Rab GTPases in Osteoclastic Bone Resorption and Autophagy
Abstract
1. Introduction
2. Major Vesicular Trafficking Pathways in Osteoclasts: Bone Resorption and Autophagy
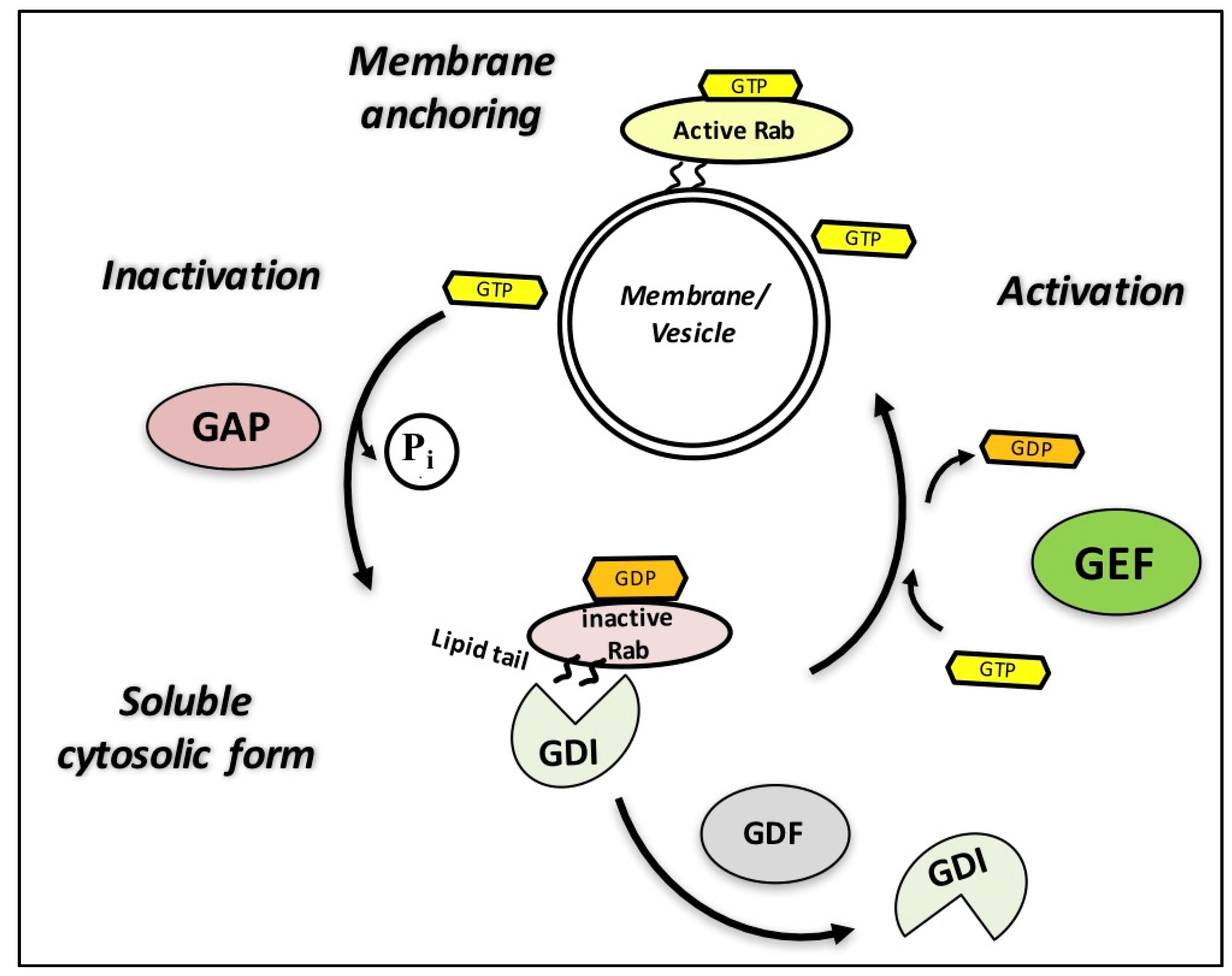
3. Rab GTPases and Their Regulators: Guanine Nucleotide Exchange Factors (GEFs), GTPase-Activating Proteins (GAPs), and Guanosine Diphosphate (GDP) Dissociation Inhibitors (GDIs)
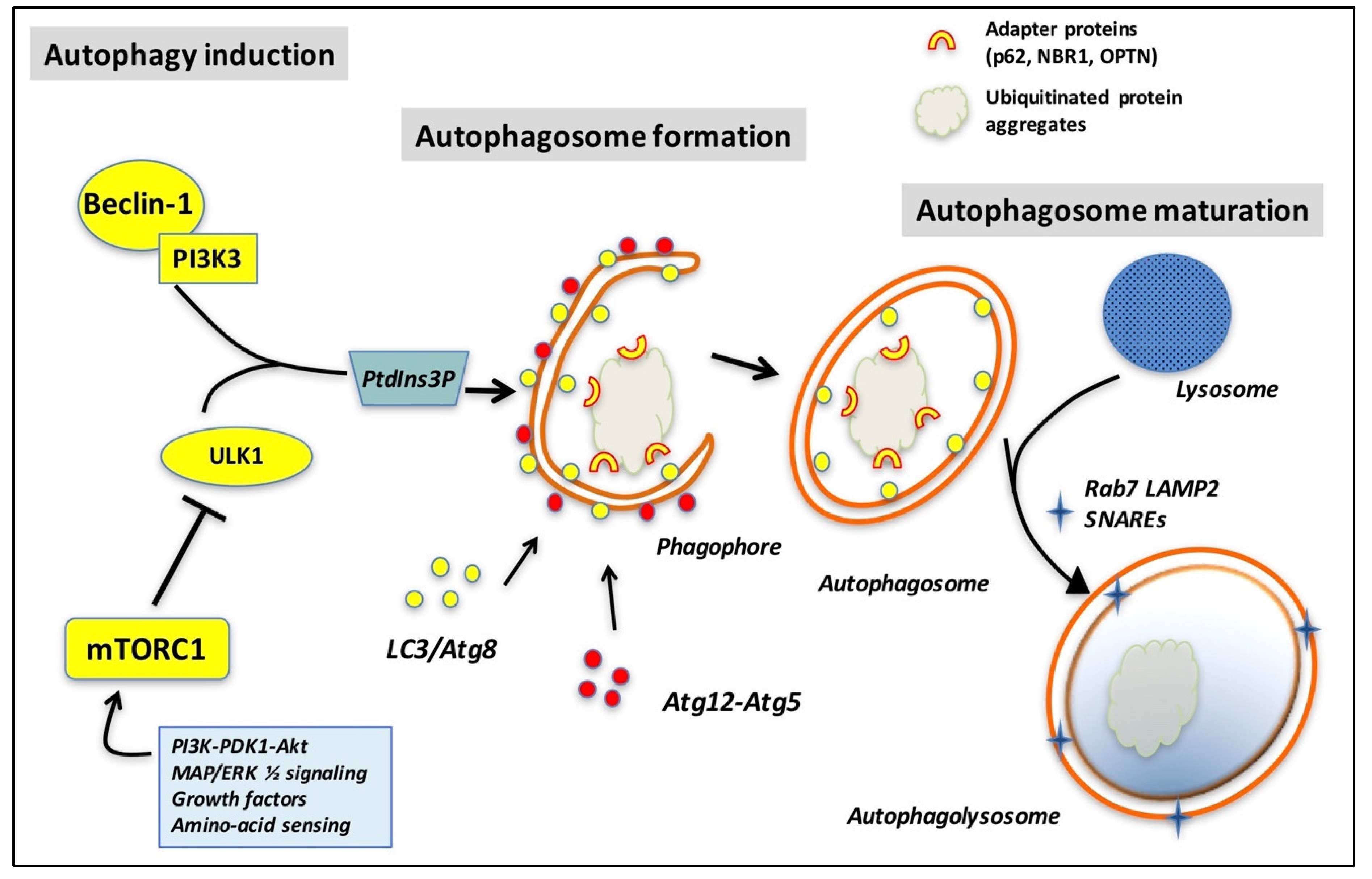
4. The Rab GTPase Network in Autophagy
4.1. Rab GTPases Are Mainly Involved in the Early Steps of Autophagosome Formation
4.2. Late Steps of Autophagolysosome Formation
4.3. GAPs in Autophagy
5. Rab GTPases in Osteoclasts
5.1. Rab7 Is Crucial for Osteoclast Bone Resorbing Activity
5.2. Other Rab GTPases Involved in Osteoclast Activities
6. Rab GTPases in Human Bone Diseases
6.1. Osteopetrosis
6.2. Paget’s Disease of Bone (PDB)
7. Rab GTPases as Therapeutic Targets
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Beclin 1 | BCL-2 interacting myosin/moesin-like coiled-coil protein 1 |
| DENN | differentially expressed in normal and neoplastic cells |
| FPP | farnesyl pyrophosphate |
| FYCO1 | FYVE-coiled coil containing 1 (protein) |
| GAP | GTPase-activating protein |
| GDI | GDP dissociation inhibitor |
| GDF | GDI displacement factor |
| GDP/GTP | guanosine diphosphate (GDP)/guanosine triphosphate (GTP) |
| GEF | guanine–nucleotide exchange factor |
| GGPP | geranylgeranyl pyrophosphate (GGPP) |
| HOPS | homotypic fusion and protein sorting complex |
| LAMP2 | lysosome-associated membrane protein 2 |
| LC3 | light chain 3 [Atg8 (yeast) is called LC3 in mammals] |
| MCSF | macrophage colony-stimulating factor |
| mTORC1 | mammalian target of rapamycin complex 1 |
| PDB | Paget’s disease of bone |
| Phafin1 | a FYVE and PH (Pleckstrin homology) domain containing protein |
| PLEKHF1 | pleckstrin homology domain-containing family F member 1 (a.k.a. Phafin1) |
| PLEKHM1 | pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| PtdIns3P | phosphatidylinositol 3-phosphate |
| RANKL | receptor activator of NF-κB ligand |
| REP | Rab escort protein |
| RIN3 | Ras and Rab Interactor 3 |
| RGGT | Rab Geranyl-Geranyl transferase |
| SNAREs | N-ethylmaleimide-sensitive factor attachment protein receptors |
| TBC | Tre-2/Bub2/Cdc16 |
| TBK1 | TANK Binding Kinase1 |
| TRAPP | Transport protein particle |
| ULK1 | UNC51-like kinase 1 |
References
- Zhao, H.; Ettala, O.; Vaananen, H.K. Intracellular membrane trafficking pathways in bone-resorbing osteoclasts revealed by cloning and subcellular localization studies of small GTP-binding rab proteins. Biochem. Biophys. Res. Commun. 2002, 293, 1060–1065. [Google Scholar] [CrossRef]
