Abstract
Gastric cancer (GC) represents a notable amount of morbidity and mortality worldwide. Understanding the molecular basis of CG will offer insight into its pathogenesis in an attempt to identify new molecular biomarkers to early diagnose this disease. Therefore, studies involving small non-coding RNAs have been widely explored. Among these, PIWI-interacting RNAs (piRNAs) are an emergent class that can play important roles in carcinogenesis. In this study, small-RNA sequencing was used to identify the global piRNAs expression profile (piRNome) of gastric cancer patients. We found 698 piRNAs in gastric tissues, 14 of which were differentially expressed (DE) between gastric cancer (GC), adjacent to gastric cancer (ADJ), and non-cancer tissues (NC). Moreover, three of these DE piRNAs (piR-48966*, piR-49145, piR-31335*) were differently expressed in both GC and ADJ samples in comparison to NC samples, indicating that the tumor-adjacent tissue was molecularly altered and should not be considered as a normal control. These three piRNAs are potential risk biomarkers for GC, especially piR-48966* and piR-31335*. Furthermore, an in-silico search for mRNAs targeted by the differentially expressed piRNAs revealed that these piRNAs may regulate genes that participate in cancer-related pathways, suggesting that these small non-coding RNAs may be directly and indirectly involved in gastric carcinogenesis.
1. Introduction
Gastric cancer (GC) is the fifth most common malignancy in the world and the third leading cause of cancer-related death worldwide [1]. Due to the lack of specific symptoms, most gastric cancer patients are diagnosed in advanced-stage disease with a poor prognosis [2,3]. Unfortunately, the molecular mechanism of gastric cancer formation and progression has not been completely elucidated [4,5].
GC exhibits a wide range of molecular alterations, including pathways involved in mitosis, immune signaling, cell adhesion, and migration [6,7]. These alterations have been described both in genetic and epigenetic levels [8,9] and many studies have deeply investigated these epigenetic alterations [10,11]. Particularly, the function of non-coding RNAs (ncRNAs) in cancer development and progression [12,13,14], mostly the role of miRNAs in post-transcriptional regulation, has been very well characterized [12,15,16].
PIWI-interacting RNAs (piRNAs) are a type of small ncRNAs of 26 to 31 nucleotides that act as a sequence-specific guide for PIWI proteins and have been shown to be implicated in the maintenance of genome integrity by acting as methylation regulators and gene controllers at transcriptional and posttranscriptional levels [17,18,19,20].
There are more than 30,000 piRNA isoforms described in the human genome, and it is believed that these molecules compose the most abundant and diverse regulatory noncoding RNA class [21,22,23]. Although for several years the functions to piRNAs were focused only in the genome integrity and development of germinal stem cells [24,25,26], the functions of piRNAs and PIWI proteins as epigenetic regulators have started to emerge, and a subset of piRNAs has also been implicated in the regulation of protein-coding genes [27,28,29,30].
Several studies have shown that the piRNAs are broadly expressed in germline cells. More recently, it has been described by high throughput sequencing that their signaling pathway is also active in somatic cells, especially in human cancers [30,31,32,33,34], suggesting their possible role in carcinogenesis. So far, piRNAs have been associated with some hallmarks of cancer, such as sustaining proliferative signaling, evading growth suppressors, activating invasion, and metastasis [35,36,37,38]. Furthermore, some cancer-associated piRNAs were found in a patient’s bloodstream [39], meaning that piRNAs may be potential biomarkers of the disease, although there are some discrepancies.
However, their involvement in gastric carcinogenesis is still poorly investigated and more studies are needed in attempting to elucidate it. This study aimed to investigate and compare the expression profile of piRNAs in gastric samples from patients without cancer, gastric cancer samples, and samples from the matched tumor-adjacent tissue and, moreover, to identify potential piRNA biomarkers of GC.
2. Results
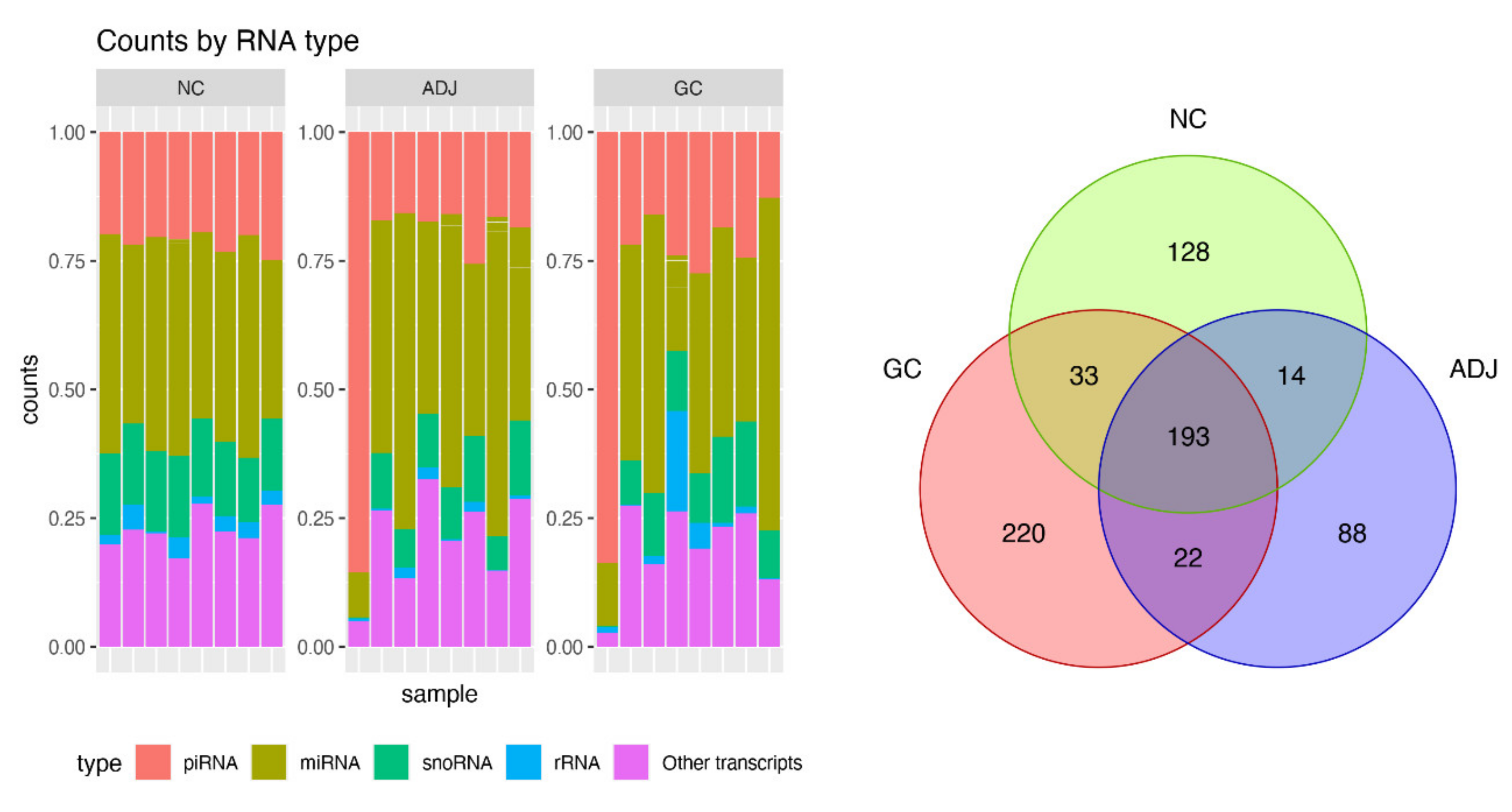
A high throughput small RNA-sequencing experiment was conducted in 24 gastric tissue samples. After trimming the adapters, quality control, alignment to human genome (hg19), and transcripts quantitation, we obtained, on average, 1.2 million reads per sample. Several small Non-coding RNAs and other transcripts fragments were identified (Figure 1—left panel) and from them, about 20% (~5 million reads) were recognized as piRNA reads, representing 698 different piRNAs. The global statistics on the reads are presented in Supplementary Materials Table S1.
