Abstract
The demyelinating diseases of the central nervous system involve myelin abnormalities, oligodendrocyte damage, and consequent glia activation. Neurotoxicant cuprizone (CPZ) was used to establish a mouse model of demyelination. However, the effects of CPZ on microRNA (miRNA) expression and behavior have not been clearly reported. We analyzed the behavior of mice administered a diet containing 0.2% CPZ for 6 weeks, followed by 6 weeks of recovery. Rotarod analysis demonstrated that the treated group had poorer motor coordination than control animals. This effect was reversed after 6 weeks of CPZ withdrawal. Open-field tests showed that CPZ-treated mice exhibited significantly increased anxiety and decreased exploratory behavior. CPZ-induced demyelination was observed to be alleviated after 4 weeks of CPZ treatment, according to luxol fast blue (LFB) staining and myelin basic protein (MBP) expression. miRNA expression profiling showed that the expression of 240 miRNAs was significantly changed in CPZ-fed mice compared with controls. Furthermore, miR-155-5p and miR-20a-5p upregulations enhanced NgR induction through Smad 2 and Smad 4 suppression in demyelination. Taken together, our results demonstrate that CPZ-mediated demyelination induces behavioral deficits with apparent alterations in miRNA expression, suggesting that differences in miRNA expression in vivo may be new potential therapeutic targets for remyelination.
1. Introduction
Demyelination of the central nervous system (CNS) is a typical feature of diseases such as multiple sclerosis (MS) and contributes to axon injury and cerebral atrophy, which are characteristic of late stages of the disease [1,2]. MS lesions are pathologically divided into four distinct patterns (I–IV) based on complement activation, IgG deposition, and loss of myelin-associated glycoproteins [3,4]. Spontaneous remyelination comes after demyelination, but often fails to complete [5,6]. Thus, the challenges of MS research are analyzing the causes of remyelination failure and developing methods to restore myelin. The research includes using animal models to help understand the demyelination and remyelination mechanisms, facilitating the study of cellular responses taking place in this process, and providing a robust platform for elucidating putative therapeutic targets. However, there is the limitation that no current animal model faithfully replicates the myriad of symptoms seen in the clinical condition of MS [2,7].
Cuprizone (bis-cyclohexanone-oxaldihydrazone, CPZ) is a copper chelating reagent that can be mixed with a rodent’s normal diet. Continuous feeding of CPZ results in a pathologic pattern similar to that of MS III lesions in white matter. It is, therefore, useful for studying the pathogenesis of primary demyelination due to mitochondrial dysfunction [4,8,9,10]. Completion of demyelination depends on mouse strain [11,12,13], anatomic location [14,15], age [16], dose [17], and sex [12], as in other animal models. However, the pathologic response that follows the CPZ intoxication of C57BL/6 mice is highly reproducible and well-characterized. In this model, demyelination after 4–6 weeks of intoxication is evident in multiple structures, including the hippocampus [18], external capsule [19], rostral cerebellar peduncles [20,21], cerebellum [22,23], striatum [19], cerebral cortex [11,14], and most notably, the corpus callosum [24]. Interestingly, spontaneous remyelination begins at the apex of demyelination and proceeds with the removal of the intoxicant CPZ, resulting in almost complete remyelination within a week [24,25]. As remyelination protects against neurodegeneration and promotes functional recovery [26,27,28], many studies have used the CPZ model to analyze the molecular mechanisms involved in remyelination [29,30,31]. However, the exact mechanisms underlying CPZ-induced demyelination and remyelination, the changes in miRNA expression, and the effects of CPZ treatment on behavior remain to be elucidated. To use the analysis of the miRNA expression in the demyelination model, we rigorously validated several behavioral tests. Here, we observed behavioral changes in mice subjected to a diet containing 0.2% CPZ for 6 weeks, followed by 6 weeks of recovery. Moreover, the expression of 240 miRNAs was significantly changed in CPZ-fed mice compared with control mice. Our results about the change of miRNA expression in vivo might suggest new potential remyelination therapeutic targets.
2. Results
2.1. Cuprizone Causes Weight Loss Which Subsides after Returning to a Normal Diet
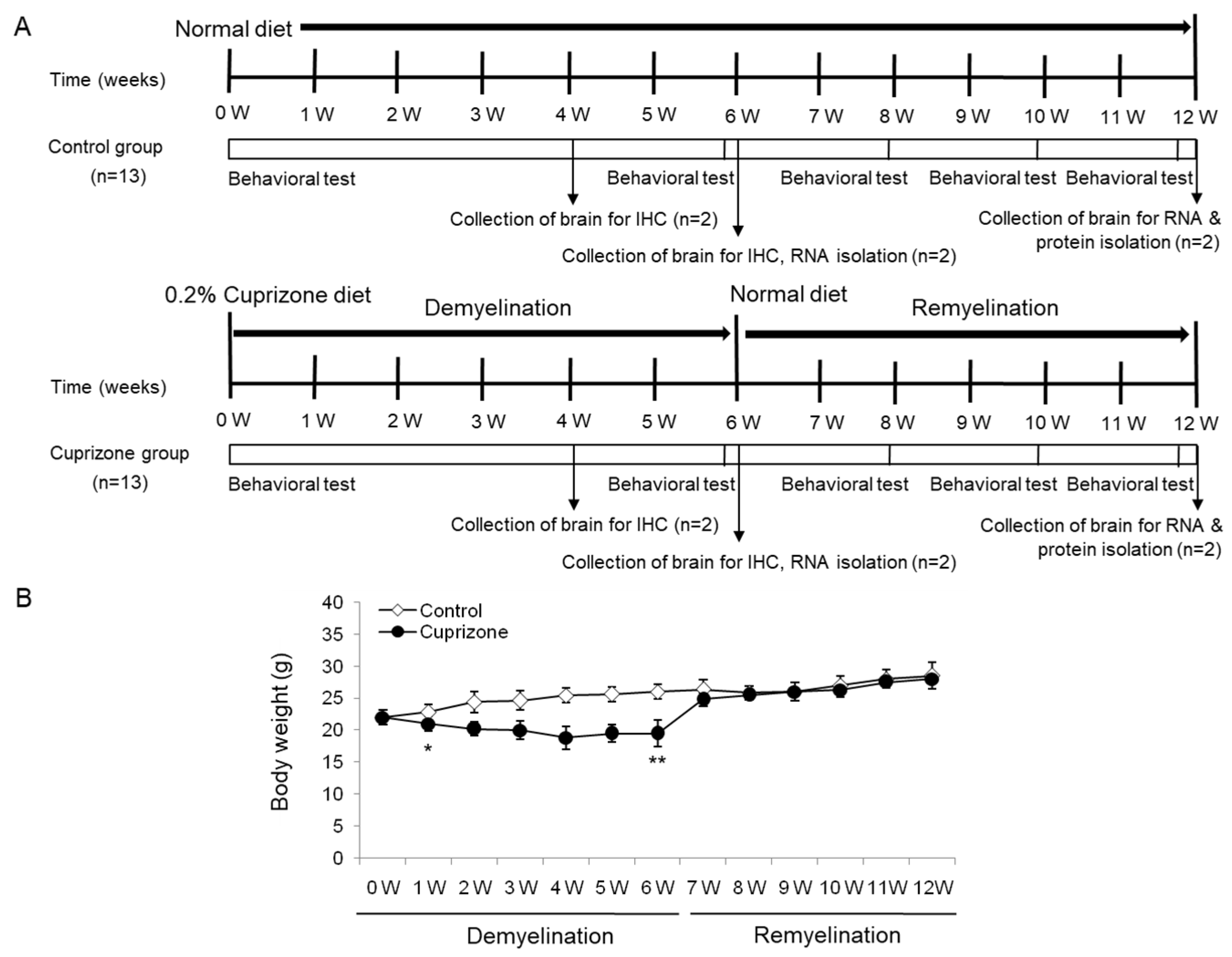
The body weight of the mice was measured weekly throughout the experimental period (Figure 1A). After 1 week of CPZ administration, the treated animals had lost 4.55% of their body weight, whereas those on a normal diet had gained 3.64% (Figure 1B). The difference between the control and the treated mice was statistically significant (p < 0.05). After 6 weeks of treatment, despite an overall increase in weight, CPZ-fed mice weighed significantly less than control mice (Figure 1B; p < 0.01). Upon returning the CPZ-fed mice to a normal diet, they gained weight rapidly, and after 1 week, were no longer different from the control mice. At the end of the recovery period with normal chow, the treated animals had an overall increase in body weight of 27.27% with respect to the initial weight, while the weight of the control animals increased by 28.5% (treated animals versus controls, p = 0.4759, Figure 1B). No deaths occurred during the experimental procedures.
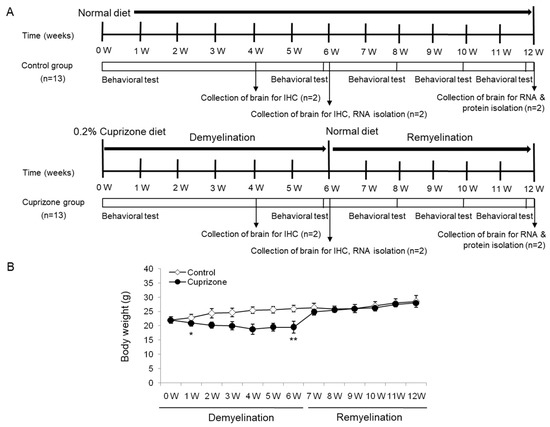
Figure 1.
Schematic representation of the experimental protocols and the measurement of body weight. (A) The control group was fed normal chow, and two of the thirteen mice were sacrificed after 4, 6, and 12 weeks. The cuprizone group was fed a 0.2% cuprizone (CPZ) diet, and two of the thirteen mice were sacrificed after 4 and 6 weeks, respectively. The CPZ group was fed the CPZ-containing diet for 6 weeks and then allowed to recover for 6 weeks on normal chow without CPZ. Behavior assessments were conducted in the two groups at 0, 6, 8, 10, and 12 weeks during the experimental period, and two of the nine mice were sacrificed at 6 weeks after CPZ withdrawal, respectively. (B) The body weight of the mice was measured weekly throughout the experimental period. The final data was made with the results of the total individuals minus the number of sacrificed mice. N = 7 for each group, and * p < 0.05, ** p < 0.01.