- Yarwood, R.; Hellicar, J.; Woodman, P.G.; Lowe, M. Membrane trafficking in health and disease. Dis. Model. Mech. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Wang, J.; Ferro-Novick, S. Crosstalk between the Secretory and Autophagy Pathways Regulates Autophagosome Formation. Dev. Cell 2017, 41, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.; Simpson, J.; Fontana, R.; Kishi-Itakura, C.; Ktistakis, N.T.; Gammoh, N. Targeting of early endosomes by autophagy facilitates EGFR recycling and signalling. EMBO Rep. 2019, 20, e47734. [Google Scholar] [CrossRef]
- Mathew, R.; Rios-Barrera, L.D.; Machado, P.; Schwab, Y.; Leptin, M. Transcytosis via the late endocytic pathway as a cell morphogenetic mechanism. EMBO J. 2020, 39, e105332. [Google Scholar] [CrossRef] [PubMed]
- Herve, J.C.; Bourmeyster, N. Rab GTPases, master controllers of eukaryotic trafficking. Small GTPases 2018, 9, 1–4. [Google Scholar] [CrossRef]
- Parfitt, A.M.; Mundy, G.R.; Roodman, G.D.; Hughes, D.E.; Boyce, B.F. A new model for the regulation of bone resorption, with particular reference to the effects of bisphosphonates. J. Bone Miner. Res. 1996, 11, 150–159. [Google Scholar] [CrossRef]
- Mulari, M.T.; Zhao, H.; Lakkakorpi, P.T.; Vaananen, H.K. Osteoclast ruffled border has distinct subdomains for secretion and degraded matrix uptake. Traffic 2003, 4, 113–125. [Google Scholar] [CrossRef]
- Zhao, H. Membrane trafficking in osteoblasts and osteoclasts: New avenues for understanding and treating skeletal diseases. Traffic 2012, 13, 1307–1314. [Google Scholar] [CrossRef]
- Ory, S.; Brazier, H.; Pawlak, G.; Blangy, A. Rho GTPases in osteoclasts: Orchestrators of podosome arrangement. Eur. J. Cell Biol. 2008, 87, 469–477. [Google Scholar] [CrossRef]
- Coxon, F.P.; Taylor, A. Vesicular trafficking in osteoclasts. Semin. Cell Dev. Biol. 2008, 19, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.Y.; Brigitte Patricia Ribet, A.; Pavlos, N.J. Membrane trafficking in osteoclasts and implications for osteoporosis. Biochem. Soc. Trans. 2019, 47, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Roux, S. New treatment targets in osteoporosis. Joint Bone Spine 2010, 77, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Roux, S. Rab GTPases in Osteoclastic Endomembrane Systems. Biomed. Res. Int. 2018, 2018, 4541538. [Google Scholar] [CrossRef] [PubMed]
- Kern, A.; Dikic, I.; Behl, C. The integration of autophagy and cellular trafficking pathways via RAB GAPs. Autophagy 2015, 11, 2393–2397. [Google Scholar] [CrossRef]
- Wang, K.; Niu, J.; Kim, H.; Kolattukudy, P.E. Osteoclast precursor differentiation by MCPIP via oxidative stress, endoplasmic reticulum stress, and autophagy. J. Mol. Cell Biol. 2011, 3, 360–368. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, G.; Zhang, W.; Xu, N.; Zhu, J.Y.; Jia, J.; Sun, Z.J.; Wang, Y.N.; Zhao, Y.F. Autophagy regulates hypoxia-induced osteoclastogenesis through the HIF-1alpha/BNIP3 signaling pathway. J. Cell Physiol. 2012, 227, 639–648. [Google Scholar] [CrossRef]