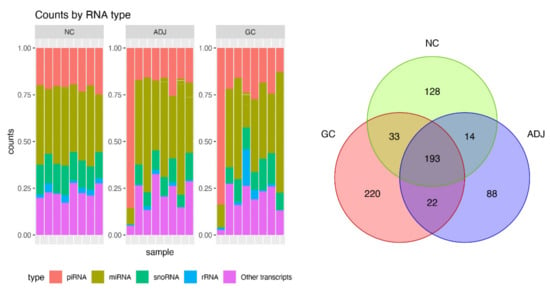
Figure 1.
Summary of small RNA sequencing data. Barplot of percent of small non-coding RNA (sncRNA) transcripts identified per sample (left panel) and Venn diagram for number of PIWI-interacting RNAs (piRNAs) detected by small RNA-Seq in three groups of gastric tissue samples. Gastric cancer (CG), matched tumor-adjacent gastric tissue (ADJ) and non-cancer (NC) (right panel).
Approximately 90% of recognized piRNAs had between 30 and 35 bp (Supplementary Figure S1), and 83% of the piRNAs reads were concentrated on the 20 most expressed piRNAs. As shown in Figure 1 (right panel), there were 193 piRNAs expressed in all sample groups and several that were group-exclusive.
2.1. Differential Expression Analysis
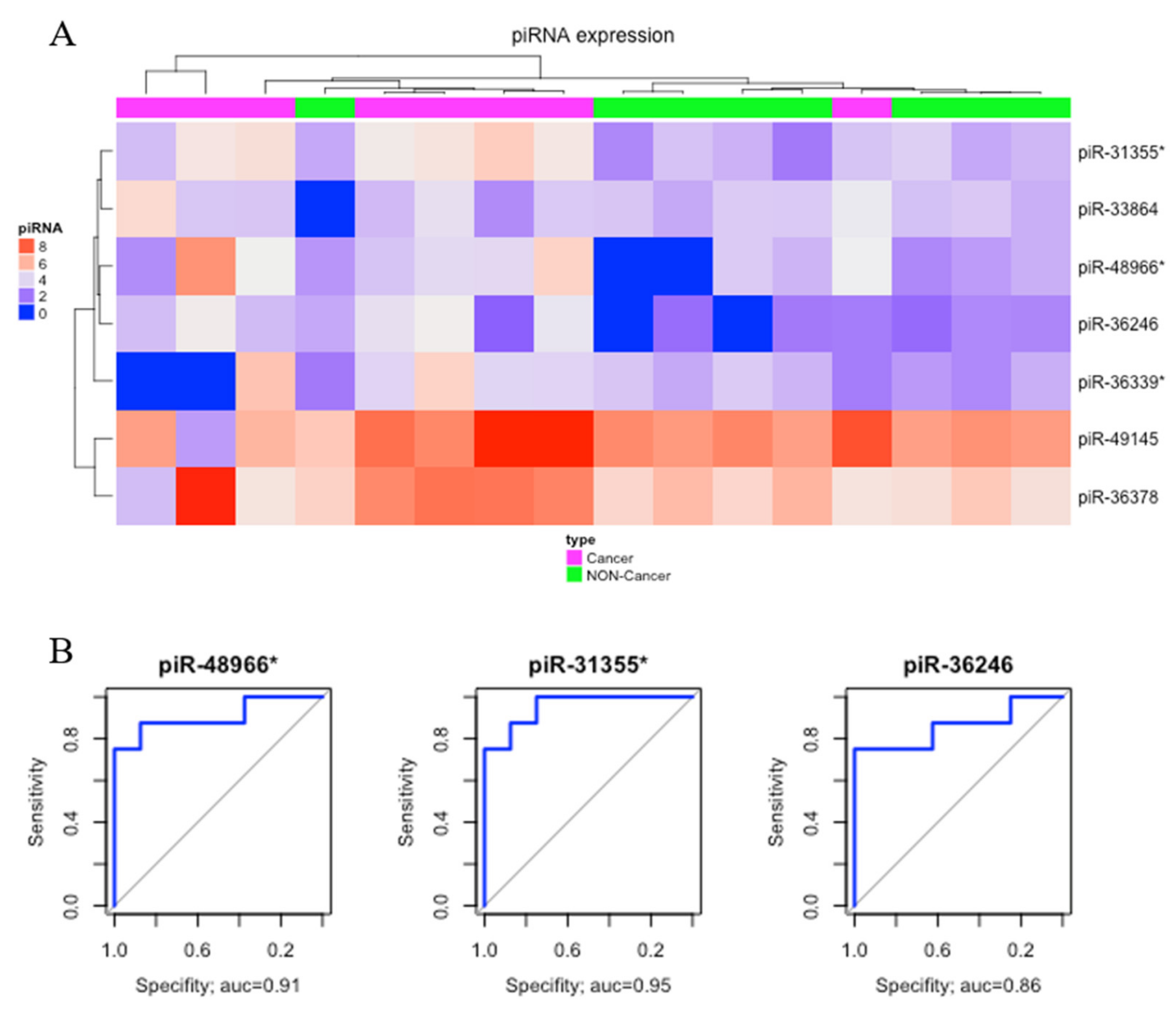
Comparing gastric cancer (GC) samples with gastric non-cancer (NC) samples, we found seven significantly upregulated piRNAs (Table 1). The heatmap obtained from this comparison is presented in Figure 2A. Three of these differentially expressed (DE) piRNAs (piR-48966*, piR31355*, and piR36246) showed AUC > 0.85 and were selected as potential biomarkers of gastric cancer (Figure 2B).

Table 1.
Differentially expressed piRNAs (adjusted p-value < 0.05; |fold-change| > 3) in each comparison.
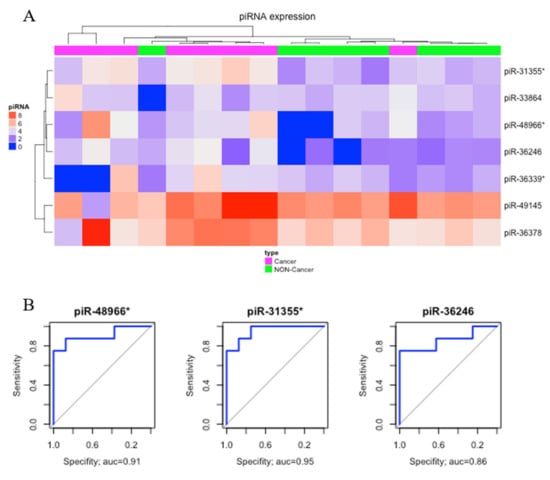
Figure 2.
Differentially expressed piRNAs between gastric cancer and non-cancer. (A) Hierarchic clustering heat map of differentially expressed piRNAs (p-value < 0.05; |fold-change| > 3). (B) The receiver operating characteristic (ROC) curve analysis of piRNAs that presented AUC > 0.85.