2.2. Cuprizone Increases the Number of Falls in the Rotarod Test
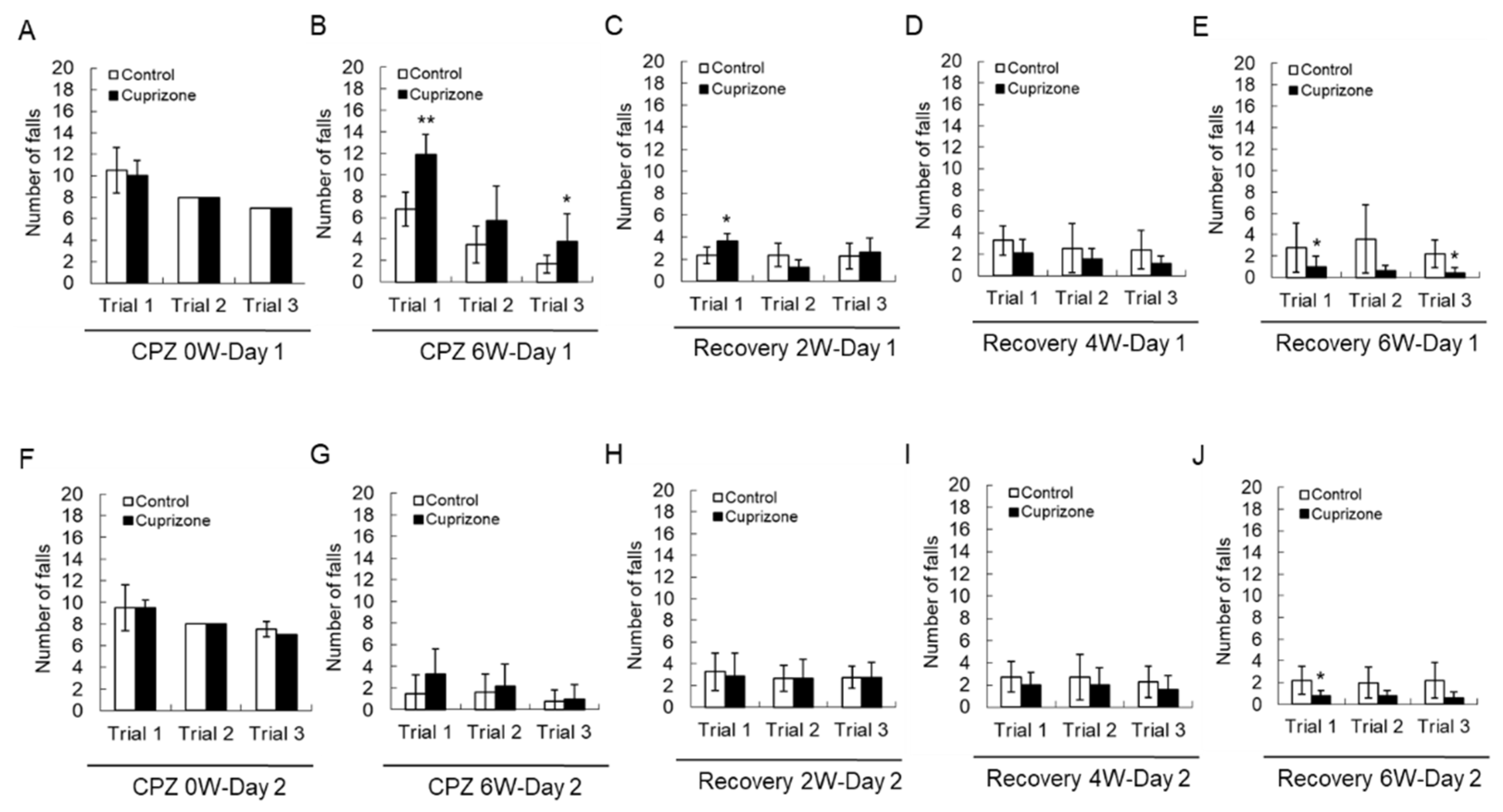
Motor coordination was evaluated using a Rotarod apparatus. All animals improved their skills and learned to stay on the rotating rod. At the week before CPZ feeding, there was no difference in the number of falls between the two groups before CPZ feeding (Figure 2A,F). At 6 weeks after CPZ feeding, a two-way repeated-measures analysis of variance showed that there was an interaction between the trial and treatment groups in the number of falls on the first day of training (Figure 2B, p < 0.01), indicating different rhythms of learning for the different groups. Animals treated for 6 weeks fell more than control animals on the first trial (Figure 2B,G). Upon returning the CPZ-fed mice to a normal diet, they fell less often than the control mice, and this effect depended on the recovery time (Figure 2C–E,H–J)). These results suggest that 6 weeks of CPZ treatment impaired motor coordination, which was fully recovered after 6 weeks of CPZ withdrawal.
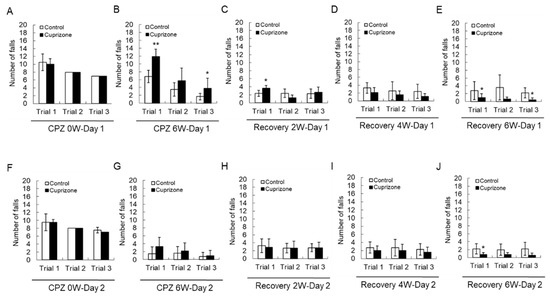
Figure 2.
Number of falls in the Rotarod test. (A–E) The graphs show the number of falls in three different trials on the first day. (F–J) The graphs show the number of falls in three different trials on the second day. Data are expressed as mean ± standard deviation (SD). The final data was made with the results of the total individuals minus the number of sacrificed mice. N = 7 for each group, and differences between groups are expressed as * p < 0.05, ** p < 0.01.
2.3. Cuprizone Decreases the Grip Strength of Mice
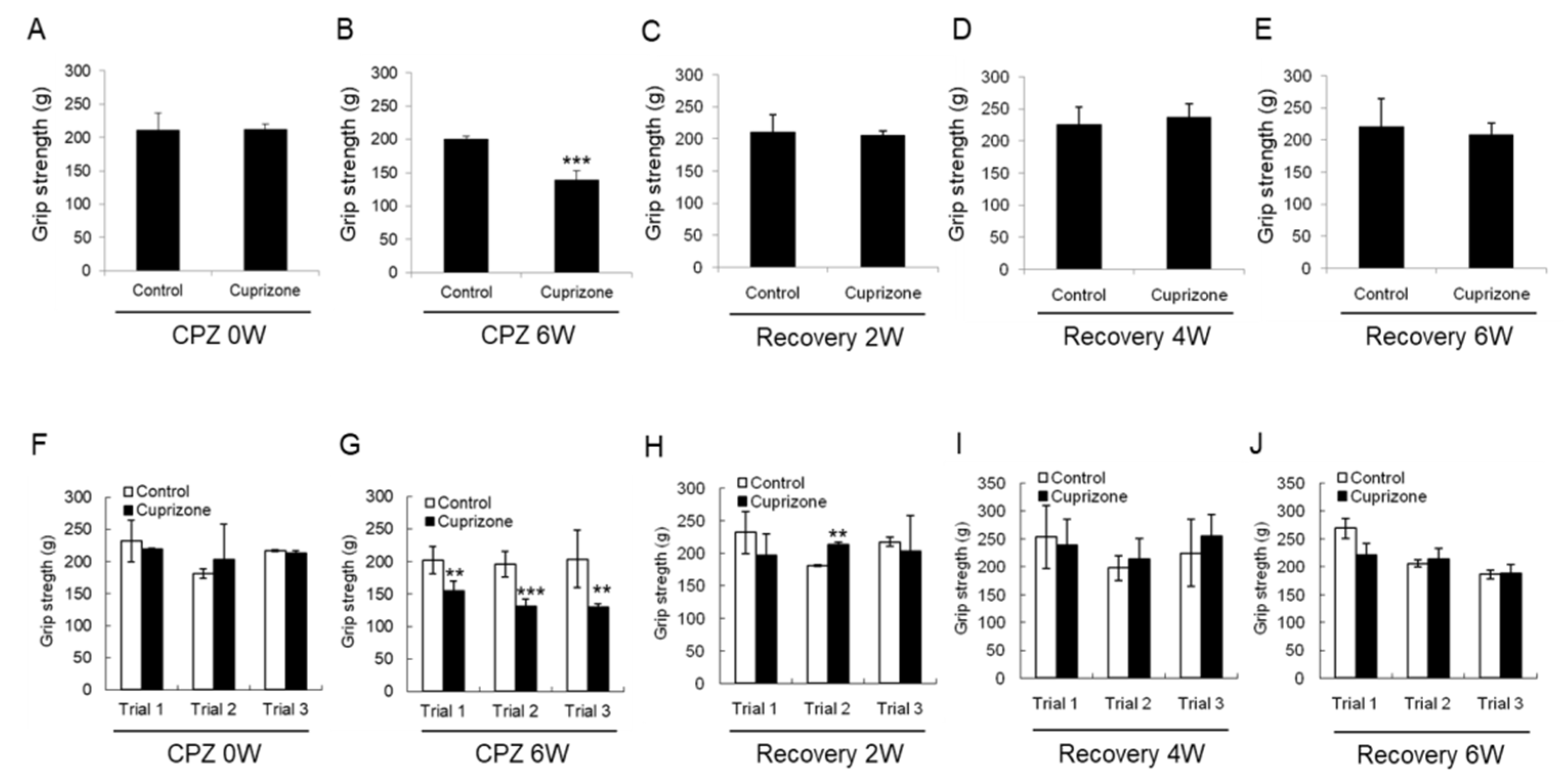
To evaluate the effect of CPZ on neuromuscular functions, we performed the grip strength test included in the functional observational battery (FOB) commonly used to screen for neurobehavioral toxicity. At the week before CPZ feeding, the grip strength was no different between the control group and CPZ group (Figure 3A,F). The grip strength of mice fed CPZ for 6 weeks was significantly decreased compared to that of control mice (Figure 3B,G). Returning the CPZ-fed mice to normal chow led to recovery of the grip strength to levels similar to those in the control group (Figure 3C–E,H–J). These results suggest that 6 weeks of CPZ treatment induced neuromuscular weakness, which was overcome after 6 weeks of CPZ withdrawal.
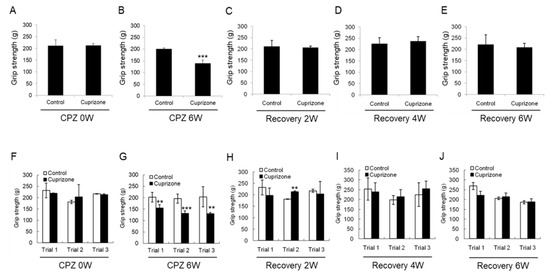
Figure 3.
Results of the grip strength test. (A–E) The graphs show the average grip strength of three different trials in the control or cuprizone group. Data are expressed as mean ± standard error of the mean (SEM). (F–J) The graphs show the grip strength in three different trials. Data are expressed as mean ± standard deviation (SD). The final data was made with the results of the total individuals minus the number of sacrificed mice. N = 7 for each group, and differences between groups are expressed as ** p < 0.01, *** p < 0.001.