- Glantschnig, H.; Fisher, J.E.; Wesolowski, G.; Rodan, G.A.; Reszka, A.A. M-CSF, TNFalpha and RANK ligand promote osteoclast survival by signaling through mTOR/S6 kinase. Cell Death Differ. 2003, 10, 1165–1177. [Google Scholar] [CrossRef]
- Sugatani, T.; Hruska, K.A. Akt1/Akt2 and mammalian target of rapamycin/Bim play critical roles in osteoclast differentiation and survival, respectively, whereas Akt is dispensable for cell survival in isolated osteoclast precursors. J. Biol. Chem. 2005, 280, 3583–3589. [Google Scholar] [CrossRef]
- Chung, Y.H.; Yoon, S.Y.; Choi, B.; Sohn, D.H.; Yoon, K.H.; Kim, W.J.; Kim, D.H.; Chang, E.J. Microtubule-associated protein light chain 3 regulates Cdc42-dependent actin ring formation in osteoclast. Int. J. Biochem. Cell Biol. 2012, 44, 989–997. [Google Scholar] [CrossRef]
- DeSelm, C.J.; Miller, B.C.; Zou, W.; Beatty, W.L.; van Meel, E.; Takahata, Y.; Klumperman, J.; Tooze, S.A.; Teitelbaum, S.L.; Virgin, H.W. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell 2011, 21, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, Y.; Wang, L.; Han, J. Autophagy promotes osteoclast podosome disassembly and cell motility athrough the interaction of kindlin3 with LC3. Cell. Signal. 2020, 67, 109505. [Google Scholar] [CrossRef] [PubMed]
- Cherfils, J.; Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef] [PubMed]
- Colicelli, J. Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, 2004, RE13. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R. Rab GTPases: Master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell 2017, 28, 712–715. [Google Scholar] [CrossRef]
- Bhuin, T.; Roy, J.K. Rab proteins: The key regulators of intracellular vesicle transport. Exp. Cell Res. 2014, 328, 1–19. [Google Scholar] [CrossRef]
- Schwartz, S.L.; Cao, C.; Pylypenko, O.; Rak, A.; Wandinger-Ness, A. Rab GTPases at a glance. J. Cell Sci. 2007, 120, 3905–3910. [Google Scholar] [CrossRef]
- Hirvonen, M.J.; Mulari, M.T.; Buki, K.G.; Vihko, P.; Harkonen, P.L.; Vaananen, H.K. Rab13 is upregulated during osteoclast differentiation and associates with small vesicles revealing polarized distribution in resorbing cells. J. Histochem. Cytochem. 2012, 60, 537–549. [Google Scholar] [CrossRef]
- Muller, M.P.; Goody, R.S. Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases 2018, 9, 5–21. [Google Scholar] [CrossRef]
- Pfeffer, S.R. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 2013, 25, 414–419. [Google Scholar] [CrossRef]
- Szatmari, Z.; Sass, M. The autophagic roles of Rab small GTPases and their upstream regulators: A review. Autophagy 2014, 10, 1154–1166. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Oguchi, M.E.; Fukuda, M. Multiple Types of Guanine Nucleotide Exchange Factors (GEFs) for Rab Small GTPases. Cell Struct. Funct. 2016, 41, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Frasa, M.A.; Koessmeier, K.T.; Ahmadian, M.R.; Braga, V.M. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, A.; Bekbulat, F.; Huesmann, H.; Ulbrich, S.; Tatzelt, J.; Behl, C.; Kern, A. The RAB GTPase RAB18 modulates macroautophagy and proteostasis. Biochem. Biophys. Res. Commun. 2017, 486, 738–743. [Google Scholar] [CrossRef]