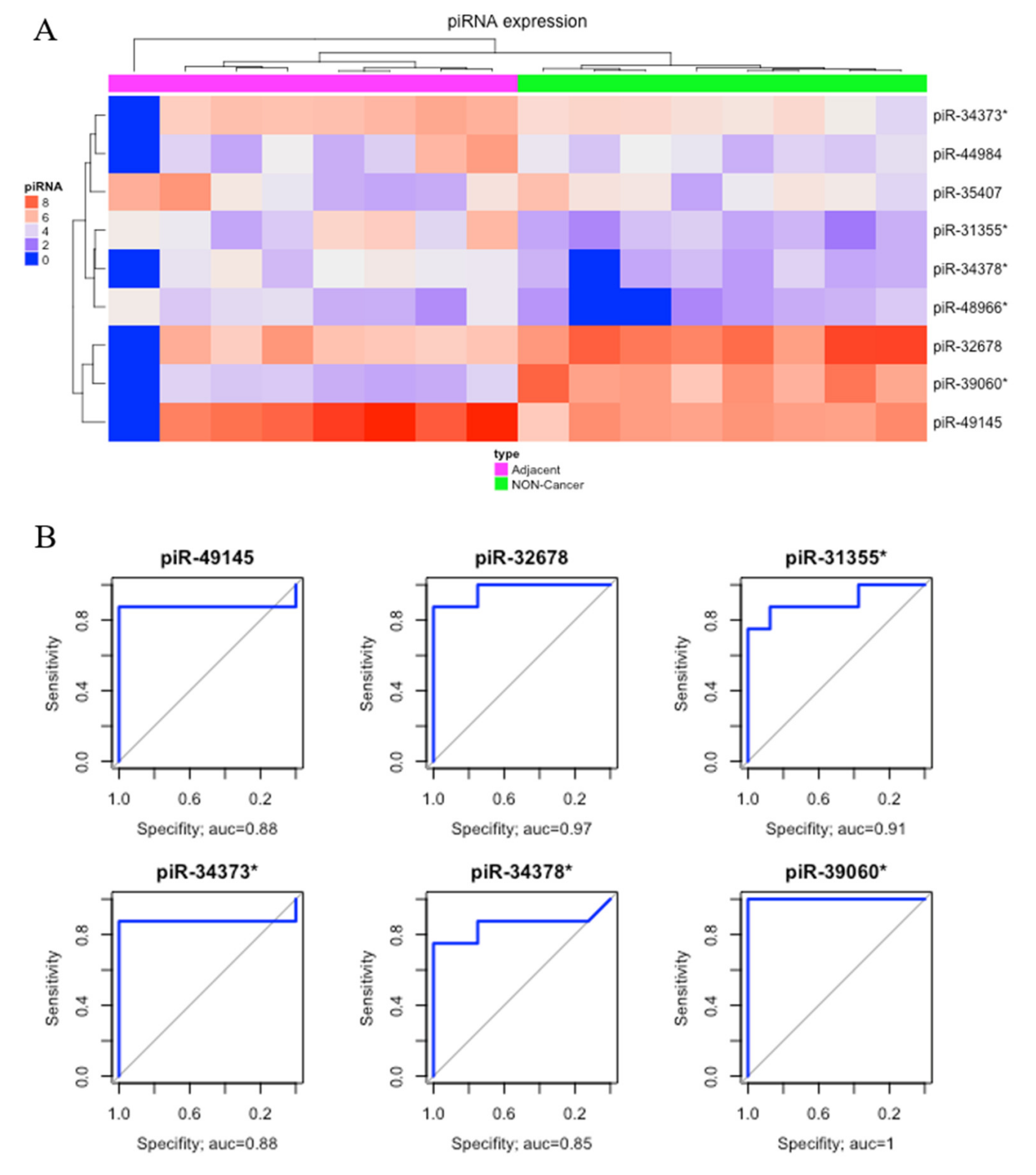
Comparing matched tumor-adjacent gastric tissue samples (ADJ) with NC, we found nine DE piRNAs, two of which were down-regulated and seven were up-regulated (Table 1). The heatmap obtained from this comparison was able to perfectly cluster ADJ and NC samples (Figure 3A). Six of these DE piRNAs (four up-regulated, piR-49145, piR-31355*, piR-34373*, and piR-34378*; and two down-regulated, piR-39060* and piR-32678) had the best sensibility/specificity relationship (AUC > 0.85) and were selected as potential biomarkers to allow early diagnosis of GC (Figure 3B).
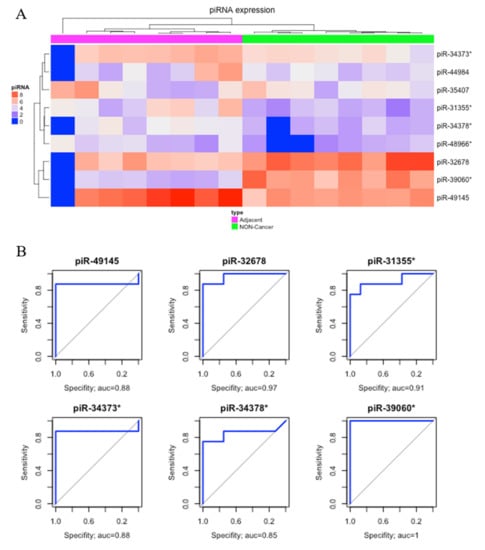
Figure 3.
Differentially expressed piRNAs between tumor-adjacent gastric tissue and non-cancer. (A) Hierarchical clustering heat map of differentially expressed piRNAs (p-value < 0.05; |fold-change| > 3). (B) The ROC analysis of differentially expressed piRNAs identified potential earlier gastric cancer biomarkers with the best sensibility/specificity relation (AUC > 0.85).
Since sample ADJ.100927_1B (first sample from left to right in Figure 3A) presented a different expression pattern, which might have been caused by technical issues, we double-checked the sample features, realized that it had a reasonable read count, and found that the identified pattern could also be related to a biological peculiarity. Aiming to verify the consequences of removing the sample, we re-analyzed the clustering steps, and no significant modification of results among the groups was found (Supplementary Figures S2 and S3).
Comparing GC with ADJ, we found three piRNAs (piR-36378, piR-33534*, and piR-39060*) that were significantly upregulated in GC (Table 1). However, the hierarchic clustering was not able to cluster GC and ADJ samples. The piRNA piR-39060* was also differentially expressed in the ADJ vs. NC analysis. piR-33534* and piR-36378 were exclusively differentially expressed in the GC vs. ADJ analysis, although none of these DE piRNAs presented high sensibility/specificity relation in the receiver operating characteristic (ROC) analysis (AUC < 0.85).
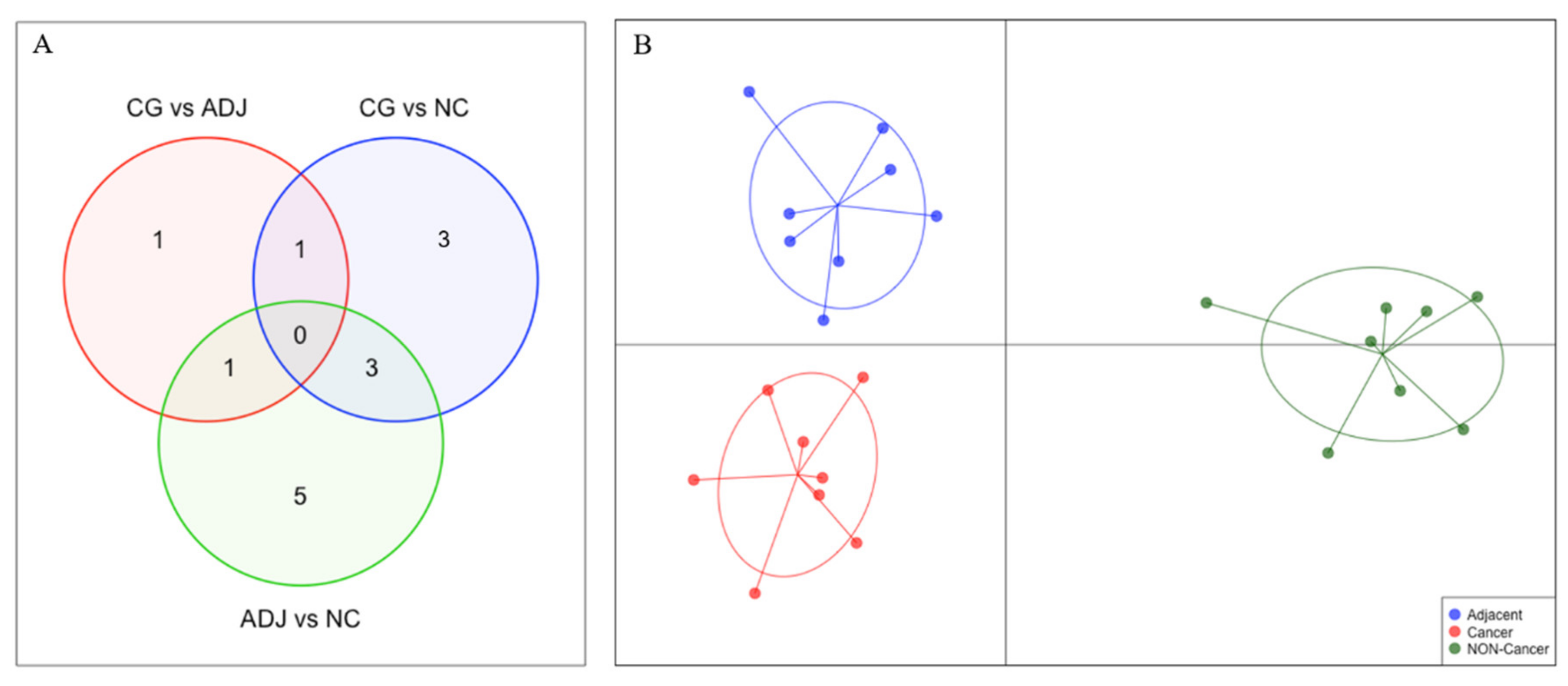
The small number of DE piRNAs in GC vs. ADJ suggests that these tissues have similar piRNA expression profiles. Using the discriminant analysis of principal components (DAPC) plot with all DE piRNAs, we observed that GC, ADJ, and NC samples generated distinct clusters, indicating expression differences among these tissues (Figure 4).
