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 2017, 29, 960–983. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinforma. 2019, 68, e86. [Google Scholar] [CrossRef]
- Treutler, H.; Tsugawa, H.; Porzel, A.; Gorzolka, K.; Tissier, A.; Neumann, S.; Balcke, G.U. Discovering Regulated Metabolite Families in Untargeted Metabolomics Studies. Anal. Chem. 2016, 88, 8082–8090. [Google Scholar] [CrossRef]
- Goeman, J.J.; Buhlmann, P. Analyzing gene expression data in terms of gene sets: Methodological issues. Bioinformatics 2007, 23, 980–987. [Google Scholar] [CrossRef]
- Isaacson, T.; Damasceno, C.M.B.; Saravanan, R.S.; He, Y.; Catala, C.; Rose, J.K.C. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef]
- Greifenhagen, U.; Frolov, A.; Blüher, M.; Hoffmann, R. Plasma proteins modified by advanced glycation end products (AGEs) reveal site-specific susceptibilities to glycemic control in patients with type 2 diabetes. J. Biol. Chem. 2016, 291, 9610–9616. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]




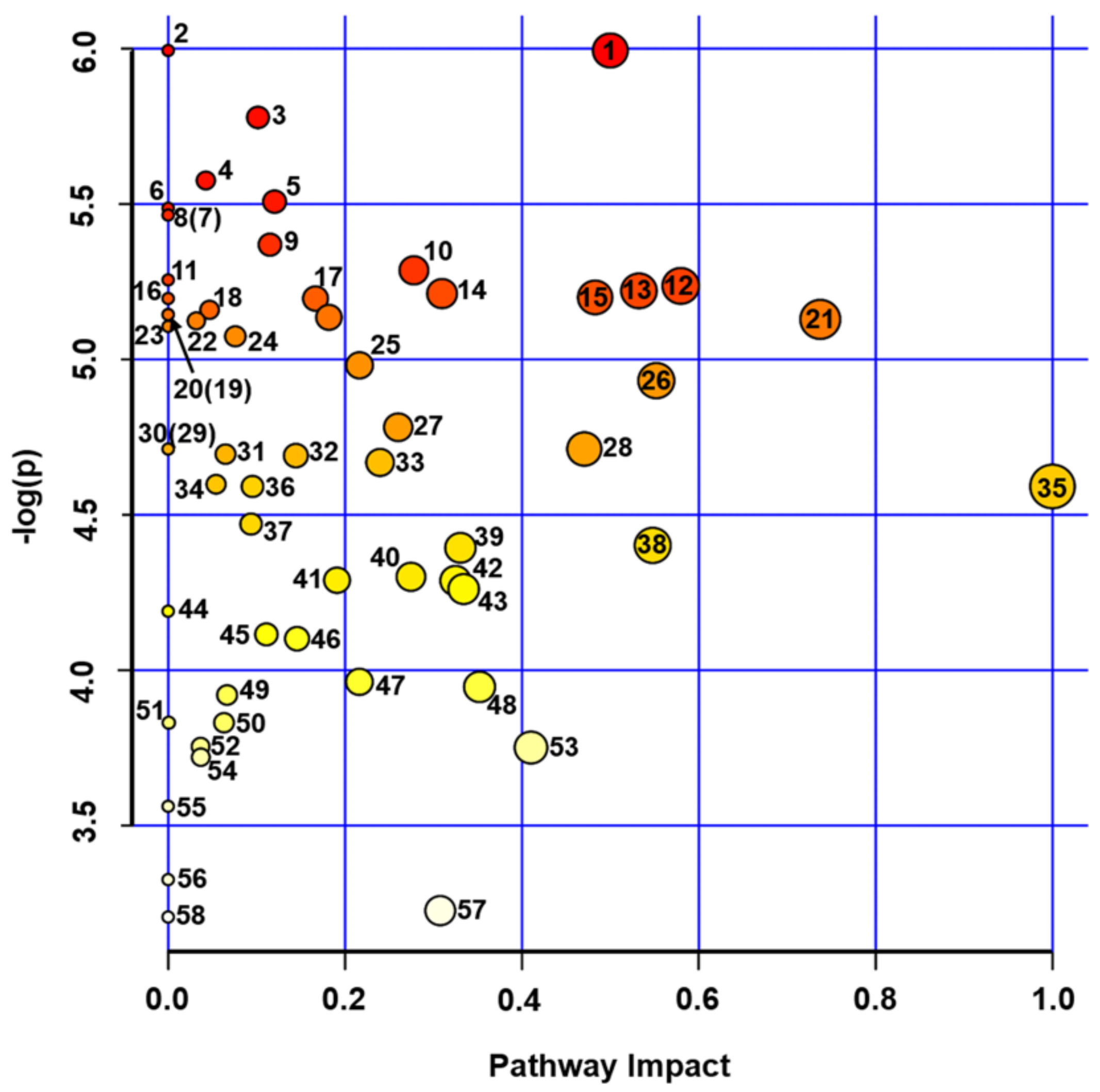

| # a | Metabolic Pathway b | Total Cmpd c | Hits d | Raw p e | -LOG (p) | Holm Adjust f | FDR g | Impact h |
|---|---|---|---|---|---|---|---|---|
| 1 | Isoquinoline alkaloid biosynthesis | 6 | 1 | 2.49 × 10−3 | 5.99 | 1.47 × 10−1 | 1.48 × 10−2 | 0.5 |
| 2 | Ubiquinone and other terpenoid-quinone biosynthesis | 38 | 1 | 2.49 × 10−3 | 5.99 | 1.47 × 10−1 | 1.48 × 10−2 | 0 |
| 3 | Phenylalanine, tyrosine and tryptophan biosynthesis | 22 | 6 | 3.09 × 10−3 | 5.78 | 1.76 × 10−1 | 1.48 × 10−2 | 0.1 |
| 4 | Histidine metabolism | 15 | 2 | 3.79 × 10−3 | 5.58 | 2.12 × 10−1 | 1.48 × 10−2 | 0.04 |
| 5 | Tryptophan metabolism | 28 | 2 | 4.06 × 10−3 | 5.51 | 2.23 × 10−1 | 1.48 × 10−2 | 0.12 |
| 6 | Indole alkaloid biosynthesis | 4 | 1 | 4.14 × 10−3 | 5.49 | 2.24 × 10−1 | 1.48 × 10−2 | 0 |
| 7 | Sphingolipid metabolism | 17 | 1 | 4.24 × 10−3 | 5.46 | 2.25 × 10−1 | 1.48 × 10−2 | 0 |
| 8 | Sulfur metabolism | 15 | 1 | 4.24 × 10−3 | 5.46 | 2.25 × 10−1 | 1.48 × 10−2 | 0 |
| 9 | Glycerophospholipid metabolism | 37 | 2 | 4.66 × 10−3 | 5.37 | 2.38 × 10−1 | 1.48 × 10−2 | 0.12 |
| 10 | Arginine and proline metabolism | 34 | 3 | 5.06 × 10−3 | 5.29 | 2.53 × 10−1 | 1.48 × 10−2 | 0.28 |