- Song, Y.; Shang, D.; Cheng, H.; Zhou, R. The small GTPase RAB37 functions as an organizer for autophagosome biogenesis. Autophagy 2018, 14, 727–729. [Google Scholar] [CrossRef]
- Amaya, C.; Fader, C.M.; Colombo, M.I. Autophagy and proteins involved in vesicular trafficking. FEBS Lett. 2015, 589, 3343–3353. [Google Scholar] [CrossRef]
- Ao, X.; Zou, L.; Wu, Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014, 21, 348–358. [Google Scholar] [CrossRef]
- Zhou, F.; Zou, S.; Chen, Y.; Lipatova, Z.; Sun, D.; Zhu, X.; Li, R.; Wu, Z.; You, W.; Cong, X.; et al. A Rab5 GTPase module is important for autophagosome closure. PLoS Genet. 2017, 13, e1007020. [Google Scholar] [CrossRef]
- Xu, J.; Fotouhi, M.; McPherson, P.S. Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep. 2015, 16, 709–718. [Google Scholar] [CrossRef]
- Longatti, A.; Lamb, C.A.; Razi, M.; Yoshimura, S.; Barr, F.A.; Tooze, S.A. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J. Cell Biol. 2012, 197, 659–675. [Google Scholar] [CrossRef]
- Lindqvist, L.M.; Simon, A.K.; Baehrecke, E.H. Current questions and possible controversies in autophagy. Cell Death Discov. 2015, 1, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y.; Arakawa, S.; Fujitani, K.; Yamaguchi, H.; Mizuta, T.; Kanaseki, T.; Komatsu, M.; Otsu, K.; Tsujimoto, Y.; Shimizu, S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009, 461, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.; Cox, S.; Nassari, S.; Kiger, A.A. Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep. 2015, 16, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Yla-Anttila, P.; Eskelinen, E.L. Roles for RAB24 in autophagy and disease. Small GTPases 2018, 9, 57–65. [Google Scholar] [CrossRef]
- Wang, T.; Ming, Z.; Xiaochun, W.; Hong, W. Rab7: Role of its protein interaction cascades in endo-lysosomal traffic. Cell. Signal. 2011, 23, 516–521. [Google Scholar] [CrossRef]
- Hyttinen, J.M.; Niittykoski, M.; Salminen, A.; Kaarniranta, K. Maturation of autophagosomes and endosomes: A key role for Rab7. Biochim. Biophys. Acta 2013, 1833, 503–510. [Google Scholar] [CrossRef]
- McEwan, D.G.; Popovic, D.; Gubas, A.; Terawaki, S.; Suzuki, H.; Stadel, D.; Coxon, F.P.; Miranda de Stegmann, D.; Bhogaraju, S.; Maddi, K.; et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 2015, 57, 39–54. [Google Scholar] [CrossRef]
- Carroll, B.; Mohd-Naim, N.; Maximiano, F.; Frasa, M.A.; McCormack, J.; Finelli, M.; Thoresen, S.B.; Perdios, L.; Daigaku, R.; Francis, R.E.; et al. The TBC/RabGAP Armus coordinates Rac1 and Rab7 functions during autophagy. Dev. Cell 2013, 25, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Sidjanin, D.J.; Park, A.K.; Ronchetti, A.; Martins, J.; Jackson, W.T. TBC1D20 mediates autophagy as a key regulator of autophagosome maturation. Autophagy 2016, 12, 1759–1775. [Google Scholar] [CrossRef]