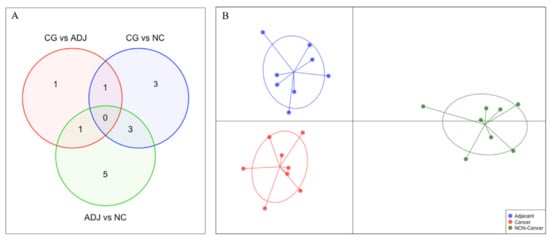
Figure 4.
Comparison of the differentially expressed piRNAs (|fold-change| > 3 and p-value < 0.05) among all three analyses. (A) Venn Diagram identifying the number of differentially expressed piRNAs in each comparison. (B) Discriminant analysis of principal components (DAPC) plot clustering samples based on differentially expressed piRNAs.
Table 1 data unveiled a group of piRNAs similarly altered in both GC and ADJ samples compared to NC samples, which may be involved in the process of field cancerization in the stomach (piR-48966* piR-49145 and piR-31355*).
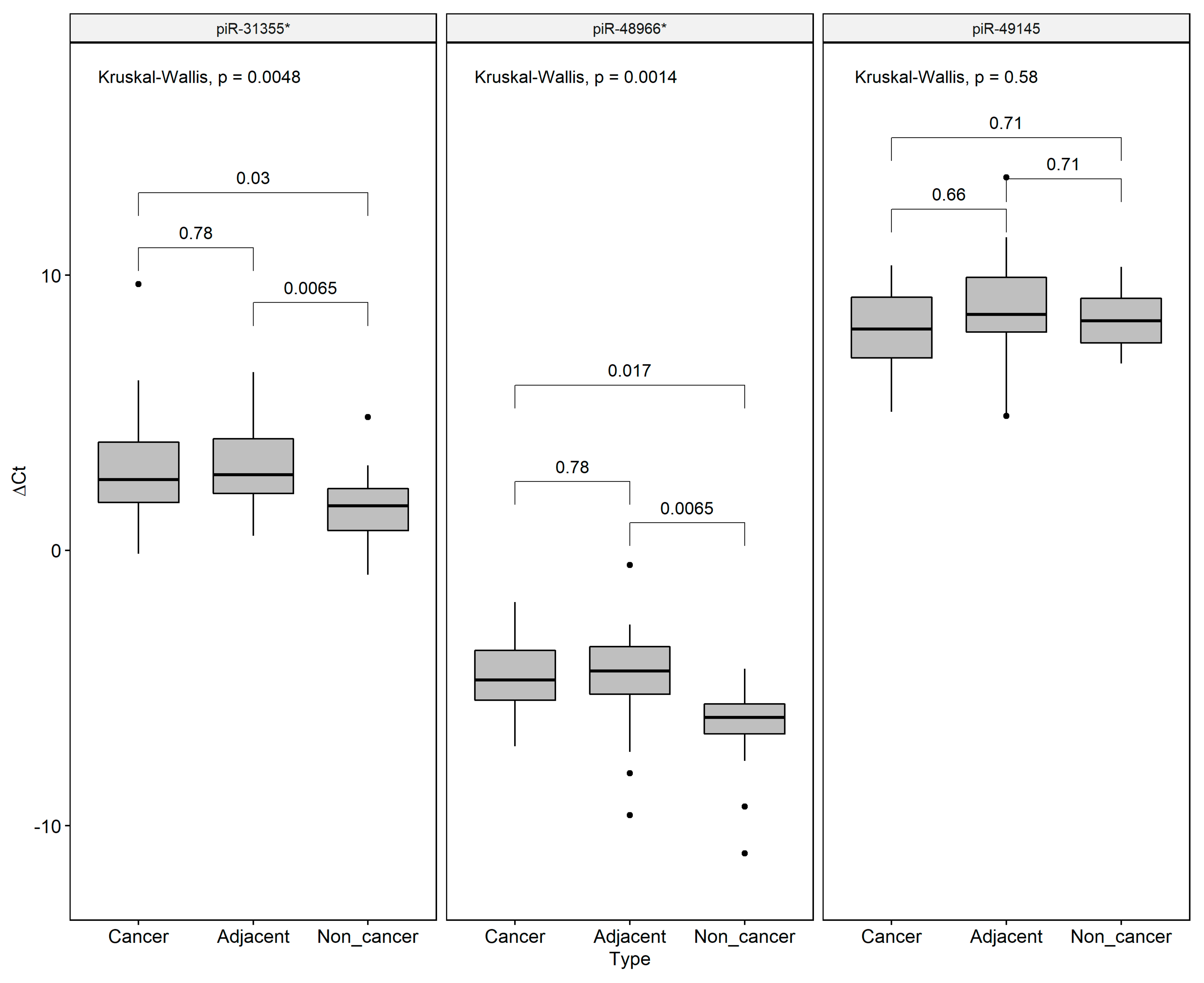
We decided to validate the field effect-related piRNAs by qRT-PCR, revealing that piR-31355* was overexpressed in both GC and ADJ when compared to NC samples, corroborating our RNA-Seq results. We also found a significant difference in piR-48966* expression between ADJ and NC samples. No difference was found for pir-49145 expression among the groups (Figure 5).
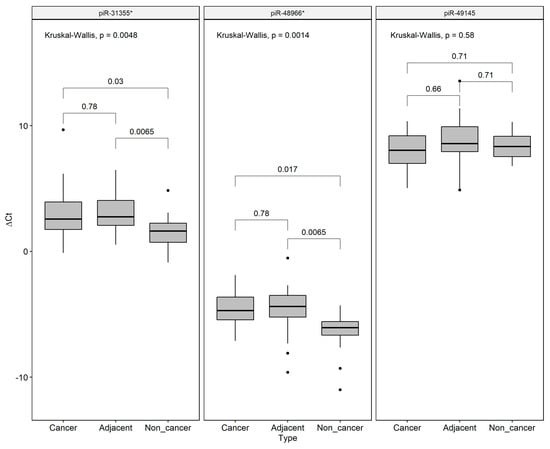
Figure 5.
Expression values (∆Ct) of the field effect-related piRNAs (piR-48966* piR-49145 and piR-31355*) in gastric cancer, matched tumor-adjacent gastric tissue, and non-cancer samples, respectively. U6 Small Nuclear RNA (RNU6) was used as an endogenous control. *p-values were adjusted to multiple comparisons by Bonferroni’s correction. Individual black points (•) are outliers.
2.2. Identification of the Cancer-Related piRNAs Differentially Expressed Target Genes
We performed two approaches to identify target genes of the differentially expressed piRNAs. First, considering that mature piRNAs in cytoplasm form a complex with PIWI proteins and migrate back into the nucleus, reaching their target transcripts and mobilizing the silencing machinery to block the transcription of DNA, we performed a BLAST search using a mature piRNA sequence as a query. We identified 33 genes that exhibited at least 13 contiguous nucleotides complementary matches (supplementary Table S3). Among these 33 genes found, we found eight cancer-related genes, which seem to be regulated by five DE piRNAs.