2.4. Cuprizone Decreases the Total Distance Traveled by Mice in the Open-Field Test
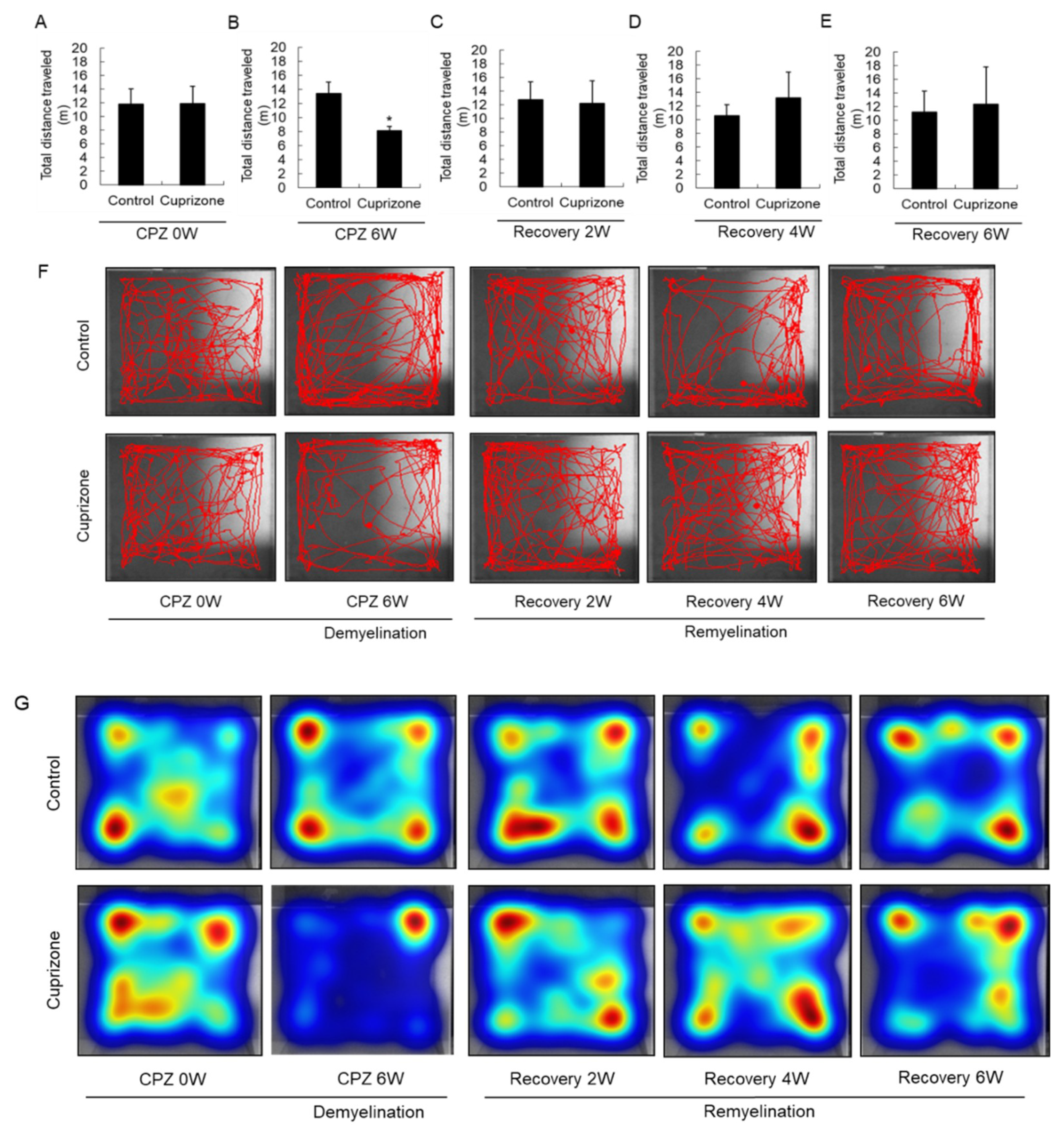
Experimental animals were assessed in the open-field test to evaluate their level of activity and their behavioral response to a novel environment. At the week before CPZ feeding, the total distance traveled was similar between the control group and the CPZ group (Figure 4A). In mice fed CPZ for 6 weeks, the total distance traveled, a measure of general locomotor activity, decreased compared to that in control mice fed normal chow (Figure 4B). The total distance traveled by the treated animals recovered 2 weeks after returning to normal chow (Figure 4C), and increased until 6 weeks after CPZ withdrawal (Figure 4D,E). The habituation pattern was measured for 5 min in the open field (Figure 4F). The habituation patterns of CPZ-fed mice were different from those of the control animals, as shown by the significant interaction between CPZ treatment and the distance traveled. In addition, 6 weeks after returning the CPZ-fed mice to a normal diet, they traveled in the central area of the open field more than the control mice, indicating diminished anxiety (Figure 4F). Furthermore, the heatmap visualizations were similar to the above results (Figure 4G).
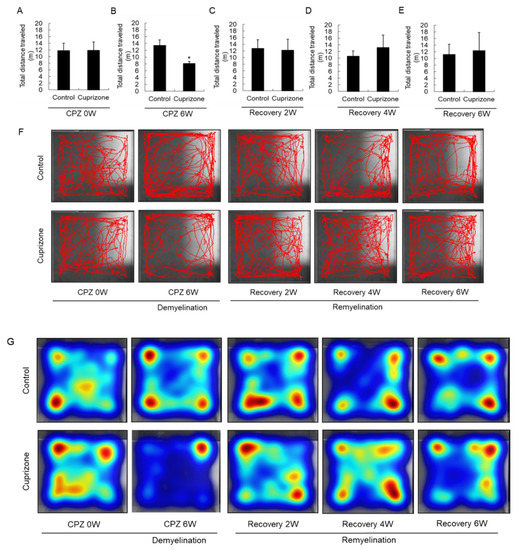
Figure 4.
Open-field behavior. (A–E) The graphs show the total distance traveled in an open field during 5 min. Data are expressed as mean ± standard deviation (SD). The final data was made with the results of the total individuals minus the number of sacrificed mice. N = 7 for each group, and differences between groups are expressed as * p < 0.05. (F) Representative images showing typical examples of exploratory behavior in the open-field test in the control and cuprizone groups. (G) The heatmap visualizations were similar to the above results.
2.5. Cuprizone Induces a Loss of Myelin in the Corpus Callosum
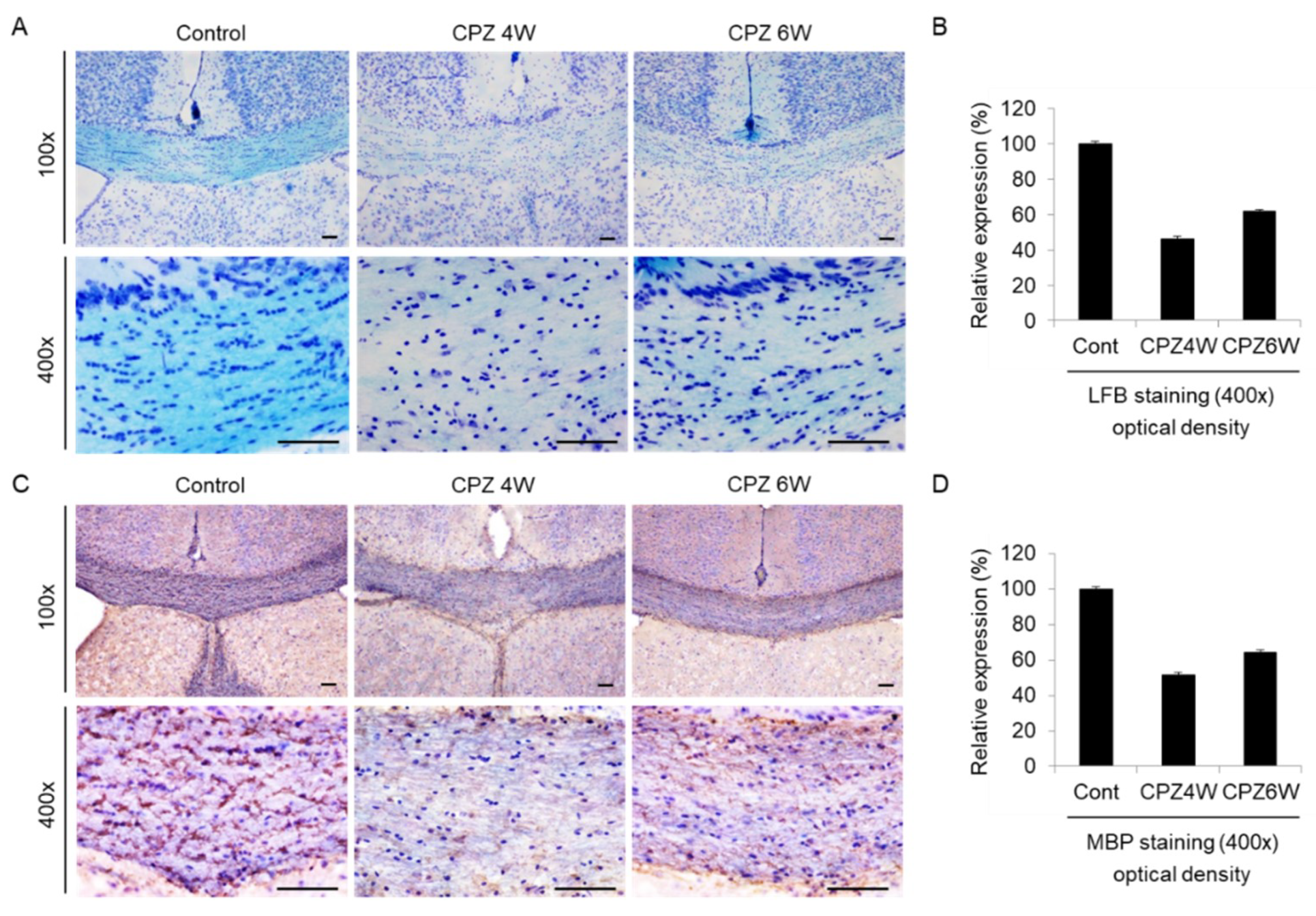
The corpus callosum (CC) is the largest white matter (WM) structure in the brain, connecting the homologous cortical areas of the two cerebral hemispheres and playing a critical role in the transfer of sensory, cognitive, and motor information [32]. We then focused on the corpus callosum region as one of the most widely studied regions in this model of demyelination. To analyze the loss of myelin in the corpus callosum of CPZ-fed mice, luxol fast blue (LFB) staining, and myelin basic protein (MBP) immunohistochemistry were performed. Brain sections of each group were stained with LFB, which stains the lipid-rich myelin blue (Figure 5A), and with antibodies against MBP, which bind to the protein myelin (Figure 5C). At the beginning of the experiment, the corpus callosums of mice was well-myelinated, with characteristic rows of interfascicular oligodendrocytes (Figure 5A, left panels). Strong loss of myelin was detected in the mice fed the CPZ diet at 4 weeks (Figure 5A, middle panels), which was slightly overcome at 6 weeks of CPZ treatment (Figure 5A, right panels). Figure 5B revealed these integral optical densities (IOD). A similar demyelination trend was observed on MBP immunohistochemical analysis (Figure 5C) and IOD (Figure 5D). These results demonstrate that CPZ alters the myelin lipid and protein structure, resulting in motor neurobehavioral defects.
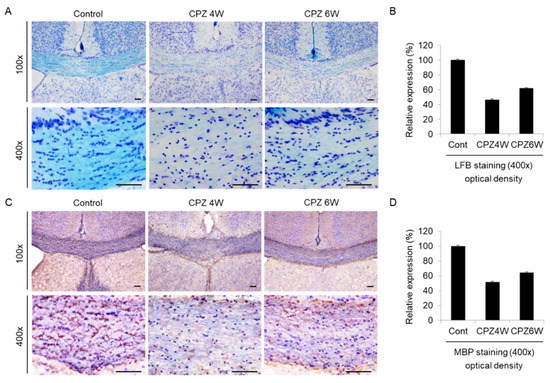
Figure 5.
Induction of demyelination in cuprizone-fed mice. (A) Luxol fast blue (LFB)/Cresyl Echt Violet staining in coronal slices of the cuprizone-fed and control groups in the corpus callosum. Myelinated fibers are shown in blue, and Nissil substance and nerve cells in violet. Lithium carbonate indicates differentiation, and Cresyl Echt Violet is a counterstain. Scale bar: 50 μm. (C) Myelin basic protein (MBP) immunostaining in coronal slices of the cuprizone-fed and control groups in the corpus callosum. Control mice display a normally myelinated corpus callosum. Maximal demyelination is observed after 4 weeks on a cuprizone diet, and the recovery of demyelination is weakly observed after 6 weeks on a cuprizone diet. Scale bar: 50 μm. For each staining, three sections per animal were used and a total of two mice were tested each group. The staining was done at least twice each. (B,D) The integral optical density of LFB or MBP revealed using i-Solution software.
2.6. Diverse Expression Patterns of microRNAs in Cuprizone-Induced Demyelination
Demyelination of the central nervous system (CNS) is the hallmark of diseases such as multiple sclerosis (MS), an immune-mediated inflammatory disease of the central nervous system. miRNAs play important roles in autoimmune diseases, including MS. Although miRNA expression profiles of MS patients were recently reported [33,34], the role of these molecules in MS remains to be elucidated.