- Itoh, T.; Kanno, E.; Uemura, T.; Waguri, S.; Fukuda, M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J. Cell Biol. 2011, 192, 839–853. [Google Scholar] [CrossRef]
- Klinck, R.; Laberge, G.; Bisson, M.; McManus, S.; Michou, L.; Brown, J.P.; Roux, S. Alternative splicing in osteoclasts and Paget’s disease of bone. BMC Med. Genet. 2014, 15, 98. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vallet, M.; Soares, D.C.; Wani, S.; Sophocleous, A.; Warner, J.; Salter, D.M.; Ralston, S.H.; Albagha, O.M. Targeted sequencing of the Paget’s disease associated 14q32 locus identifies several missense coding variants in RIN3 that predispose to Paget’s disease of bone. Hum. Mol. Genet. 2015, 24, 3286–3295. [Google Scholar] [CrossRef]
- Charles, J.F.; Coury, F.; Sulyanto, R.; Sitara, D.; Wu, J.; Brady, N.; Tsang, K.; Sigrist, K.; Tollefsen, D.M.; He, L.; et al. The collection of NFATc1-dependent transcripts in the osteoclast includes numerous genes non-essential to physiologic bone resorption. Bone 2012, 51, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Ha, B.G.; Hong, J.M.; Park, J.Y.; Ha, M.H.; Kim, T.H.; Cho, J.Y.; Ryoo, H.M.; Choi, J.Y.; Shin, H.I.; Chun, S.Y.; et al. Proteomic profile of osteoclast membrane proteins: Identification of Na+/H+ exchanger domain containing 2 and its role in osteoclast fusion. Proteomics 2008, 8, 2625–2639. [Google Scholar] [CrossRef] [PubMed]
- Itzstein, C.; Coxon, F.P.; Rogers, M.J. The regulation of osteoclast function and bone resorption by small GTPases. Small GTPases 2011, 2, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Shimada-Sugawara, M.; Sakai, E.; Okamoto, K.; Fukuda, M.; Izumi, T.; Yoshida, N.; Tsukuba, T. Rab27A regulates transport of cell surface receptors modulating multinucleation and lysosome-related organelles in osteoclasts. Sci. Rep. 2015, 5, 9620. [Google Scholar] [CrossRef]
- Taylor, A.; Mules, E.H.; Seabra, M.C.; Helfrich, M.H.; Rogers, M.J.; Coxon, F.P. Impaired prenylation of Rab GTPases in the gunmetal mouse causes defects in bone cell function. Small GTPases 2011, 2, 131–142. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Sakai, E.; Okamoto, K.; Kajiya, H.; Okabe, K.; Naito, M.; Kadowaki, T.; Tsukuba, T. Rab44, a novel large Rab GTPase, negatively regulates osteoclast differentiation by modulating intracellular calcium levels followed by NFATc1 activation. Cell Mol. Life Sci. 2018, 75, 33–48. [Google Scholar] [CrossRef]
- Palokangas, H.; Mulari, M.; Vaananen, H.K. Endocytic pathway from the basal plasma membrane to the ruffled border membrane in bone-resorbing osteoclasts. J. Cell Sci. 1997, 110, 1767–1780. [Google Scholar]