We used a Miranda tool to predict biologically relevant RNA–RNA interactions between each DE piRNA and the cancer-related genes. Interestingly, we found more than one match between the DE piRNAs and their target genes in most of the cases (Table 2), suggesting that these genes may be regulated by the pi-RISC complex at the transcriptional level. Using RNA-Seq and small RNA-Seq data from 64 paired tissues from ENCODE, we correlated the expression of DE piRNAs and their possible target genes, revealing a negative correlation between PIKFYVE and piR-31355* (r = −0.27, p = 0.028) (Supplementary Figure S4).

Table 2.
Complementarity between the DE piRNAs and some cancer-related genes at the transcriptional level.
Second, to identify the possible targets of the DE piRNAs at the post-transcriptional level, we considered that piRNAs can form specific RNA silencing complexes (pi-RISC) capable of promoting mRNA repression via imperfect base-pairing between the two RNAs, by a mechanism that closely resembles the regulation by miRNAs. Thus, we searched, within UTR3′, UTR5′, and CDS transcript databases, all RNAs that, according to this mechanism, may be potential targets for the 14 DE piRNAs. We found 2248 RNA–RNA interactions between piRNA and mRNAs that corresponded to 967 genes from the human genome. To each piRNA, we found a complementarity to range from 2 to 252 genes (Table 3).

Table 3.
Complementarity of DE piRNAs with mRNAs of human genes at the post-transcriptional level.
We performed the functional analysis by grouping all post-transcriptional target genes of the DE piRNAs. This approach revealed that several proteins encoded by these putative piRNA-target mRNAs are involved in key cellular processes in cancer, including cell adhesion, signaling and interaction, and cell morphogenesis (Supplementary Tables S4 and S5). Using the genes involved in these biological processes, we made a Kyotto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway analysis. Interestingly, we found that several piRNA target genes participate in important molecular pathways in cancer (Table 4).

Table 4.
Kyotto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway to target genes of piRNAs.
3. Discussion
Studies in association with small non-coding RNAs have been widely explored in cancer research, aiming to reveal novel tumor biomarkers. Among these species of RNAs, the piRNAs are an emergent class, which need further studies to elucidate their roles in carcinogenesis [40,41,42,43]. PIWI-interacting RNAs (piRNAs) are known to be related to the maintenance of genomic integrity, and, for a long time, their expression was associated with restriction to the stem cells [24,25,27,44] and reproductive cells in mammals [33,45,46]. However, an increased number of studies have demonstrated that piRNAs are also found in somatic cells and cancer, regulating gene expression at transcriptional and post-transcriptional levels by epigenetic mechanisms [23,38,47,48]. Using deep sequencing technology to analyze the piRNA expression pattern in gastric cancer, we found 698 expressed piRNAs, confirming their presence and abundance in gastric somatic cells, and 14 of them are associated with gastric cancer.
Although piRNA expression had been previously demonstrated, its role in gastric cancer is still unclear [23,40]. Cheng et al. (2011) demonstrated the deregulation of piRNAs in gastric cancer and in vitro modulation of these isoforms leads to cell cycle arrest in gastric tumor cells [35,49]. Martinez et al. (2016) described the expression pattern of 273 of the ~20,000 known piRNAs in tumor tissues, reporting that most of the deregulated piRNAs were inserted into protein-coding regions and that some piRNA isoforms are associated with recurrence-free patient survival [38]. Recently, Lin et al. (2019) reported 50 piRNA isoforms associated with gastric cancer across the whole transcriptome, 60% of which were mapping within protein-coding regions [50]. In the present study, we compared three types of gastric tissue (NC, ADJ, and GC) and identified 14 differentially expressed piRNAs.
There are not many studies concerning piRNA expression in cancer, especially in gastric tumors [23,50,51]. However, one of the DE piRNAs found in the present study, piR-35407, was once described as down-expressed in breast cancer [36]. In gastric cancer, this piRNA was found overexpressed in comparison to NC and ADJ, but no differences in its expression were found between GC and ADJ. Most of the DE piRNAs described here were not yet described as cancer-related in the literature. This may be due to our study design, which was the first to include the three types of gastric tissue.
Considering that epigenetic alterations could potentially be detected in the early stages of carcinogenesis [9,40,52], our analysis, comparing the NC and ADJ tissues, allows for better comprehension of tumor progression and highlights alterations that could be used as prognostic biomarkers. Although a few studies have demonstrated some discrepancies of piRNA expression between blood and cancer tissues, their potential as cancer biomarkers is remarkable [43,53]. In this study, we found eight possible piRNAs and some that are exclusive to GC and ADJ, such as piR-31355* (Figure 2B and Figure 3B).
When searching for biomarkers, it is important to consider that cancer is a complex disease that involves epigenetic microenvironment factors and somatic and hereditary mutations. A few studies have demonstrated that, besides their normal histological features, the adjacent tissue, which is the tissue surrounding the tumor, presents molecular alterations, which are very similar to those observed in cancer tissues, suggesting the presence of a field-effect [54,55]. Here, using next generation sequencing (NGS) data, we found three piRNAs that suggest the field effect in gastric cancer (piR-48966, piR-49145, and piR-31335). Their expressions between ADJ and GC are different in comparison to NC, but are similar between each other, indicating that piRNA expression levels are aberrant in adjacent-tissue samples. We further confirmed the involvement of piR-48966 and piR-31335 in the field-effect by qRT-PCR. As the literature observes [56,57,58], the field-effect has crucial implications in research and clinical practices as it impacts the matching adjacent-to-tumor tissue in such a way that it cannot be considered as a normal tissue. Corroborating our study, similar findings were observed in investigations involving other non-coding RNAs [9,12,52]. The combination of these data with our results may provide further knowledge about gastric carcinogenic transformation, which may be used as an informative tool to improve clinical practice.
Although piRNA deregulation was described in the literature, its impact in tumor progression remains poorly understood due to the lack of functional annotation data [23]. In an attempt to better comprehend the piRNA functions, we aligned mature piRNAs and human genome sequences and found some potential target genes. Among these, we identified eight possible cancer-related genes, which may be regulated by five of the eight DE piRNAs, and among those eight genes, FGFR2 and PEAK1 were described as cancer driver genes [40] (Table 2). Interestingly, we found a negative correlation between PIKFYVE and piR-31355* in healthy tissues. This gene was described as being related to cellular homeostasis and migration by regulating phosphatidylinositol 5-monophosphate (PtdIns5P), which was associated with the expression of oncogenes [59,60].
Several studies have shown that the piRNA biogenesis can be reactivated in somatic and cancerous cells, dysregulating their expression, resulting in downregulation of tumor suppressor genes and overexpression of oncogenes at the post-transcription level [43,61,62]. It was also demonstrated that piRNAs may act as mRNAs regulators, as well as the miRNAs, by repressing mRNAs via imperfect base-pairing [36,42,63]. This approach allowed us to find a list of genes (Supplementary Tables S2–S5) that were enriched in cancer-related pathways, such as cellular differentiation, cellular adhesion, and apoptosis.
Interestingly, the DE piRNA targets found in the comparison between ADJ and GC are genes involved in cellular morphogenesis and structure, which reflect the normal histological features of the ADJ tissue and the dedifferentiation typically found in tumor cells.
A group of 18 target genes was enriched in four signaling pathways in cancer (Table 4). Two of them, cellular adhesion and expression of cell adhesion molecules (CAMs), are crucial processes to improve metastasis and cellular inflammation [59,64,65]. Among the DE piRNAs, piR-36339* had the highest number of gene targets. Its overexpression in GC in comparison to NC, hypothetically, indicates that this piRNA may silence CAMs and may become a potential biomarker of metastasis. In this way, we expect that CAM piRNA regulators are overexpressed, also in ADJ tissue.