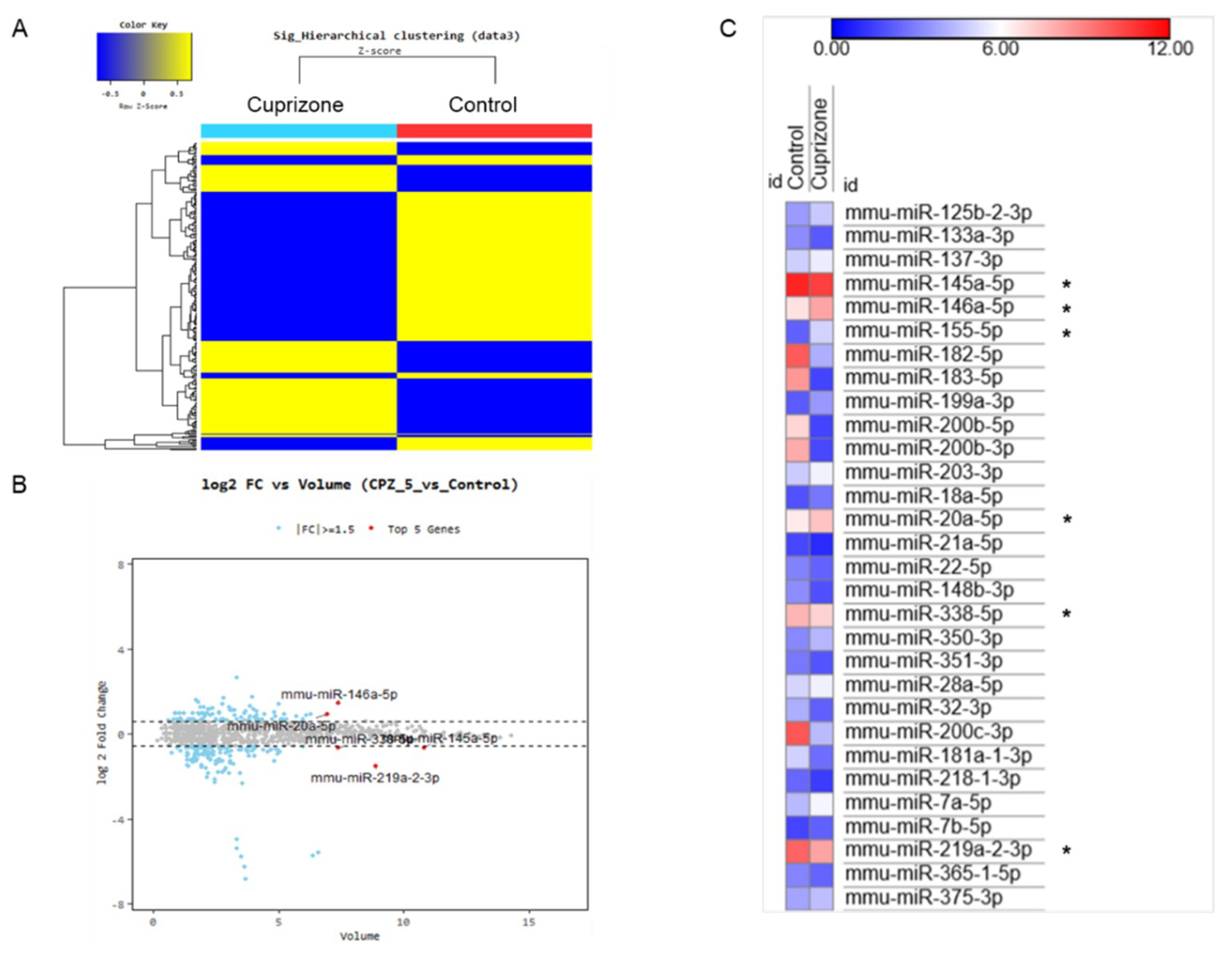
To evaluate the miRNA expression pattern in CPZ-induced demyelination, we used the Affymetrix GeneChip® miRNA 4.0 array. We identified 1348 and 1253 expressed miRNAs in control and CPZ-treated mice, respectively. Furthermore, the expression of 240 miRNAs was significantly changed in CPZ-treated mice compared with controls—these were clustered by hierarchical clustering to create the heatmap shown in Figure 6A. The top five miRNAs by fold change vs. the average expression in CPZ-fed mice compared with control mice were mmu-miR 146a-5p, mmu-miR-20a-5p, mmu-miR-338-5p, mmu-miR-145a-5p, and mmu-miR-219a-2-3p (Figure 6B). Figure 6C represents the miRNA expression heatmap, including the top five miRNAs, using the MORPHEUS tutorial program. The expressions of miR 146a-5p, miR 155-5p, and miR 20a-5p were upregulated in CPZ-treated mice compared with control mice. However, the miR 145a-5p, miR 338-5p, and miR 219a-2-3p were downregulated in CPZ-treated mice (Figure 6C). We selected the 39 (30) miRNAs that were upregulated (downregulated) in CPZ-fed mice with a fold change (FC) of ≥1.5 compared with control mice and predicted their respective target genes using the micro-RNA database (miRDB) (Table 1 and Table 2, respectively). This analysis demonstrated the diverse expression patterns of miRNAs in CPZ-induced demyelination. It could be used to clarify the relationship between miRNA modulation and the molecular cascade induced during CPZ-induced demyelination.
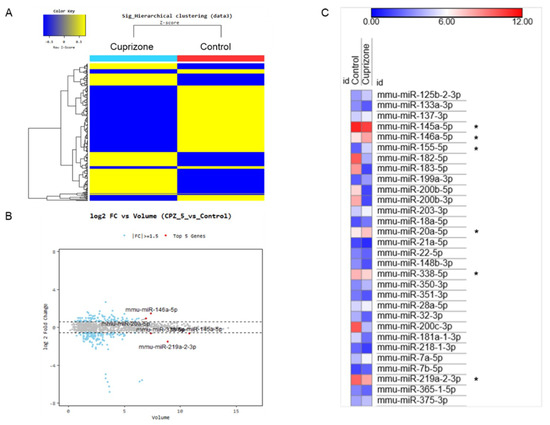
Figure 6.
The miRNA expression analysis of cuprizone-fed mice and control mice. (A) Hierarchical clustering map of miRNA expression levels in cuprizone-fed mice versus control mice. Color key is Z-score (−0.5~0.5). (B) The graph shows the top five miRNAs evaluated using logarithmic fold change (log 2) vs. average expression in cuprizone-fed mice compared with control mice. (C) The expression heatmap shows miRNA expression changes (color key is expression score, 0–12), including the top five miRNAs (*) using the MORPHEUS tutorial program.

Table 1.
Upregulated miRNA in a cuprizone-induced demyelination mouse model (Fold Change ≥ 1.5).

Table 2.
Downregulated miRNA in a cuprizone-induced demyelination mouse model (Fold Change ≥ 1.5).
2.7. Validation of miRNAs Expression Using the miRNA Quantitation PCR
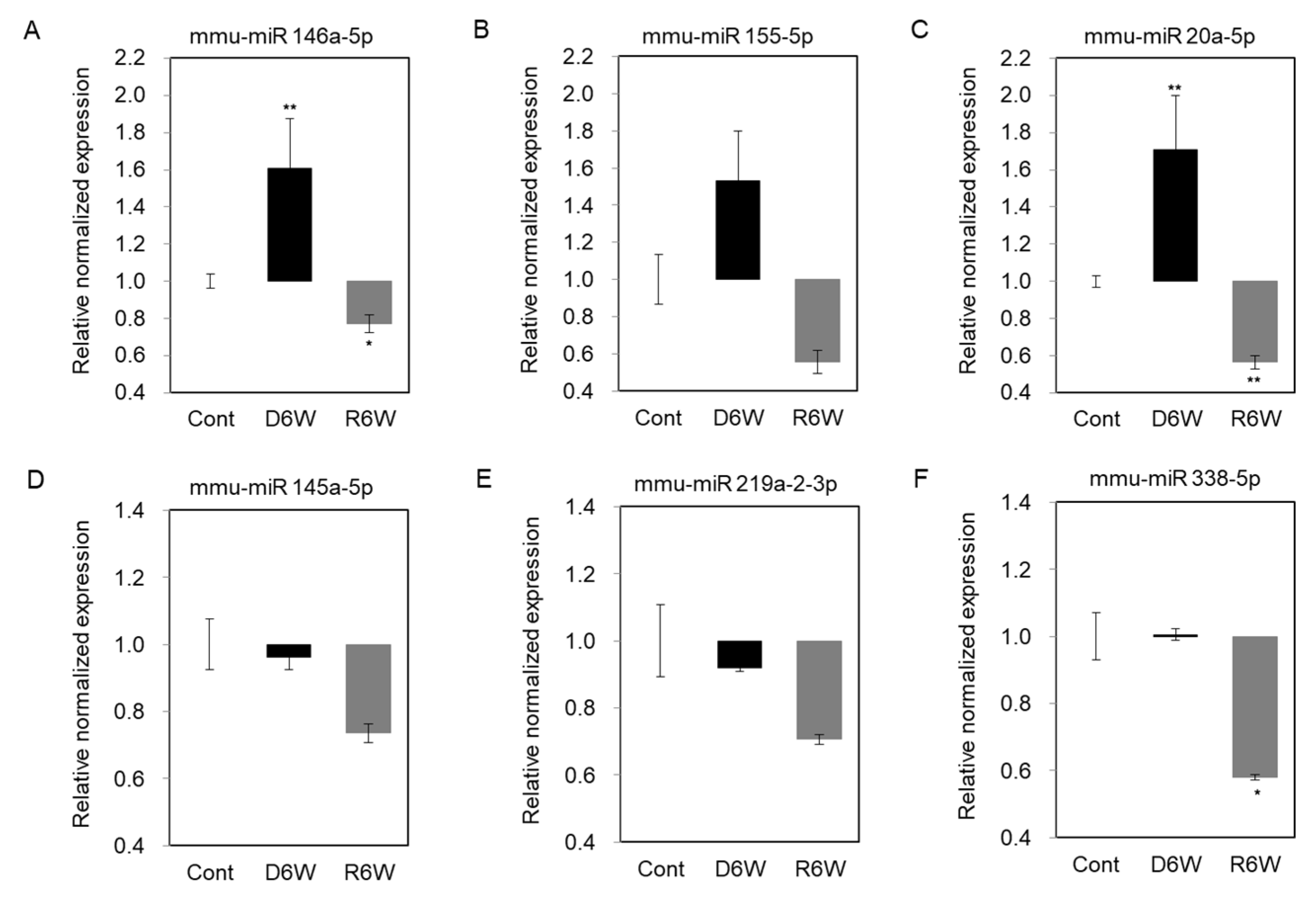
To validate the changes of miRNA expression in cuprizone-induced demyelination mice, we performed the miRNA quantification polymerase chain reaction (qPCR), including the top five miRNAs by fold change vs. the average expression in CPZ-fed mice compared with control mice and miR 155-5p (Figure 7). As shown in Figure 7A–C, the expressions of miR 146a-5p, miR 155-5p, and miR 20a-5p were upregulated in demyelinated mice (D6W) as in the results by miRNA microarray, and then downregulated at 6 weeks after cuprizone withdrawal (R6W) compared to control mice (Cont). The miR 145a-5p and miR 219a-2-3p were downregulated in D6W, and confirmed miRNA microarray (Figure 7D,E), but not miR 338-5p (Figure 7F); however, three miRNAs were downregulated R6W compared to Cont. Then, to focus on the regulation mechanism of de- and remyelination, we analyzed the expressions of the miRNAs’ predicted target gene (Table 1 and Table 2) using real-time PCR (Figure S1) and western blotting (Figure 8).