- Zhao, H.; Laitala-Leinonen, T.; Parikka, V.; Vaananen, H.K. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J. Biol. Chem. 2001, 276, 39295–39302. [Google Scholar] [CrossRef]
- Matsumoto, N.; Sekiya, M.; Tohyama, K.; Ishiyama-Matsuura, E.; Sun-Wada, G.H.; Wada, Y.; Futai, M.; Nakanishi-Matsui, M. Essential Role of the a3 Isoform of V-ATPase in Secretory Lysosome Trafficking via Rab7 Recruitment. Sci. Rep. 2018, 8, 6701. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.Q.; Feng, S.; Chen, W.; Zhao, H.; Paulson, C.; Li, Y.P. V-ATPase subunit ATP6AP1 (Ac45) regulates osteoclast differentiation, extracellular acidification, lysosomal trafficking, and protease exocytosis in osteoclast-mediated bone resorption. J. Bone Miner. Res. 2012, 27, 1695–1707. [Google Scholar] [CrossRef] [PubMed]
- Rawet-Slobodkin, M.; Elazar, Z. PLEKHM1: A multiprotein adaptor for the endolysosomal system. Mol. Cell 2015, 57, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Ye, S.; Castro-Gomes, T.; Winchell, C.G.; Andrews, N.W.; Voth, D.E.; Varughese, K.I.; Mackintosh, S.G.; Feng, Y.; Pavlos, N.; et al. PLEKHM1/DEF8/RAB7 complex regulates lysosome positioning and bone homeostasis. JCI Insight 2016, 1, e86330. [Google Scholar] [CrossRef]
- Witwicka, H.; Jia, H.; Kutikov, A.; Reyes-Gutierrez, P.; Li, X.; Odgren, P.R. TRAFD1 (FLN29) Interacts with Plekhm1 and Regulates Osteoclast Acidification and Resorption. PLoS ONE 2015, 10, e0127537. [Google Scholar] [CrossRef]
- Van Wesenbeeck, L.; Odgren, P.R.; Coxon, F.P.; Frattini, A.; Moens, P.; Perdu, B.; MacKay, C.A.; Van Hul, E.; Timmermans, J.P.; Vanhoenacker, F.; et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J. Clin. Investig. 2007, 117, 919–930. [Google Scholar] [CrossRef]
- Sun, Y.; Buki, K.G.; Ettala, O.; Vaaraniemi, J.P.; Vaananen, H.K. Possible role of direct Rac1-Rab7 interaction in ruffled border formation of osteoclasts. J. Biol. Chem. 2005, 280, 32356–32361. [Google Scholar] [CrossRef]
- Hirvonen, M.J.; Buki, K.G.; Sun, Y.; Mulari, M.T.; Harkonen, P.L.; Vaananen, K.H. Novel interaction of Rab13 and Rab8 with endospanins. FEBS Open Bio 2013, 3, 83–88. [Google Scholar] [CrossRef]
- Abu-Amer, Y.; Teitelbaum, S.L.; Chappel, J.C.; Schlesinger, P.; Ross, F.P. Expression and regulation of RAB3 proteins in osteoclasts and their precursors. J. Bone Miner. Res. 1999, 14, 1855–1860. [Google Scholar] [CrossRef]
- Pavlos, N.J.; Xu, J.; Riedel, D.; Yeoh, J.S.; Teitelbaum, S.L.; Papadimitriou, J.M.; Jahn, R.; Ross, F.P.; Zheng, M.H. Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption. Mol. Cell Biol. 2005, 25, 5253–5269. [Google Scholar] [CrossRef]
- Pavlos, N.J.; Cheng, T.S.; Qin, A.; Ng, P.Y.; Feng, H.T.; Ang, E.S.; Carrello, A.; Sung, C.H.; Jahn, R.; Zheng, M.H.; et al. Tctex-1, a novel interaction partner of Rab3D, is required for osteoclastic bone resorption. Mol. Cell Biol. 2011, 31, 1551–1564. [Google Scholar] [CrossRef]