According to previous studies, piRNAs may be reactivated during the process of tumorigenesis. Fagegaltier et al. (2016) demonstrated that Hippo signaling inhibition may contribute to this activation [47]. Here, we identified six genes from this pathway as piRNA potential targets (Table 4), suggesting that the aberrant expression of piRNAs in cancer may silence the genes involved in this pathway, indicating a possible complex process of regulation in loop.
Our functional analysis also revealed that the deregulated piRNAs may affect the TGF-beta pathway, specifically by targeting the genes BMP5, BMPR1A, SMAD2, and TGFB3. This pathway plays key roles in carcinogenesis and its deregulation is involved in a cascade of aberrant events that trigger cell growth and proliferation pathways, such as JNK and ERK, contributing to tumor progression, cell invasion, and metastasis [66]. It also promotes epithelial-mesenchymal transition (EMT) in late stages of GC, enhancing tumor cell stemness, cell invasion, chemo-resistance, and metastasis with CAMs [67,68].
Recent studies have shown piRNAs expression in many diseases, suggesting their potential as biomarkers. Here, we report the differential expression of piRNAs in non-cancer, gastric cancer, and adjacent tissues, corroborating studies that suggest the field effect in GC. Our functional analyses revealed the potential involvement of piRNAs in key processes of early gastric carcinogenesis, tumor progression, and metastasis. Particularly, our study suggests a set of three piRNAs that may become potential biomarkers of gastric cancer. However, several aspects of the biology of piRNAs in gastric cancer remain to be investigated. Hence, further studies are needed not only to improve our knowledge about the piRNA’s epigenetic mechanisms of gene regulation, but also to provide their validation as biomarkers in gastric cancer.
4. Materials and Methods
4.1. Biological Material and Clinical Data Collection
A total of 84 samples of the stomach antrum region were collected: i) 28 samples without cancer that were collected during the procedure of upper digestive endoscopy (non-cancer(NC) samples); ii) 28 samples of gastric cancer tissue (GC), and iii) 28 samples of matched tumor-adjacent gastric tissue histopathologically diagnosed as non-tumoral (ADJ). Sample sizing was according to patient availability.
All samples were obtained prior to antibiotic, chemotherapeutic, and/or radiotherapeutic treatments from patients from the João de Barros Barreto University Hospital (HUJBB), located in the city of Belém (Pará, Brazil). Immediately after collection, all samples were frozen and stored in liquid nitrogen until analysis.
All cases investigated in this study were reviewed and confirmed by a pathologist. It should be noted that the histopathological analysis of the tumor fragments was performed according to Lauren’s classification (30).
All research procedures were carried out in accordance with the Declaration of Helsinki and the Nuremberg Code, in compliance with the National Health Council’s Research Guidelines Involving Human Beings (Res. CNS 466/12), and approved (protocol number CAAE 43961215.9.0000.5634). Informed consent was obtained from all individual participants included.
4.2. Total RNA Extraction
Total RNA was extracted using TRIzol® Reagent (Thermo Fisher Scientific, Waltham, MA, USA). After isolation, total RNA was stored at −80 °C until further analysis. Total RNA amount and integrity were determined using the Qubit 2.0 (Life Technologies, Foster City, CA, USA) and Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA), according to manufacturer’s specification.
4.3. piRNA Library Preparation
Library preparation was performed using an input concentration of 1/5 µL per sample. We synthesized 24 libraries (8 NC, 8 GC, and 8 ADJ samples) using a TruSeq Small RNA Library Preparation kit (Illumina®, San Diego, CA, USA), according to the manufacturer’s instructions. The RNA library pool was quantified on ABI 7500 equipment (Applied Biosystem, Foster City, CA, USA) by using a KAPA Library Quantification Kit (KAPA Biosystems, Woburn, MA, USA). The libraries were sequenced on the MiSeq Sequencing System (Illumina®, San Diego, CA, USA) using the MiSeq Reagent Kit v3 150 cycle (Illumina®, San Diego, CA, USA).
4.4. NGS Reads Alignment and Quality Control
After sequencing, the sequencing adapters were removed using trimmomatic software [69] and the reads were trimmed and quality filtered (QV > 25). STAR Aligner (v. 2.4.0.1) [70] was used to map the reads to human genomes (v. hg19).
Once the reads were aligned, the HTseq-count [71] was used to quantify sncRNA expression. mirBase [72] and piRbase annotations [73] were used to estimate microRNA and piRNA expression, respectively. For the identification of other transcripts, we used the ENSEMBL annotation (www.ensembl.org). Before piRNA quantitation, we grouped the co-localized piRNAs to avoid ambiguous counting (Supplementary Table S6).
4.5. Differential Expression Analysis
To identify the differentially expressed (DE) piRNAs, three comparisons were made: i) gastric cancer (CG) vs. non-cancer (NC) samples, ii) matched tumor-adjacent gastric tissue (ADJ) vs. NC samples, and iii) GC vs. ADJ samples. For these analyses, raw read count (read counts ≥10) was performed with DESeq2 library [74].
piRNAs satisfying the following criteria were tagged as differentially expressed, |fold-change| > 3 and p-value < 0.05. For graphical analysis of piRNAs, expression data were normalized to reads per kilobase million (RPKM). The area under the curve (AUC > 0.85) from the receiver operating characteristic (ROC) analysis was used to identify potential biomarkers. A discriminant analysis of principal components (DAPC) was made to identify clusters of expressed piRNAs between the tissues. All statistical analyses were performed in an R platform (version 3.2.1).
4.6. Validation of DE piRNAs by qRT-PCR
We further validated piR-48966*, piR-31355*, and piR-49145 expression by qRT-PCR in 60 tissue samples (20 NC, 20 GC, and 20 ADJ). For the detection of piRNA levels, the TaqMan MicroRNA Reverse Transcription Kit (Thermo Scientific) was used to generate cDNA, according to the manufacturer’s instructions. Real-time PCR was performed using a TaqMan Universal Master Mix II (Thermo Scientific) and custom probes (Custom TaqMan Small RNA Assays - Thermo Scientific). The probe sequences were: 5′-TGGGGCGAAGCTACCATC-3′ (piR-48966*), 5′-GGCCGTGATCGTATAGTGGTTAGTAC-3′ (pirR31355), and 5′-TGAGGTAGTAGGTTGTATGGTTTAG-3′ (piR49145). We used RNU6B as endogenous to allow the comparative Ct method. Assays were performed in triplicate on ABI 7500 (Applied Biosystems, Foster City, CA, USA).
Statistical analysis was performed by the Shapiro–Wilk test (Supplementary Table S7) followed by Kruskal–Wallis tests for multiple comparison and the Wilcoxon Mann–Whitney test to paired comparisons. The results were adjusted for multiple comparisons using Bonferroni’s correction. Adjusted p-values of <0.05 were considered statistically significant.
4.7. Identification of DE piRNA Target RNAs
Potential RNA targets of differentially expressed piRNAs were identified by using mature sequences of piRNAs as a query to run a BLAST search. Results that exhibited at least 13 contiguous nucleotide complementary matches were considered potential targets of piRNA regulation [63]. We also correlated the expression of the DE piRNAs to the genes by analyzing 64 paired tissues from the ENCODE database (Supplementary Table S8).
PiRNA target genes were identified by predicting the complementarity sequence between each piRNA and the 5′-UTRs, coding sequences (CDSs), or 3′-UTRs of all known human mRNAs (RefSeq gene annotations, Human genome assembly, GRCh38/ hg18), with miRanda [v3.3a], applying a stringent alignment score (sc ≥170), energy threshold (free energy ≤ −30.0 kcal/mol) (27), and at least 80% of complementarity. Functional analysis was performed using DAVID Bioinformatics Resources [v 6.8] [75,76] to identify all biological processes significantly associated (p value ≤ 0.05) with piRNA-targeted mRNAs.
4.8. Dataset Availability
The datasets used during the present study are available at European Nucleotide Archive (EBI-ENA) under accession number PRJEB27213.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/20/7656/s1.