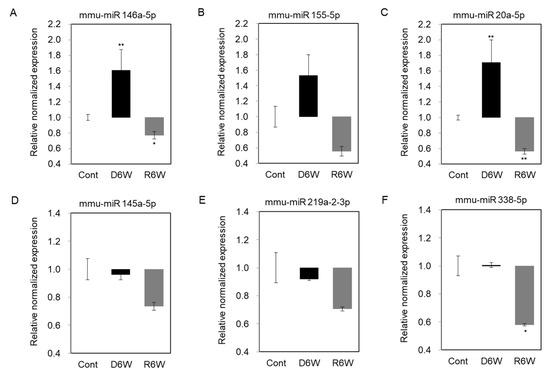
Figure 7.
Validation of miRNA expression using miRNA quantitation Polymerization Chain Reation (PCR) in cuprizone-induced demyelination and remyelination mice. (A–C) The expression levels of miR 146a-5p, miR 155-5p, and miR 20a-5p were significantly upregulated in demyelinated mice (D6W) compared to control mice (Cont) by real-time PCR analysis. (D,E) The expression levels of miR 145a-5p and miR 219a-2-3p were weakly downregulated in D6W compared to Cont., but not miR 338-5p (F). n = 2, in triplicate for each group. The differences between groups are expressed as * p < 0.05, ** p < 0.01.
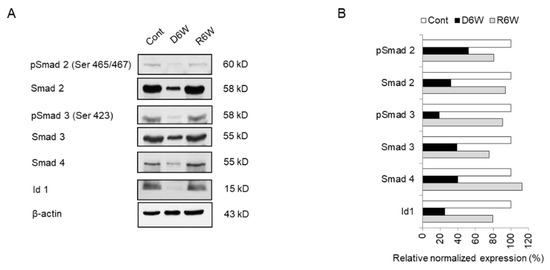
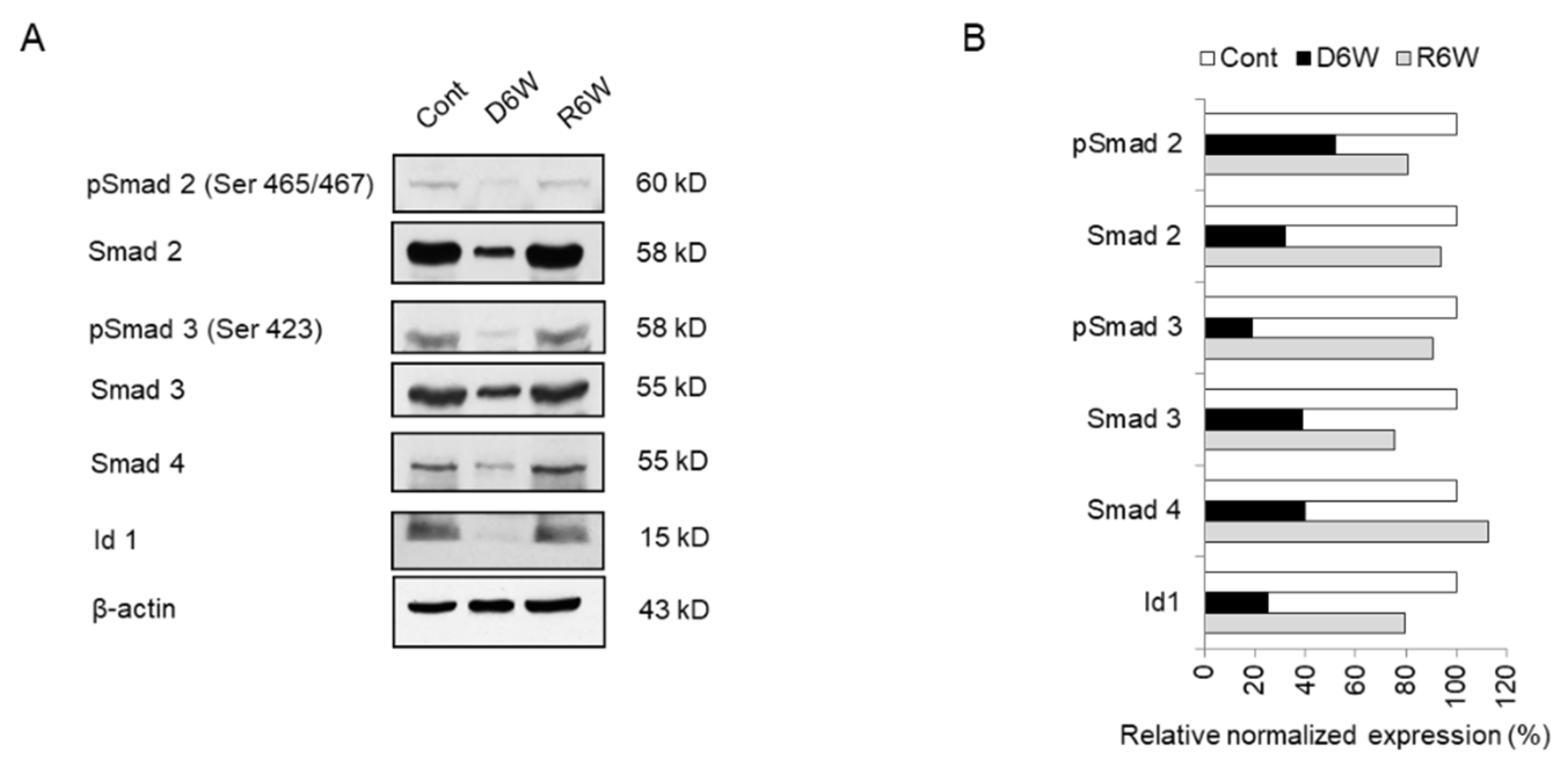
Figure 8.
Analysis of miRNA target genes expression in cuprizone-fed mice and control mice. (A) Western blot analysis shows the phosphorylated and total Smad 2, Smad 3, Smad 4, and Id1 in demyelinated mice (D6W) and remyelinated mice (R6W) compared to control mice (Cont). n = 2. (B) The graph shows the relative normalized expression of these in D6W and R6W with Cont. The uncut original western blot images are in Supplementary Figure S2.
2.8. Analysis of miRNA Target Genes and Signal Cascades in Cuprizone-Induced Demyelination
In our previous study, we showed the activation of Smad 2, 3, and 4 via the TGFβ pathway in glioblastoma multiforme and primary glioma stem cells and discussed the importance of the smad pathway in brain diseases [35]. We hypothesized that the smad pathway should be one of the regulation pathways of demyelination. Therefore, we focused on the upregulated miR 155-5p and miR 20a-5p, which predicted to target Smad 2 and Smad 4/TGFβR2 in cuprizone-fed mice (Table 1).
To evaluate the TGFβ pathway such as TGFβR1, which was a predicted target of miR 20a-5p, TGFβR2, and TGFβ1 and its signal cascade genes, Id1 and Nogo receptor (NgR) in CPZ-induced demyelination and remyelination after CPZ withdrawal, we carried out the real-time PCR (Figure S1). The transcriptional levels of TGFβ1, TGFβR1, and TGFβR2 were not significantly altered in CPZ-induced demyelination (Figure S1A–C), but NgR was significantly increased (Figure S1E). These results showed that the upregulated miR 20a-5p could not affect upstream molecules of smad signal cascades, but finally regulated the downstream molecules of smad signal cascades.
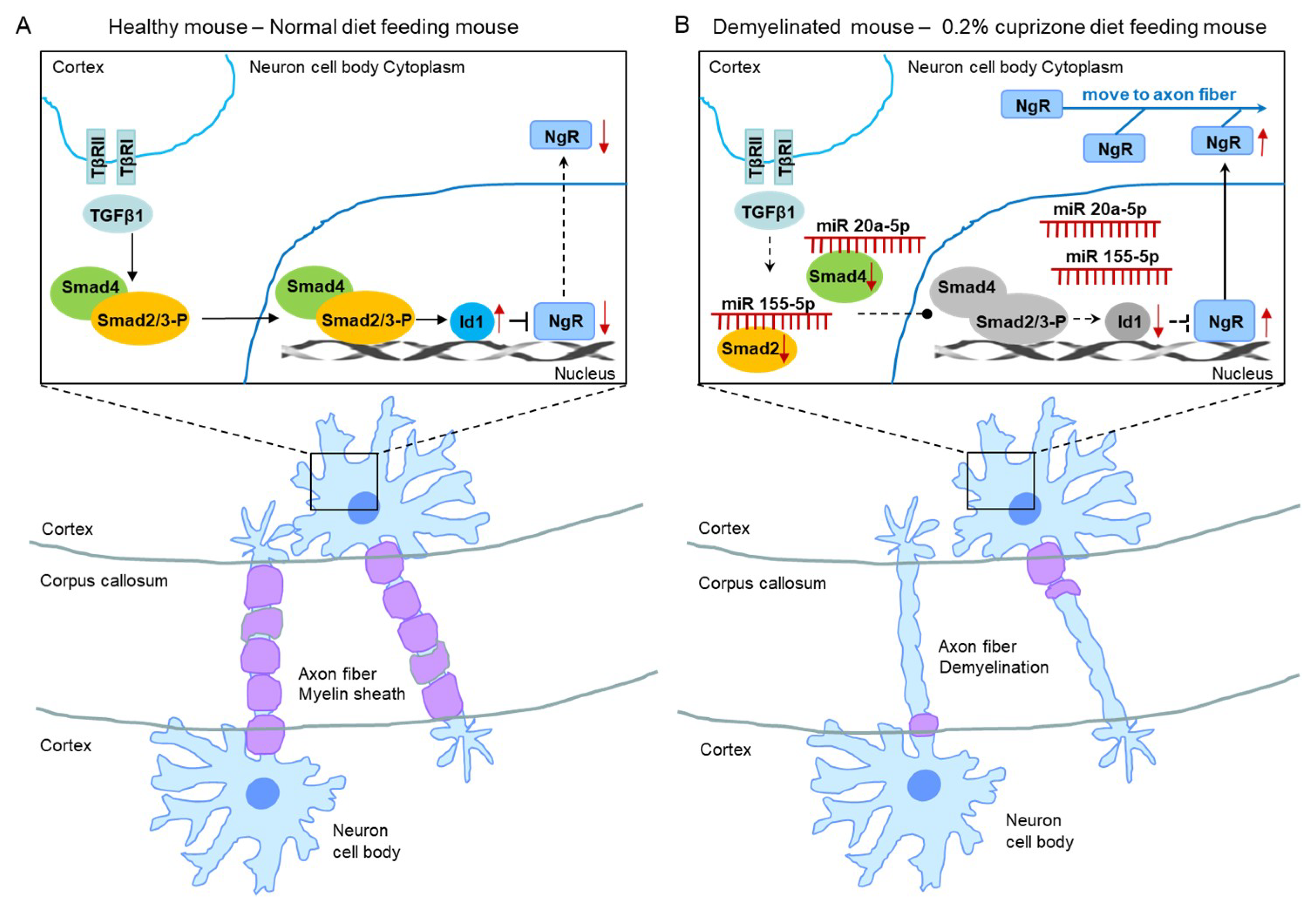
Therefore, we evaluated the smad family and Id1 protein, the predicted targets of upregulated miR 155-5p and miR 20a-5p using western blot analysis (Figure 8). As shown in Figure 8, the expression and phosphorylation of Smad 2 and 3 were dramatically decreased in CPZ-induced demyelination mice compared to control mice (Figure 8A,B). Also, Smad 4 and Id1 expression were downregulated in demyelination (Figure 8A,B). Thus, these results demonstrate that miR 155-5p and miR 20a-5p upregulation in demyelination leads to the induction of NgR expression through the suppression of Smad signal cascades. We then proposed this as one of the modulated mechanisms of demyelination (Figure 9).