- Tang, L.; Yin, Y.; Liu, J.; Li, Z.; Lu, X. MiR-124 Attenuates Osteoclastogenic Differentiation of Bone Marrow Monocytes Via Targeting Rab27a. Cell Physiol. Biochem. 2017, 43, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Tokuhisa, M.; Kadowaki, T.; Ogawa, K.; Yamaguchi, Y.; Kido, M.A.; Gao, W.; Umeda, M.; Tsukuba, T. Expression and localisation of Rab44 in immune-related cells change during cell differentiation and stimulation. Sci. Rep. 2020, 10, 10728. [Google Scholar] [CrossRef] [PubMed]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef]
- Tolar, J.; Teitelbaum, S.L.; Orchard, P.J. Osteopetrosis. N. Engl. J. Med. 2004, 351, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Pangrazio, A.; Caldana, M.E.; Lo Iacono, N.; Mantero, S.; Vezzoni, P.; Villa, A.; Sobacchi, C. Autosomal recessive osteopetrosis: Report of 41 novel mutations in the TCIRG1 gene and diagnostic implications. Osteoporos. Int. 2012, 23, 2713–2718. [Google Scholar] [CrossRef]
- Kornak, U.; Kasper, D.; Bosl, M.R.; Kaiser, E.; Schweizer, M.; Schulz, A.; Friedrich, W.; Delling, G.; Jentsch, T.J. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 2001, 104, 205–215. [Google Scholar] [CrossRef]
- Lange, P.F.; Wartosch, L.; Jentsch, T.J.; Fuhrmann, J.C. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature 2006, 440, 220–223. [Google Scholar] [CrossRef]
- Pandruvada, S.N.; Beauregard, J.; Benjannet, S.; Pata, M.; Lazure, C.; Seidah, N.G.; Vacher, J. Role of Ostm1 Cytosolic Complex with Kinesin 5B in Intracellular Dispersion and Trafficking. Mol. Cell Biol. 2016, 36, 507–521. [Google Scholar] [CrossRef]
- Stattin, E.L.; Henning, P.; Klar, J.; McDermott, E.; Stecksen-Blicks, C.; Sandstrom, P.E.; Kellgren, T.G.; Ryden, P.; Hallmans, G.; Lonnerholm, T.; et al. SNX10 gene mutation leading to osteopetrosis with dysfunctional osteoclasts. Sci. Rep. 2017, 7, 3012. [Google Scholar] [CrossRef]
- Teasdale, R.D.; Collins, B.M. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. Biochem. J. 2012, 441, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Bo, T.; Yan, F.; Guo, J.; Lin, X.; Zhang, H.; Guan, Q.; Wang, H.; Fang, L.; Gao, L.; Zhao, J.; et al. Characterization of a Relatively Malignant Form of Osteopetrosis Caused by a Novel Mutation in the PLEKHM1 Gene. J. Bone Miner. Res. 2016, 31, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Sobacchi, C.; Schulz, A.; Coxon, F.; Villa, A.; Helfrich, M. Osteopetrosis: Genetics, treatment and new insights into osteoclast function. Nat. Rev. Endocrinol. 2013, 9, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Roodman, G.D.; Windle, J.J. Paget disease of bone. J. Clin. Investig. 2005, 115, 200–208. [Google Scholar] [CrossRef]
- Chamoux, E.; Couture, J.; Bisson, M.; Morissette, J.; Brown, J.P.; Roux, S. The p62 P392L mutation linked to Paget’s disease induces activation of human osteoclasts. Mol. Endocrinol. 2009, 23, 1668–1680. [Google Scholar] [CrossRef]