Author Contributions
Conceptualization, T.V.-S. and A.F.V.; methodology, T.V.-S. and A.F.V.; gastric samples collection and preparation, S.D. and A.K.M.A.; RNAseq experimental analyses T.V.-S., A.F.V., and P.P.; validation, T.V.-S., G.F.C., and R.L.S.C.; bioinformatical and statistical analyses T.V.-S., F.C.M., A.M.R.-d.-S., K.d.P.L., and A.R.-d.-S.; writing—original draft preparation, T.V.-S. and F.M.; writing—review and editing, T.V.-S., A.F.V., and G.F.C.; supervision and project administration, Â.R.-d.-S., P.P.d.A., and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study received financial support by CAPES (Coordenacão de Aperfeicoamento Pessoal de Nível Superior), Rede de Pesquisa em Genômica Populacional Humana (RPGPH)—3381/2013 CAPES BioComputacional, CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), and PROPESP/UFPA (Universidade Federal do Pará). A.R.S. is supported by CNPq/Produtividade (CNPQ 304413/2015-1); S.S. is supported by CNPq/Produtividade (CNPq 305258/2013-3). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We would like to thank the patients (and their relatives) who initially volunteered for participating and provided the follow-up for some months, contributing substantially to the piRNA expression profile in this series.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ADJ | Samples of matched non-tumoral tumor-adjacent gastric tissue |
| AUC | Area Under the Curve (ROC analysis) |
| CAMs | Cell Adhesion Molecules |
| DE | Differentially Expressed |
| EMT | Epithelial-Mesenchymal Transition |
| GC | Gastric Cancer |
| KEGG | Kyotto Encyclopedia of Genes and Genomes |
| miRNA | MicroRNA |
| NC | Non-Cancer tissue |
| NGS | Next Generation Sequencing |
| piRNA | Piwi-Interacting RNA |
| pi-RISC | piRNA-mediated RNA silencing complex |
| qRT-PCR | Quantitative Real-Time PCR |
| sncRNA | Small non-coding RNA |
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Lyons, K.; Le, L.C.; Pham, Y.T.-H.; Borron, C.; Park, J.Y.; Tran, C.T.D.; Tran, T.V.; Tran, H.T.-T.; Vu, K.T.; Do, C.D.; et al. Gastric cancer: epidemiology, biology, and prevention. Eur. J. of Cancer Prev. 2019, 28, 397–412. [Google Scholar] [CrossRef] [PubMed]
- McLean, M.H.; El-Omar, E.M. Genetics of gastric cancer. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 664–674. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, Z.; Guo, W.; Li, J. Gene mutations in gastric cancer: a review of recent next-generation sequencing studies. Tumor Biol. 2015, 36, 7385–7394. [Google Scholar] [CrossRef]
- Pan, X.; Ji, X.; Zhang, R.; Zhou, Z.; Zhong, Y.; Peng, W.; Sun, N.; Xu, X.; Xia, L.; Li, P.; et al. Landscape of somatic mutations in gastric cancer assessed using next-generation sequencing analysis. Oncol. Lett. 2018. [Google Scholar] [CrossRef]
- Strand, M.S.; Lockhart, A.C.; Fields, R.C. Genetics of Gastric Cancer. Surg. Clin. of N. Am. 2017, 97, 345–370. [Google Scholar] [CrossRef]
- Astudillo, P. Wnt5a Signaling in Gastric Cancer. Front Cell. Dev. Biol. 2020, 8, 110. [Google Scholar] [CrossRef]
- Baba, Y.; Ishimoto, T.; Kurashige, J.; Iwatsuki, M.; Sakamoto, Y.; Yoshida, N.; Watanabe, M.; Baba, H. Epigenetic field cancerization in gastrointestinal cancers. Cancer Lett. 2016, 375, 360–366. [Google Scholar] [CrossRef]
- Pereira, A.L.; Magalhães, L.; Moreira, F.C.; Reis-das-Mercês, L.; Vidal, A.F.; Ribeiro-dos-Santos, A.M.; Demachki, S.; Anaissi, A.K.M.; Burbano, R.M.R.; Albuquerque, P.; et al. Epigenetic Field Cancerization in Gastric Cancer: microRNAs as Promising Biomarkers. J. Cancer 2019, 10, 1560–1569. [Google Scholar] [CrossRef]
- Figueiredo, C.; Camargo, M.C.; Leite, M.; Fuentes-Pananá, E.M.; Rabkin, C.S.; Machado, J.C. Pathogenesis of Gastric Cancer: Genetics and Molecular Classification. In Molecular Pathogenesis and Signal Transduction by Helicobacter Pylori; Tegtmeyer, N., Backert, S., Eds.; Current Topics in Microbiology and Immunology; Springer International Publishing: Cham, Switzerland, 2017; Volume 400, pp. 277–304. ISBN 978-3-319-50519-0. [Google Scholar]
- Patel, T.N.; Roy, S.; Ravi, R. Gastric cancer and related epigenetic alterations. Ecancermedicalscience 2017, 11, 714. [Google Scholar] [CrossRef]
- Pereira, A.; Moreira, F.; Vinasco-Sandoval, T.; Cunha, A.; Vidal, A.; Ribeiro-dos-Santos, A.M.; Pinto, P.; Magalhães, L.; Assumpção, M.; Demachki, S.; et al. miRNome Reveals New Insights Into the Molecular Biology of Field Cancerization in Gastric Cancer. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, L.; Quintana, L.G.; Lopes, D.C.F.; Vidal, A.F.; Pereira, A.L.; D’Araujo Pinto, L.C.; de Jesus Viana Pinheiro, J.; Khayat, A.S.; Goulart, L.R.; Burbano, R.; et al. APC gene is modulated by hsa-miR-135b-5p in both diffuse and intestinal gastric cancer subtypes. BMC Cancer 2018, 18, 1055. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.-B.; He, Y.-F.; Li, X.-Q.; Wang, K.; Wang, R.-L. The role of miRNA and lncRNA in gastric cancer. Oncotarget 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.F.; Cruz, A.M.P.; Magalhães, L.; Pereira, A.L.; Anaissi, A.K.M.; Alves, N.C.F.; Albuquerque, P.J.B.S.; Burbano, R.M.R.; Demachki, S.; Ribeiro-dos-Santos, Â. hsa-miR-29c and hsa-miR-135b differential expression as potential biomarker of gastric carcinogenesis. World J. Gastroenterol. 2016, 22, 2060–2070. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.C.; Assumpção, M.B.; Hamoy, I.G.; Darnet, S.; Burbano, R.; Khayat, A.; Gonçalves, A.N.; Alencar, D.O.; Cruz, A.; Magalhães, L.; et al. MiRNA Expression Profile for the Human Gastric Antrum Region Using Ultra-Deep Sequencing. PLoS ONE 2014, 9, e92300. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006. [Google Scholar] [CrossRef]
- Grivna, S.T.; Beyret, E.; Wang, Z.; Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006. [Google Scholar] [CrossRef]
- Kim, V.N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006, 20, 1993–1997. [Google Scholar] [CrossRef]
- Ku, H.-Y.; Lin, H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Nat. Sci. Rev. 2014, 1, 205–218. [Google Scholar] [CrossRef]
- Chalbatani, G.M.; Dana, H.; Memari, F.; Gharagozlou, E.; Ashjaei, S.; Kheirandish, P.; Marmari, V.; Mahmoudzadeh, H.; Mozayani, F.; Maleki, A.R.; et al. Biological function and molecular mechanism of piRNA in cancer. Pract. Lab. Med. 2019, 13, e00113. [Google Scholar] [CrossRef] [PubMed]
- Fonseca Cabral, G.; Azevedo Dos Santos Pinheiro, J.; Vidal, A.F.; Santos, S.; Ribeiro-Dos-Santos, Â. piRNAs in Gastric Cancer: A New Approach Towards Translational Research. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Saito, K. The epigenetic regulation of transposable elements by PIWI-interacting RNAs in Drosophila. Genes Genet. Syst. 2013, 88, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.A.; Yin, H.; Sweeney, S.; Raha, D.; Snyder, M.; Lin, H. A Major Epigenetic Programming Mechanism Guided by piRNAs. Dev. Cell 2013, 24, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A Broadly Conserved Pathway Generates 3′UTR-Directed Primary piRNAs. Curr. Bio. 2009, 19, 2066–2076. [Google Scholar] [CrossRef]