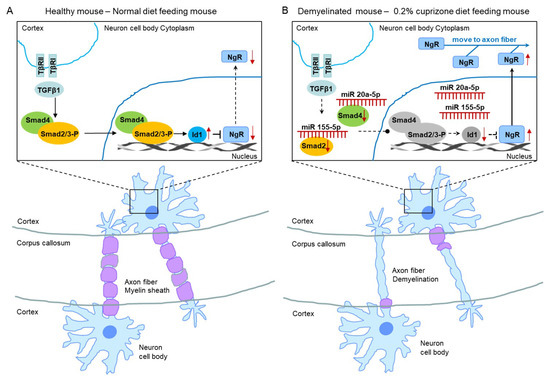
Figure 9.
The proposed mechanism of miR 155-5p and miR 20a-5p upregulations led to demyelination. (A) The illustration shows a proposed mechanism in normal diet-fed, healthy mice. (B) The illustration shows that the proposed mechanism of miR 155-5p and miR 20a-5p upregulations led to demyelination in 0.2% cuprizone diet-fed mice.
3. Discussion
The disruption of the myelin sheath of CNS is a prominent phenomenon of many clinically relevant disorders on the basis of physiopathology [36]. Several mouse models are available for studying demyelination and remyelination. The best-characterized demyelinating mouse model is the C57BL/6J with 0.2% CPZ added to the diet [24]. Several hypotheses have been proposed [17,24,37], but it is still unclear why CPZ, a copper chelator molecule, specifically affects oligodendrocytes, the cell type that synthesizes and maintains the CNS myelin sheath.
The behavior of CPZ-fed mice has not been sufficiently studied [17,37,38]. Morell et al. (1998) reported that CPZ-treated animals did not appear to be as active at the end of the first week of CPZ administration, and also during the following several weeks of CPZ exposure. The same authors reported that, during the recovery period, the animals displayed normal activity levels, and were indistinguishable from control animals [37]. However, Franco-Pons et al. (2007) reported that behavioral deficits followed the course of demyelination–remyelination induced by administration of 0.2% CPZ and that some of the changes persisted even after 6 weeks of a normal diet [38]. Our experiments showed motor neurobehavioral defects using the Rotarod, grip strength, and open-field tests after 6 weeks of 0.2% cuprizone feeding, and the recovery of such neurobehavioral defects upon returning to a normal diet for 6 weeks, together with remyelination. We believe the differences in the behavioral patterns observed in the various studies might have been caused by differences in the behavioral assessment protocols, underlining the importance of using standardized behavioral assessment protocols.
miRNAs are short, noncoding RNA molecules that are processed from larger transcripts of non-classical genes by the Drosha and Dicer nucleases [39]. Mature miRNA biogenesis is a tightly controlled, multistep process finalized in the production of a ~22-nt-long duplex [40]. miRNAs regulate gene-expression programs by reducing the translation and stability of target mRNAs [41]. It has been estimated that the expression of as many as one-third of protein-coding genes may be regulated by miRNA [42].
Many miRNAs have functioned as important regulators of the immune system [33,34,43,44,45,46]. In particular, miR-155 is known to be a crucial regulator orchestrating the role of numerous acquired and native immune cell populations. The miR-155 host gene (MIR155HG) produces miR-155-3p and miR-155-5p, being the functional form [47]. Several researchers identified increased miR-155 expression in T cells both in vivo and in vitro during the development of autoimmune responses [48], and miR-155-deficient mice have been found to be resistant to the development of experimental autoimmune encephalomyelitis (EAE) [49]. Additionally, Mycko et al. (2015) identified an EAE-specific mechanism of miR-155 expression, in which miR-155-3p drives the development of autoimmune demyelination by regulating heat shock protein 40 [46]. Furthermore, we showed that miR-155-5p, the functional form of miR-155, is significantly upregulated in mice subjected to CPZ-induced demyelination. This result is in line with those of other reports described above, and suggest that miR-155-5p upregulation may induce demyelination through upregulation of NgR via suppression of Smad signal cascades in CPZ-fed mice. However, in Table 1, there is mmu-miR-195a-3, which might suppress Id1 and increase with demyelination. Proteins Smad2, Smad3, and Smad4, as seen from the tables, are also inhibited by various miRNAs. We would not ignore such direct connections between the Smad family and various miRNAs.
One of the MS-related miRNAs, miR-146a, is differentially expressed in MS lesions and promotes the differentiation of oligodendrocyte precursor cells (OPCs) during remyelination [50]; moreover, it is upregulated during CPZ-induced de- and remyelination [51]. In particular, Martin et al. (2018) reported that experimental demyelination and axonal loss are reduced in mice deficient in microRNA-146a and that the number of OPCs is slightly higher in WT mice during remyelination, indicating the complex role of miR-146a during in vivo de- and remyelination [51]. In our results, miR-146a-5p was significantly upregulated in mice subjected to CPZ-induced demyelination, suggesting that it may be a key miRNA involved in the induction of demyelination in CPZ-fed mice. These results support the reliability of our miRNA array results, which may be useful in understanding the demyelination mechanism. However, a limitation of our study is that the miRNA expression pattern was not analyzed in remyelinated mice. Thus, we cannot thoroughly discuss the changes in miRNA expression during CPZ-induced de- and remyelination.
It is a long-held view that the motor corpus callosum is important for bimanual coordination and learning of bimanual motor skills. This notion was built mainly on bimanual coordination deficits observed in patients with extensive lesions of the corpus callosum or partial callosotomies [52]. The administration of CPZ in combination with 0.5 mg/kg of WIN55212.2 declined the expression of NgR1 and might confer neuroprotection against CPZ [53]. Actually, NgR1 participates in oligodendrocyte differentiation, myelination [54], and cell cytoskeleton reorganization [55]. Our supplementary data shows that the expression of NgR increased at 6 weeks after CPZ treatment in the corpus callosum, suggesting that the neuroprotection in axon fibers against CPZ and the participation of OPC differentiation into myelinating oligodendrocytes resulted in spontaneous remyelination in CPZ-fed mice. These results support the reliability of our predicted gene targets in miRNA microarray results and provide a better understanding of the demyelination mechanism. Based on the findings, we are looking forward to developing the new targets underlying the de- and remyelination mechanism as a useful therapeutic target for remyelination.
4. Materials and Methods
4.1. Animals and Cuprizone Administration
Twenty-six male C57BL/6 mice aged 7 weeks were purchased from KOATECH (Pyeongtaek, Korea) and allowed 1 week of acclimation to their new environment before beginning the experiment. The 0.2% CPZ diet (TD.140803) and normal diet (2018S) were purchased from ENVIGO (Madison, WI). The mice in the CPZ group were freshly fed the 0.2% CPZ diet each day for a total of 6 weeks to induce demyelination. The control mice, kept on a normal diet, were age-matched to the CPZ-fed mice. Food pellets were changed every day. After 6 weeks of CPZ treatment, the diet was changed to normal rodent chow for an additional 2, 4, and 6 weeks to examine remyelination. According to a bibliographic investigation before the start of the experiment [13,38,51,56], it was confirmed that both females and males were used to induce the cuprizone-induced model, and the test results were similar in the laboratory. Twenty-six animals were divided into two groups of 13 animals each. The final data was made with the results of the total individuals minus the number of sacrificed mice. N = 7 in each group. These were sufficient numbers for different aspects of the experiment. All animal protocols adhered to the Ministry of Food and Drug Safety (MFDS) Guidelines for Care and Use of Laboratory Animals and were approved by the Eulji University Laboratory Animal Care and Use Committee (Approval No., EUIACUC17-21; approval date, 15 December 2017).
4.2. Motor Coordination and Learning: Rotarod Test
Motor coordination and balance were evaluated in a Rotarod apparatus (BR1001, B.S Technolab INC., Seoul, Korea), which consists of a motor-driven rotating rod whose speed can be adjusted. All mice were evaluated on the Rotarod three times a day for two consecutive days, with the rotation set at 15 to 16 revolutions per minute (rpm). To test the performance, the mice were placed on the rotating cylinder at an angle of 45° with an initial rotation speed of 16 rpm, and were allowed to run for 60 s. If the animal fell, the chronometer was stopped, the animal was put back onto the cylinder, and then the timing was continued. The trial was repeated after 5–10 min. The falls and flips (when the animal hangs on to the cylinder and continues all the way around) were recorded within 60 s of each trial.
4.3. Grip Strength Test
The grip strength test allows for the assessment of neuromuscular functions by determining the maximal peak force developed by a rodent when the operator tries to pull it out of a specially designed grid, available for both the fore and hind limbs. The grip strength test is included in the functional observational battery (FOB) used to screen for neurobehavioral toxicity. In this context, changes in grip strength peak values of the rodents are interpreted as evidence of motor neurotoxicity. Forelimb strength (g force) was measured with a grip strength tester (BIO-GS3, BIOSEB, Vitrolles Cedex, France) to detect contralateral paw weakness. After both forelimbs of the mouse were loosened by pulling the tail, the maximal force was recorded. Each mouse was subjected to three trials of each test, and the mean values (g) were calculated and normalized to body weight.
4.4. Open-Field Assessment
Activity and behavioral responses to a novel environment were measured in an open-field apparatus consisting of a 30 cm × 30 cm wooden square surrounded by a dark wall 30 cm high. The mice were placed in the center of the arena at the beginning of the test period. During the test, the mice were allowed to move freely around the open field and explore the environment for 5 min. Their path was recorded by a video camera (LS903, LG Electronics, Daejeon, Korea) placed above the square. The video tracking program EthoVision XT5 (Noldus Information Technology, Wageningen, NL, Canada) was used to measure the total distance traveled.
4.5. Luxol Fast Blue Staining and Myelin Basic Protein Immunohistochemistry
All mice were deeply anesthetized by intraperitoneal injection of ketamine (80 mg/kg) and xylacine (10 mg/kg) dissolved in 0.9% saline and transcardially perfused with 4% paraformaldehyde (PFA). For Luxol fast blue staining and immunohistochemical analysis, the brains were placed in 30% sucrose/phosphate buffered solution (PBS) for 48 h at 4 °C and then snap-frozen in isopentane. The corpus callosum was chosen as a representative white matter region as it has been extensively examined in this animal model [17,37]. Coronal sections (30 μm thick) of the mouse brain, corresponding to bregma 0.62 mm to bregma -0.46 mm, were cut with a cryostat and stored at −20 °C in cryoprotection buffer (40% phosphate buffer 0.1 M, 30% glycerol, 30% ethylene glycol).
Demyelination was determined histochemically by examining coronal sections stained with Luxol fast blue (LFB). For each staining, three sections per animal were used, and a total of two mice were tested for each group. The staining was done at least twice each. All reagents were purchased from Abcam (ab150675, Cambridge, UK). Luxol fast blue staining was carried out in accordance with the manufacturer’s instructions (Abcam, Cambridge, UK). Brain tissue slides were incubated in Luxol Fast Blue Solution (0.1%) for 24 h at room temperature. The sections were rinsed thoroughly in distilled water, and differentiated by dipping in Lithium Carbonate Solution (0.05%) several times. Differentiation for the sections were then continued by repeatedly dipping in alcohol reagent (70%) until the gray matter became colorless and the white matter remained blue. Then, the sections were rinsed in distilled water, followed by incubation with Cresyl Echt Violet (0.1%) for 2–5 min. The sections were rinsed quickly in one change of distilled water, dehydrated quickly in three changes of absolute alcohol, cleared in three changes of xylene, and mounted with mounting medium.