| 11 | Cyanoamino acid metabolism | 29 | 5 | 5.22 × 10−3 | 5.26 | 2.56 × 10−1 | 1.48 × 10−2 | 0 |
| 12 | Glycine, serine and threonine metabolism | 33 | 6 | 5.32 × 10−3 | 5.24 | 2.56 × 10−1 | 1.48 × 10−2 | 0.58 |
| 13 | Pyrimidine metabolism | 38 | 16 | 5.41 × 10−3 | 5.22 | 2.56 × 10−1 | 1.48 × 10−2 | 0.53 |
| 14 | Arginine biosynthesis | 18 | 7 | 5.45 × 10−3 | 5.21 | 2.56 × 10−1 | 1.48 × 10−2 | 0.31 |
| 15 | Nicotinate and nicotinamide metabolism | 13 | 3 | 5.52 × 10−3 | 5.20 | 2.56 × 10−1 | 1.48 × 10−2 | 0.48 |
| 16 | Monobactam biosynthesis | 8 | 1 | 5.54 × 10−3 | 5.20 | 2.56 × 10−1 | 1.48 × 10−2 | 0 |
| 17 | beta-Alanine metabolism | 18 | 3 | 5.54 × 10−3 | 5.20 | 2.56 × 10−1 | 1.48 × 10−2 | 0.17 |
| 18 | Carbon fixation in photosynthetic organisms | 21 | 4 | 5.75 × 10−3 | 5.16 | 2.56 × 10−1 | 1.48 × 10−2 | 0.05 |
| 19 | Porphyrin and chlorophyll metabolism | 48 | 1 | 5.83 × 10−3 | 5.14 | 2.56 × 10−1 | 1.48 × 10−2 | 0 |
| 20 | Nitrogen metabolism | 12 | 2 | 5.84 × 10−3 | 5.14 | 2.56 × 10−1 | 1.48 × 10−2 | 0 |
| 21 | Butanoate metabolism | 17 | 4 | 5.89 × 10−3 | 5.13 | 2.56 × 10−1 | 1.48 × 10−2 | 0.18 |
| 22 | Alanine, aspartate and glutamate metabolism | 22 | 9 | 5.93 × 10−3 | 5.13 | 2.56 × 10−1 | 1.48 × 10−2 | 0.74 |
| 23 | Cysteine and methionine metabolism | 46 | 4 | 5.95 × 10−3 | 5.12 | 2.56 × 10−1 | 1.48 × 10−2 | 0.03 |
| 24 | Glucosinolate biosynthesis | 65 | 2 | 6.06 × 10−3 | 5.11 | 2.56 × 10−1 | 1.48 × 10−2 | 0 |
| 25 | Folate biosynthesis | 27 | 1 | 6.26 × 10−3 | 5.07 | 2.56 × 10−1 | 1.48 × 10−2 | 0.08 |
| 26 | Ascorbate and aldarate metabolism | 18 | 3 | 6.87 × 10−3 | 4.98 | 2.56 × 10−1 | 1.56 × 10−2 | 0.22 |
| 27 | Glutathione metabolism | 26 | 6 | 7.22 × 10−3 | 4.93 | 2.56 × 10−1 | 1.58 × 10−2 | 0.55 |
| 28 | Terpenoid backbone biosynthesis | 30 | 5 | 8.38 × 10−3 | 4.78 | 2.68 × 10−1 | 1.62 × 10−2 | 0.26 |
| 29 | Phenylalanine metabolism | 11 | 1 | 8.99 × 10−3 | 4.71 | 2.79 × 10−1 | 1.62 × 10−2 | 0.47 |
| 30 | Phenylpropanoid biosynthesis | 46 | 1 | 8.99 × 10−3 | 4.71 | 2.79 × 10−1 | 1.62 × 10−2 | 0 |
| 31 | Tropane, piperidine and pyridine alkaloid biosynthesis | 8 | 1 | 8.99 × 10−3 | 4.71 | 2.79 × 10−1 | 1.62 × 10−2 | 0 |
| 32 | Glycerolipid metabolism | 21 | 3 | 9.14 × 10−3 | 4.70 | 2.79 × 10−1 | 1.62 × 10−2 | 0.06 |