- Watanabe, T.; Lin, H. Posttranscriptional Regulation of Gene Expression by Piwi Proteins and piRNAs. Mol. Cell 2014, 56, 18–27. [Google Scholar] [CrossRef]
- Rojas-Ríos, P.; Simonelig, M. piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 2018, 145, dev161786. [Google Scholar] [CrossRef]
- Fathizadeh, H.; Asemi, Z. Epigenetic roles of PIWI proteins and piRNAs in lung cancer. Cell Biosci. 2019, 9, 102. [Google Scholar] [CrossRef]
- Guo, B.; Li, D.; Du, L.; Zhu, X. piRNAs: biogenesis and their potential roles in cancer. Cancer Metastasis Rev. 2020. [Google Scholar] [CrossRef]
- Hyun, S. Small RNA Pathways That Protect the Somatic Genome. Int. J. Mol. Sci. 2017, 18, 912. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Song, L.; Liu, C.; Lv, X.; Li, X.; Jie, J.; Zhao, D.; Li, D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumor Biol. 2016, 37, 2749–2756. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Guo, J.-M.; Xiao, B.-X.; Miao, Y.; Jiang, Z.; Zhou, H.; Li, Q. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta 2011, 412, 1621–1625. [Google Scholar] [CrossRef] [PubMed]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef]
- Huang, G.; Hu, H.; Xue, X.; Shen, S.; Gao, E.; Guo, G.; Shen, X.; Zhang, X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin. Transl. Oncol. 2013, 15, 563–568. [Google Scholar] [CrossRef]
- Martinez, V.D.; Enfield, K.S.S.; Rowbotham, D.A.; Lam, W.L. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric. Cancer 2016, 19, 660–665. [Google Scholar] [CrossRef]
- Cui, L.; Lou, Y.; Zhang, X.; Zhou, H.; Deng, H.; Song, H.; Yu, X.; Xiao, B.; Wang, W.; Guo, J. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin. Biochem. 2011, 44, 1050–1057. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Assumpção, C.B.; Calcagno, D.Q.; Araújo, T.M.T.; dos Santos, S.E.B.; dos Santos, Â.K.C.R.; Riggins, G.J.; Burbano, R.R.; Assumpção, P.P. The role of piRNA and its potential clinical implications in cancer. Epigenomics 2015, 7, 975–984. [Google Scholar] [CrossRef]
- Fu, A.; Jacobs, D.I.; Zhu, Y. Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA Biol. 2014, 11, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.D.; Vucic, E.A.; Thu, K.L.; Hubaux, R.; Enfield, K.S.S.; Pikor, L.A.; Becker-Santos, D.D.; Brown, C.J.; Lam, S.; Lam, W.L. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci. Rep. 2015, 5, 10423. [Google Scholar] [CrossRef] [PubMed]
- Weng, W.; Liu, N.; Toiyama, Y.; Kusunoki, M.; Nagasaka, T.; Fujiwara, T.; Wei, Q.; Qin, H.; Lin, H.; Ma, Y.; et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol. Cancer 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.; Odom, D.T.; Kutter, C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 2017, 8, 1411. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Hui, G.; Yuan, L.; Shi, D.; Wang, Y.; Du, M.; Zhong, D.; Ma, L.; Tong, N.; Qin, C.; et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015, 356, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Siomi, M.C. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Fagegaltier, D.; Falciatori, I.; Czech, B.; Castel, S.; Perrimon, N.; Simcox, A.; Hannon, G.J. Oncogenic transformation of Drosophila somatic cells induces a functional piRNA pathway. Genes Dev. 2016, 30, 1623–1635. [Google Scholar] [CrossRef]
- Ishizu, H.; Iwasaki, Y.W.; Hirakata, S.; Ozaki, H.; Iwasaki, W.; Siomi, H.; Siomi, M.C. Somatic Primary piRNA Biogenesis Driven by cis-Acting RNA Elements and trans-Acting Yb. Cell Rep. 2015, 12, 429–440. [Google Scholar] [CrossRef]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef]
- Lin, X.; Xia, Y.; Hu, D.; Mao, Q.; Yu, Z.; Zhang, H.; Li, C.; Chen, G.; Liu, F.; Zhu, W.; et al. Transcriptome-wide piRNA profiling in human gastric cancer. Oncol. Rep. 2019. [Google Scholar] [CrossRef]
- Zhang, M.; Du, X. Noncoding RNAs in gastric cancer: Research progress and prospects. World J. Gastroenterol. 2016, 22, 6610–6618. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.F.; Ribeiro-dos-Santos, A.M.; Vinasco-Sandoval, T.; Magalhães, L.; Pinto, P.; Anaissi, A.K.M.; Demachki, S.; Assumpção, P.P.; Santos, S.E.B.; Ribeiro-dos-Santos, Â. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with field cancerization. Sci. Rep. 2017, 7, 14551. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.W.; Anderson, C.; Marshall, E.A.; Minatel, B.C.; Enfield, K.S.S.; Saprunoff, H.L.; Lam, W.L.; Martinez, V.D. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol. Cancer 2016, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Braakhuis, B.J.M.; Tabor, M.P.; Kummer, J.A.; Leemans, C.R.; Brakenhoff, R.H. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003, 63, 1727–1730. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Brown, R.E. Field effect in cancer-an update. Ann. Clin. Lab. Sci. 2009, 39, 331–337. [Google Scholar]
- Graham, T.a.; McDonald, S.A.; Wright, N. A Field cancerization in the GI tract. Future Oncol. 2011, 7, 981–993. [Google Scholar] [CrossRef]
- Curtius, K.; Wright, N.A.; Graham, T.A. An evolutionary perspective on field cancerization. Nat. Rev. Cancer 2018, 18, 19–32. [Google Scholar] [CrossRef]
- Labelle, M.; Hynes, R.O. The initial hours of metastasis: the importance of cooperative host-tumor cell interactions during hematogenous dissemination. Cancer Discov. 2012, 2, 1091–1099. [Google Scholar] [CrossRef]
- Poli, A.; Zaurito, A.E.; Abdul-Hamid, S.; Fiume, R.; Faenza, I.; Divecha, N. Phosphatidylinositol 5 Phosphate (PI5P): From Behind the Scenes to the Front (Nuclear) Stage. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Yin, J.; Jiang, X.-Y.; Qi, W.; Ji, C.-G.; Xie, X.-L.; Zhang, D.-X.; Cui, Z.-J.; Wang, C.-K.; Bai, Y.; Wang, J.; et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef]
- Krishnan, P.; Ghosh, S.; Graham, K.; Mackey, J.R.; Kovalchuk, O.; Damaraju, S. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget 2016, 7, 37944–37956. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.E.; Firek, M.; Jliedi, A.; Amaar, Y.G. Identification and characterization of RASSF1C piRNA target genes in lung cancer cells. Oncotarget 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Sökeland, G.; Schumacher, U. The functional role of integrins during intra- and extravasation within the metastatic cascade. Mol. Cancer 2019, 18, 12. [Google Scholar] [CrossRef] [PubMed]
- Harjunpää, H.; Llort Asens, M.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Hu, Z.; Wen, J.; Wang, K.; Liu, Y. TGF- promotes invasion and metastasis of gastric cancer cells by increasing fascin1 expression via ERK and JNK signal pathways. Acta Bioch. Bioph. Sin. 2009, 41, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wu, R.-L.; Xu, A.-M. Epithelial-mesenchymal transition in gastric cancer. Am. J. Transl. Res. 2015, 7, 2141–2158. [Google Scholar]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, P.; Lu, Y.; Li, Y.; Zheng, Y.; Kan, Y.; Chen, R.; He, S. piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. 2019, 47, D175–D180. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).