The method used for myelin basic protein immunohistochemical analysis has been described in a previous article [57]. For detection of MBP expression, we used immunodetection kits (M.O.M. kit, MP-2400, Vector laboratories, Burlingame, CA, USA) which are specifically designed to localize primary mouse antibodies on mouse tissues. Subsequently, all sections were treated with 3% H2O2 for 10 min to block endogenous peroxidase activity, and then slides were incubated for 1 h in mouse Ig blocking reagent (Vector laboratories, Burlingame, CA, USA). Then, the slides were blocked with 2.5% normal horse serum (Vector laboratories, Burlingame, CA, USA) for 1 h at room temperature (RT) and then incubated with anti-MBP antibody (1:100; sc-271524, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) overnight at 4 °C. The sections were then incubated with the Immpress anti-mouse IgG reagent (made in horse; Vector laboratories, Burlingame, CA, USA) for 30 min at RT. The second Ab used in this study, horse anti-mouse IgG, had novel conjugation and micropolymer chemistries to create a highly sensitive, ready-to-use, one-step, non-biotin detection system for immunohistochemistry staining. The color was then developed with 3,3′-diaminobenzidine (DAB) and counterstained with Mayer’s hematoxylin. The DAB developing time should be determined by the investigator under a microscope, but generally, optimal staining intensity is produced in 2–10 min. DAB incubation on the slides was about 2 min in this staining. Finally, the slides were observed under an Eclipse E400 microscope (Nikon Instruments Inc., Melville, New York, NY, USA) and images were captured with a Nikon Digital Sight DS-U2 camera. The integral optical density (IOD) of LFB and MBP were analyzed by i-SOLUTION software (Image & Microscope Technology Inc., Burnaby, BC, Canada) by extracting nuclei from photos [58]. For Luxol fast blue stained sections, the area of the deep sky-blue signal and its intensity were assessed using the IOD analyzed by i-SOLUTION software. The brown coloring was regarded as a positive signal of MBP immunoreactivity, and its intensity of MBP immunoreactivity was evaluated by IOD. Nuclei were excluded from the analysis of IOD. Each section taken to obtain a mean optical density value had images at 400× magnification.
4.6. RNA Isolation and RNA Quality Check for Affymetrix miRNA Arrays
Total RNA was extracted from the cerebrum of normal/CPZ-paired mice using the easy-BLUETM Total RNA Extraction Kit (iNtRON biotechnology, Daejeon, Korea) according to the manufacturer’s instructions. RNA purity and integrity were evaluated using an ND-1000 Spectrophotometer (NanoDrop, Wilmington, NC, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), respectively.
4.7. Affymetrix miRNA Arrays
Affymetrix Genechip miRNA 4.0 array processing was performed according to the manufacturer’s protocol. RNA samples (1000 ng) were labeled with the FlashTag™ Biotin RNA Labeling Kit (Genisphere, Hatfield, PA, USA). The labeled RNA was quantified, fractionated, and hybridized in the miRNA microarray according to the standard procedures recommended by the manufacturer. The labeled RNA was heated to 99 °C for 5 min and then to 45 °C for 5 min. RNA-array hybridization was performed with agitation at 60 rotations per minute for 16 h at 48 °C on an Affymetrix® 450 Fluidics Station. The chips were washed and stained using a Genechip Fluidics Station 450 (Affymetrix, Santa Clara, CA, USA). The chips were then scanned with an Affymetrix GCS 3000 scanner (Affymetrix). Signal values were computed using the Affymetrix® GeneChip™ Command Console software.
4.8. Raw Data Preparation and Statistical Analysis of Affymetrix miRNA Arrays
Raw data were extracted automatically with the Affymetrix data extraction protocol using the Affymetrix GeneChip® Command Console® Software (AGCC) software. The CEL files and miRNA level quantified by RMA+DABG-All analysis were imported, and the results were exported using Affymetrix® Power Tools (APT) software. The array data were filtered to select probes annotated to the mouse genome. The comparative analysis between the test sample and the control sample was based on fold-change. Hierarchical cluster analysis was performed on the differentially expressed miRNAs using complete linkage and the Euclidean distance as a measure of similarity. Statistical tests and the visualization of differentially expressed genes were conducted using the R statistical language v. 3.1.2 (https://www.r-project.org/). In addition, miRNA expression was analyzed using the MORPHEUS tutorial program (MACROGEN, Seoul, Korea).
4.9. Statistical Analysis
Data are shown as mean ± standard deviation, and significant differences were assessed using the one-way analysis of variance (ANOVA) with the post hoc test and student’s t-test using SPSS for Windows, release 17.0 (SPSS Inc., Chicago, IL, USA). Repeated-measure analysis of variance was also used to evaluate accustomization in the open-field test, the rota-rod test, and grip strength test. The level of statistical significance stated in the text is based on the p-values * p < 0.01, ** p < 0.005, or *** p < 0.001 which were considered statistically significant.
4.10. Real-Time PCR for miRNA Quantitation and Target Genes of miRNAs
Oligonucleotide sequences corresponding to the miR146a-5p, miR20a-5p, miR155-5p, miR145a-5p, miR219a-2-3p, and miR338-5p were mimicked on miRDB sequences, and the TGFβ1, TGFβR1, TGFβR2, Id1, and NgR gene were designed using Primer3 software (http://frodo.wi.mit.edu). The first-strand cDNA mixture contained 1.0 μg of total RNA as a template for miRNA quantitative PCR. The Mir-XTM miRNA First-strand Synthesis and TB GreenTM kits (TaKaRa, Daejeon, Korea) were used to perform real-time PCR according to the manufacturer’s protocol. All primer sequences have been described in Supplementary Table S1. Optimized real-time PCR was carried out as a previous study [35]. Relative miRNA expression levels were nomalized to U6 expression, and relative gene expression levels were normalized to GAPDH expression.
4.11. Western Blot Analysis
Western blot analysis of miRNA target proteins was performed according to the previous report [35]. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was conducted using a Mini-PROTEIN® System (Bio-Rad, Hercules, CA., USA) and a 12% gel according to the manufacturer’s protocol. Proteins were transferred to a nitrocellulose blotting membrane and probed overnight at 4 °C with primary antibodies (1:1000, pSmad 2, Smad 2, pSmad 3, Smad 3, and Smad 4, Bioss; Id1, Santa Cruz; β-actin, Sigma-Aldrich) followed by 1 h at RT by an HRP-conjugated secondary antibody (1:2000, rabbit anti-mouse HRP and goat anti-rabbit HRP, Santa Cruz). Immunolabeled proteins were detected by incubation with enhanced chemiluminescence (ECL) substrate, followed by exposure of the membrane to autoradiography film.
5. Conclusions
In conclusion, our results indicate that behavioral deficits follow the course of demyelination induced by administration of 0.2% CPZ, and that some of the changes do not persist after 6 weeks on a normal diet. In particular, the expression of 240 miRNAs was significantly changed in CPZ-fed mice compared with control mice. In particular, miR 155-5p and miR 20a-5p upregulations may induce demyelination through upregulation of NgR via suppression of Smad signal cascades in CPZ-fed mice. Taken together, these results suggest that the changes in miRNA expression in vivo are new potential remyelination therapeutic targets.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/2/646/s1.
Author Contributions
Conceptualization, S.R.H., Y.H.K., M.-S.L. and S.-H.L.; Data curation, S.R.H., Y.H.K.; Formal analysis, S.R.H., Y.H.K., H.J., S.L., S.-J.P., D.-Y.S., S.S.M., S.-M.Y., M.-S.L. and S.-H.L.; Funding acquisition, Y.H.K., S.-M.Y., M.-S.L. and S.-H.L.; Investigation, S.R.H., Y.H.K., M.-S.L. and S.-H.L.; Methodology, S.R.H. and Y.H.K.; Validation, S.R.H. and Y.H.K.; Visualization, S.R.H. and Y.H.K.; Writing—Original draft, S.R.H. and Y.H.K.; Writing—Review & editing, Y.H.K., M.-S.L. and S.-H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Mid-Career Researcher Program and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korea government (2016R1A2B4007413, 2017R1A2B4002405, 2017R1A2B1006373, 2019R1A2C2083947, 2017R1D1A1B03031502).
Acknowledgments
We would like to thank Tai Kyung Baik (Department of Anatomy and Neuroscience, Eulji University School of Medicine) for helpful discussion of proposed illustration in this study and Sang-Hak Lee (A solution, Suwon, Korea) for help of IOD analysis.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Data Availability
The miRNA array data obtained in this study have been deposited in the NCBI’s Gene Expression Omnibus (GEO) repository and are accessible through the GEO Series accession number GSE122839 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122839). Other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Sen, M.K.; Mahns, D.A.; Coorssen, J.R.; Shortland, P.J. Behavioural phenotypes in the cuprizone model of central nervous system demyelination. Neurosci. Biobehav. Rev. 2019, 107, 23–46. [Google Scholar] [CrossRef] [PubMed]
- Lucchinetti, C.F.; Bruck, W.; Rodriguez, M.; Lassmann, H. Distinct patterns of multiple sclerosis pathology indicates heterogeneity on pathogenesis. Brain Pathol. 1996, 6, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Lucchinetti, C.; Bruck, W.; Parisi, J.; Scheithauer, B.; Rodriguez, M.; Lassmann, H. Heterogeneity of multiple sclerosis lesions: Implications for the pathogenesis of demyelination. Ann. Neurol. 2000, 47, 707–717. [Google Scholar] [CrossRef]
- Patrikios, P.; Stadelmann, C.; Kutzelnigg, A.; Rauschka, H.; Schmidbauer, M.; Laursen, H.; Sorensen, P.S.; Bruck, W.; Lucchinetti, C.; Lassmann, H. Remyelination is extensive in a subset of multiple sclerosis patients. Brain 2006, 129, 3165–3172. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, T.; Antel, J.; Konig, F.B.; Bruck, W.; Kuhlmann, T. Remyelination capacity of the MS brain decreases with disease chronicity. Neurology 2009, 72, 1914–1921. [Google Scholar] [CrossRef]
- Sen, M.K.; Almuslehi, M.S.M.; Gyengesi, E.; Myers, S.J.; Shortland, P.J.; Mahns, D.A.; Coorssen, J.R. Suppression of the Peripheral Immune System Limits the Central Immune Response Following Cuprizone-Feeding: Relevance to Modelling Multiple Sclerosis. Cells 2019, 8, 1314. [Google Scholar] [CrossRef]
- Liu, L.; Belkadi, A.; Darnall, L.; Hu, T.; Drescher, C.; Cotleur, A.C.; Padovani-Claudio, D.; He, T.; Choi, K.; Lane, T.E.; et al. CXCR2-positive neutrophils are essential for cuprizone-induced demyelination: Relevance to multiple sclerosis. Nat. Neurosci. 2010, 13, 319–326. [Google Scholar] [CrossRef]
- Pasquini, L.A.; Calatayud, C.A.; Bertone Una, A.L.; Millet, V.; Pasquini, J.M.; Soto, E.F. The neurotoxic effect of cuprizone on oligodendrocytes depends on the presence of pro-inflammatory cytokines secreted by microglia. Neurochem. Res. 2007, 32, 279–292. [Google Scholar] [CrossRef]