| 33 | Galactose metabolism | 27 | 8 | 9.19 × 10−3 | 4.69 | 2.79 × 10−1 | 1.62 × 10−2 | 0.14 |
| 34 | Purine metabolism | 63 | 14 | 9.39 × 10−3 | 4.67 | 2.79 × 10−1 | 1.62 × 10−2 | 0.24 |
| 35 | Zeatin biosynthesis | 21 | 5 | 1.01 × 10−2 | 4.60 | 2.79 × 10−1 | 1.62 × 10−2 | 0.05 |
| 36 | Synthesis and degradation of ketone bodies | 4 | 2 | 1.01 × 10−2 | 4.59 | 2.79 × 10−1 | 1.62 × 10−2 | 1 |
| 37 | Valine, leucine and isoleucine degradation | 37 | 2 | 1.01 × 10−2 | 4.59 | 2.79 × 10−1 | 1.62 × 10−2 | 0.1 |
| 38 | Pentose and glucuronate interconversions | 16 | 2 | 1.14 × 10−2 | 4.47 | 2.79 × 10−1 | 1.78 × 10−2 | 0.09 |
| 39 | Starch and sucrose metabolism | 22 | 6 | 1.23 × 10−2 | 4.40 | 2.79 × 10−1 | 1.82 × 10−2 | 0.55 |
| 40 | Glyoxylate and dicarboxylatemetabolism | 29 | 8 | 1.24 × 10−2 | 4.39 | 2.79 × 10−1 | 1.82 × 10−2 | 0.33 |
| 41 | Inositol phosphate metabolism | 28 | 5 | 1.36 × 10−2 | 4.30 | 2.79 × 10−1 | 1.88 × 10−2 | 0.27 |
| 42 | Phosphatidylinositol signaling system | 26 | 5 | 1.37 × 10−2 | 4.29 | 2.79 × 10−1 | 1.88 × 10−2 | 0.19 |
| 43 | Citrate cycle (TCA cycle) | 20 | 5 | 1.37 × 10−2 | 4.29 | 2.79 × 10−1 | 1.88 × 10−2 | 0.32 |
| 44 | Fatty acid degradation | 37 | 2 | 1.41 × 10−2 | 4.26 | 2.79 × 10−1 | 1.89 × 10−2 | 0.33 |
| 45 | Thiamine metabolism | 22 | 2 | 1.52 × 10−2 | 4.19 | 2.79 × 10−1 | 1.99 × 10−2 | 0 |
| 46 | Aminoacyl-tRNA biosynthesis | 46 | 13 | 1.63 × 10−2 | 4.12 | 2.79 × 10−1 | 2.08 × 10−2 | 0.11 |
| 47 | Pantothenate and biosynthesis CoA | 23 | 2 | 1.66 × 10−2 | 4.10 | 2.79 × 10−1 | 2.08 × 10−2 | 0.15 |
| 48 | Tyrosine metabolism | 16 | 2 | 1.90 × 10−2 | 3.96 | 2.79 × 10−1 | 2.33 × 10−2 | 0.22 |
| 49 | Amino sugar and nucleotide sugar metabolism | 50 | 7 | 1.93 × 10−2 | 3.95 | 2.79 × 10−1 | 2.33 × 10−2 | 0.35 |
| 50 | Riboflavin metabolism | 11 | 3 | 1.98 × 10−2 | 3.92 | 2.79 × 10−1 | 2.34 × 10−2 | 0.07 |
| 51 | Propanoate metabolism | 20 | 1 | 2.17 × 10−2 | 3.83 | 2.79 × 10−1 | 2.46 × 10−2 | 0.06 |
| 52 | Fatty acid biosynthesis | 56 | 1 | 2.17 × 10−2 | 3.83 | 2.79 × 10−1 | 2.46 × 10−2 | 0 |
| 53 | Glycolysis/Gluconeogenesis | 26 | 4 | 2.34 × 10−2 | 3.75 | 2.79 × 10−1 | 2.57 × 10−2 | 0.04 |
| 54 | Pentose phosphate pathway | 19 | 6 | 2.35 × 10−2 | 3.75 | 2.79 × 10−1 | 2.57 × 10−2 | 0.41 |