- Suzuki, K.; Kikkawa, Y. Status spongiosus of CNS and hepatic changes induced by cuprizone (biscyclohexanone oxalyldihydrazone). Am. J. Pathol. 1969, 54, 307–325. [Google Scholar]
- Skripuletz, T.; Lindner, M.; Kotsiari, A.; Garde, N.; Fokuhl, J.; Linsmeier, F.; Trebst, C.; Stangel, M. Cortical demyelination is prominent in the murine cuprizone model and is strain-dependent. Am. J. Pathol. 2008, 172, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.C.; Gilmore, W.; Matsushima, G.K. SJL mice exposed to cuprizone intoxication reveal strain and gender pattern differences in demyelination. Brain Pathol. 2009, 19, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.C.; Gilmore, W.; Ting, J.P.; Matsushima, G.K. Cuprizone induces similar demyelination in male and female C57BL/6 mice and results in disruption of the estrous cycle. J. Neurosci. Res. 2010, 88, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Gudi, V.; Moharregh-Khiabani, D.; Skripuletz, T.; Koutsoudaki, P.N.; Kotsiari, A.; Skuljec, J.; Trebst, C.; Stangel, M. Regional differences between grey and white matter in cuprizone induced demyelination. Brain Res. 2009, 1283, 127–138. [Google Scholar] [CrossRef]
- Stidworthy, M.F.; Genoud, S.; Suter, U.; Mantei, N.; Franklin, R.J. Quantifying the early stages of remyelination following cuprizone-induced demyelination. Brain Pathol. 2003, 13, 329–339. [Google Scholar] [CrossRef]
- Kipp, M.; Clarner, T.; Dang, J.; Copray, S.; Beyer, C. The cuprizone animal model: New insights into an old story. Acta Neuropathol. 2009, 118, 723–736. [Google Scholar] [CrossRef]
- Hiremath, M.M.; Saito, Y.; Knapp, G.W.; Ting, J.P.; Suzuki, K.; Matsushima, G.K. Microglial/macrophage accumulation during cuprizone-induced demyelination in C57BL/6 mice. J. Neuroimmunol. 1998, 92, 38–49. [Google Scholar] [CrossRef]
- Koutsoudaki, P.N.; Skripuletz, T.; Gudi, V.; Moharregh-Khiabani, D.; Hildebrandt, H.; Trebst, C.; Stangel, M. Demyelination of the hippocampus is prominent in the cuprizone model. Neurosci. Lett. 2009, 451, 83–88. [Google Scholar] [CrossRef]
- Pott, F.; Gingele, S.; Clarner, T.; Dang, J.; Baumgartner, W.; Beyer, C.; Kipp, M. Cuprizone effect on myelination, astrogliosis and microglia attraction in the mouse basal ganglia. Brain Res. 2009, 1305, 137–149. [Google Scholar] [CrossRef]
- Blakemore, W.F. Demyelination of the superior cerebellar peduncle in the mouse induced by cuprizone. J. Neurol. Sci. 1973, 20, 63–72. [Google Scholar] [CrossRef]
- Ludwin, S.K. Central nervous system demyelination and remyelination in the mouse: An ultrastructural study of cuprizone toxicity. Lab. Investig. J. Tech. Methods Pathol. 1978, 39, 597–612. [Google Scholar]
- Groebe, A.; Clarner, T.; Baumgartner, W.; Dang, J.; Beyer, C.; Kipp, M. Cuprizone treatment induces distinct demyelination, astrocytosis, and microglia cell invasion or proliferation in the mouse cerebellum. Cerebellum 2009, 8, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Skripuletz, T.; Bussmann, J.H.; Gudi, V.; Koutsoudaki, P.N.; Pul, R.; Moharregh-Khiabani, D.; Lindner, M.; Stangel, M. Cerebellar cortical demyelination in the murine cuprizone model. Brain Pathol. 2010, 20, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, G.K.; Morell, P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001, 11, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Blakemore, W.F. Observations on oligodendrocyte degeneration, the resolution of status spongiosus and remyelination in cuprizone intoxication in mice. J. Neurocytol. 1972, 1, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Bando, Y.; Takakusaki, K.; Ito, S.; Terayama, R.; Kashiwayanagi, M.; Yoshida, S. Differential changes in axonal conduction following CNS demyelination in two mouse models. Eur. J. Neurosci. 2008, 28, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Duncan, I.D.; Brower, A.; Kondo, Y.; Curlee, J.F., Jr.; Schultz, R.D. Extensive remyelination of the CNS leads to functional recovery. Proc. Natl. Acad. Sci. USA 2009, 106, 6832–6836. [Google Scholar] [CrossRef]
- Irvine, K.A.; Blakemore, W.F. Remyelination protects axons from demyelination-associated axon degeneration. Brain 2008, 131, 1464–1477. [Google Scholar] [CrossRef]
- Arnett, H.A.; Mason, J.; Marino, M.; Suzuki, K.; Matsushima, G.K.; Ting, J.P. TNF alpha promotes proliferation of oligodendrocyte progenitors and remyelination. Nat. Neurosci. 2001, 4, 1116–1122. [Google Scholar] [CrossRef]
- Mi, S.; Miller, R.H.; Tang, W.; Lee, X.; Hu, B.; Wu, W.; Zhang, Y.; Shields, C.B.; Zhang, Y.; Miklasz, S.; et al. Promotion of central nervous system remyelination by induced differentiation of oligodendrocyte precursor cells. Ann. Neurol. 2009, 65, 304–315. [Google Scholar] [CrossRef]
- Plant, S.R.; Arnett, H.A.; Ting, J.P. Astroglial-derived lymphotoxin-alpha exacerbates inflammation and demyelination, but not remyelination. Glia 2005, 49, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, P.; Liang, F.; Huang, W. The microstructural status of the corpus callosum is associated with the degree of motor function and neurological deficit in stroke patients. PLoS ONE 2015, 10, e0122615. [Google Scholar] [CrossRef] [PubMed]
- Ghadiri, N.; Emamnia, N.; Ganjalikhani-Hakemi, M.; Ghaedi, K.; Etemadifar, M.; Salehi, M.; Shirzad, H.; Nasr-Esfahani, M.H. Analysis of the expression of mir-34a, mir-199a, mir-30c and mir-19a in peripheral blood CD4+T lymphocytes of relapsing-remitting multiple sclerosis patients. Gene 2018, 659, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Vistbakka, J.; Sumelahti, M.L.; Lehtimaki, T.; Elovaara, I.; Hagman, S. Evaluation of serum miR-191-5p, miR-24-3p, miR-128-3p, and miR-376c-3 in multiple sclerosis patients. Acta Neurol. Scand. 2018, 138, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Han, S.R.; Jeon, H.; Lee, S.; Lee, J.; Yoo, S.M.; Park, J.B.; Park, M.J.; Kim, J.T.; Lee, H.G.; et al. Nogo receptor-vimentin interaction: A novel mechanism for the invasive activity of glioblastoma multiforme. Exp. Mol. Med. 2019, 51, 125. [Google Scholar] [CrossRef] [PubMed]
- Prineas, J.W. Pathology of inflammatory demyelinating neuropathies. Bailliere’s Clin. Neurol. 1994, 3, 1–24. [Google Scholar]
- Morell, P.; Barrett, C.V.; Mason, J.L.; Toews, A.D.; Hostettler, J.D.; Knapp, G.W.; Matsushima, G.K. Gene expression in brain during cuprizone-induced demyelination and remyelination. Mol. Cell Neurosci. 1998, 12, 220–227. [Google Scholar] [CrossRef]
- Franco-Pons, N.; Torrente, M.; Colomina, M.T.; Vilella, E. Behavioral deficits in the cuprizone-induced murine model of demyelination/remyelination. Toxicol. Lett. 2007, 169, 205–213. [Google Scholar] [CrossRef]
- Xiao, C.; Rajewsky, K. MicroRNA control in the immune system: Basic principles. Cell 2009, 136, 26–36. [Google Scholar] [CrossRef]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Wardle, G.S.; Tuschl, T.; Patel, D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 2009, 461, 754–761. [Google Scholar] [CrossRef]
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009, 457, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Talebi, F.; Ghorbani, S.; Chan, W.F.; Boghozian, R.; Masoumi, F.; Ghasemi, S.; Vojgani, M.; Power, C.; Noorbakhsh, F. MicroRNA-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J. Neuroinflamm. 2017, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.B.; Chen, L.X.; Qu, X.B.; Ren, C.L.; Wu, X.X.; Dong, F.X.; Zhang, B.L.; Gao, D.S.; Yao, R.Q. Transplanted miR-219-overexpressing oligodendrocyte precursor cells promoted remyelination and improved functional recovery in a chronic demyelinated model. Sci. Rep. 2017, 7, 41407. [Google Scholar] [CrossRef] [PubMed]
- Hoye, M.L.; Archambault, A.S.; Gordon, T.M.; Oetjen, L.K.; Cain, M.D.; Klein, R.S.; Crosby, S.D.; Kim, B.S.; Miller, T.M.; Wu, G.F. MicroRNA signature of central nervous system-infiltrating dendritic cells in an animal model of multiple sclerosis. Immunology 2018, 155, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Mycko, M.P.; Cichalewska, M.; Cwiklinska, H.; Selmaj, K.W. miR-155-3p Drives the Development of Autoimmune Demyelination by Regulation of Heat Shock Protein 40. J. Neurosci. Off. J. Soc. Neurosci. 2015, 35, 16504–16515. [Google Scholar] [CrossRef]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef]
- Murugaiyan, G.; Beynon, V.; Mittal, A.; Joller, N.; Weiner, H.L. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 2011, 187, 2213–2221. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.G.; Lu, M.; Wang, X.; Shang, X.; Elias, S.B.; Chopp, M. MiR-146a promotes remyelination in a cuprizone model of demyelinating injury. Neuroscience 2017, 348, 252–263. [Google Scholar] [CrossRef]
- Martin, N.A.; Molnar, V.; Szilagyi, G.T.; Elkjaer, M.L.; Nawrocki, A.; Okarmus, J.; Wlodarczyk, A.; Thygesen, E.K.; Palkovits, M.; Gallyas, F., Jr.; et al. Experimental Demyelination and Axonal Loss Are Reduced in MicroRNA-146a Deficient Mice. Front. Immunol. 2018, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.; Lauterbach-Soon, B.; Hattingen, E.; Jung, P.; Singer, O.; Volz, S.; Klein, J.C.; Steinmetz, H.; Ziemann, U. Human motor corpus callosum: Topography, somatotopy, and link between microstructure and function. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 12132–12138. [Google Scholar] [CrossRef] [PubMed]
- Tomas-Roig, J.; Wirths, O.; Salinas-Riester, G.; Havemann-Reinecke, U. The Cannabinoid CB1/CB2 Agonist WIN55212.2 Promotes Oligodendrocyte Differentiation In Vitro and Neuroprotection During the Cuprizone-Induced Central Nervous System Demyelination. CNS Neurosci. Ther. 2016, 22, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Miller, R.H.; Lee, X.; Scott, M.L.; Shulag-Morskaya, S.; Shao, Z.; Chang, J.; Thill, G.; Levesque, M.; Zhang, M.; et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat. Neurosci. 2005, 8, 745–751. [Google Scholar] [CrossRef]
- Petratos, S.; Azari, M.F.; Ozturk, E.; Papadopoulos, R.; Bernard, C.C. Novel therapeutic targets for axonal degeneration in multiple sclerosis. J. Neuropathol. Exp. Neurol. 2010, 69, 323–334. [Google Scholar] [CrossRef]
- Steelman, A.J.; Thompson, J.P.; Li, J. Demyelination and remyelination in anatomically distinct regions of the corpus callosum following cuprizone intoxication. Neurosci. Res. 2012, 72, 32–42. [Google Scholar] [CrossRef]
- Jeon, H.; Han, S.R.; Lee, S.; Park, S.J.; Kim, J.H.; Yoo, S.M.; Lee, M.S. Activation of the complement system in an osteosarcoma cell line promotes angiogenesis through enhanced production of growth factors. Sci. Rep. 2018, 8, 5415. [Google Scholar] [CrossRef]
- Underhill, H.R.; Rostomily, R.C.; Mikheev, A.M.; Yuan, C.; Yarnykh, V.L. Fast bound pool fraction imaging of the in vivo rat brain: Association with myelin content and validation in the C6 glioma model. NeuroImage 2011, 54, 2052–2065. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).