| 55 | Fructose and mannose metabolism | 20 | 1 | 2.42 × 10−2 | 3.72 | 2.79 × 10−1 | 2.60 × 10−2 | 0.04 |
| 56 | Lysine biosynthesis | 9 | 3 | 2.84 × 10−2 | 3.56 | 2.79 × 10−1 | 2.99 × 10−2 | 0 |
| 57 | Valine, leucine and isoleucine biosynthesis | 22 | 1 | 3.59 × 10−2 | 3.33 | 2.79 × 10−1 | 3.72 × 10−2 | 0 |
| 58 | Pyruvate metabolism | 22 | 4 | 3.97 × 10−2 | 3.23 | 2.79 × 10−1 | 4.04 × 10−2 | 0.31 |
| 59 | Lysine degradation | 18 | 2 | 4.05 × 10−2 | 3.21 | 2.79 × 10−1 | 4.05 × 10−2 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonova, T.; Popova, V.; Tsarev, A.; Henning, C.; Antonova, K.; Rogovskaya, N.; Vikhnina, M.; Baldensperger, T.; Soboleva, A.; Dinastia, E.; et al. Does Protein Glycation Impact on the Drought-Related Changes in Metabolism and Nutritional Properties of Mature Pea (Pisum sativum L.) Seeds? Int. J. Mol. Sci. 2020, 21, 567. https://doi.org/10.3390/ijms21020567
Leonova T, Popova V, Tsarev A, Henning C, Antonova K, Rogovskaya N, Vikhnina M, Baldensperger T, Soboleva A, Dinastia E, et al. Does Protein Glycation Impact on the Drought-Related Changes in Metabolism and Nutritional Properties of Mature Pea (Pisum sativum L.) Seeds? International Journal of Molecular Sciences. 2020; 21(2):567. https://doi.org/10.3390/ijms21020567
Chicago/Turabian StyleLeonova, Tatiana, Veronika Popova, Alexander Tsarev, Christian Henning, Kristina Antonova, Nadezhda Rogovskaya, Maria Vikhnina, Tim Baldensperger, Alena Soboleva, Ekaterina Dinastia, and et al. 2020. "Does Protein Glycation Impact on the Drought-Related Changes in Metabolism and Nutritional Properties of Mature Pea (Pisum sativum L.) Seeds?" International Journal of Molecular Sciences 21, no. 2: 567. https://doi.org/10.3390/ijms21020567
APA StyleLeonova, T., Popova, V., Tsarev, A., Henning, C., Antonova, K., Rogovskaya, N., Vikhnina, M., Baldensperger, T., Soboleva, A., Dinastia, E., Dorn, M., Shiroglasova, O., Grishina, T., Balcke, G. U., Ihling, C., Smolikova, G., Medvedev, S., Zhukov, V. A., Babakov, V., ... Frolov, A. (2020). Does Protein Glycation Impact on the Drought-Related Changes in Metabolism and Nutritional Properties of Mature Pea (Pisum sativum L.) Seeds? International Journal of Molecular Sciences, 21(2), 567. https://doi.org/10.3390/ijms21020567