- Helfrich, M.H.; Hocking, L.J. Genetics and aetiology of Pagetic disorders of bone. Arch. Biochem. Biophys. 2008, 473, 172–182. [Google Scholar] [CrossRef]
- McManus, S.; Bisson, M.; Chamberland, R.; Roy, M.; Nazari, S.; Roux, S. Autophagy and 3-Phosphoinositide-Dependent Kinase 1 (PDK1)-Related Kinome in Pagetic Osteoclasts. J. Bone Miner. Res. 2016, 31, 1334–1343. [Google Scholar] [CrossRef]
- Usategui-Martin, R.; Gestoso-Uzal, N.; Calero-Paniagua, I.; De Pereda, J.M.; Del Pino-Montes, J.; Gonzalez-Sarmiento, R. A mutation in p62 protein (p. R321C), associated to Paget’s disease of bone, causes a blockade of autophagy and an activation of NF-kB pathway. Bone 2020, 133, 115265. [Google Scholar] [CrossRef]
- Sun, Q.; Sammut, B.; Wang, F.M.; Kurihara, N.; Windle, J.J.; Roodman, G.D.; Galson, D.L. TBK1 mediates critical effects of measles virus nucleocapsid protein (MVNP) on pagetic osteoclast formation. J. Bone Miner. Res. 2014, 29, 90–102. [Google Scholar] [CrossRef]
- Pilli, M.; Arko-Mensah, J.; Ponpuak, M.; Roberts, E.; Master, S.; Mandell, M.A.; Dupont, N.; Ornatowski, W.; Jiang, S.; Bradfute, S.B.; et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012, 37, 223–234. [Google Scholar] [CrossRef]
- Itoh, T.; Satoh, M.; Kanno, E.; Fukuda, M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells 2006, 11, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Coxon, F.P.; Thompson, K.; Rogers, M.J. Recent advances in understanding the mechanism of action of bisphosphonates. Curr. Opin. Pharmacol. 2006, 6, 307–312. [Google Scholar] [CrossRef]
- Russell, R.G. Bisphosphonates: From bench to bedside. Ann. N. Y. Acad. Sci. 2006, 1068, 367–401. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.J.; Monkkonen, J.; Munoz, M.A. Molecular mechanisms of action of bisphosphonates and new insights into their effects outside the skeleton. Bone 2020, 139, 115493. [Google Scholar] [CrossRef]
- Coxon, F.P.; Helfrich, M.H.; Van’t Hof, R.; Sebti, S.; Ralston, S.H.; Hamilton, A.; Rogers, M.J. Protein geranylgeranylation is required for osteoclast formation, function, and survival: Inhibition by bisphosphonates and GGTI-298. J. Bone Miner. Res. 2000, 15, 1467–1476. [Google Scholar] [CrossRef]
- Coxon, F.P.; Helfrich, M.H.; Larijani, B.; Muzylak, M.; Dunford, J.E.; Marshall, D.; McKinnon, A.D.; Nesbitt, S.A.; Horton, M.A.; Seabra, M.C.; et al. Identification of a novel phosphonocarboxylate inhibitor of Rab geranylgeranyl transferase that specifically prevents Rab prenylation in osteoclasts and macrophages. J. Biol. Chem. 2001, 276, 48213–48222. [Google Scholar] [CrossRef]
- Agola, J.O.; Jim, P.A.; Ward, H.H.; Basuray, S.; Wandinger-Ness, A. Rab GTPases as regulators of endocytosis, targets of disease and therapeutic opportunities. Clin. Genet. 2011, 80, 305–318. [Google Scholar] [CrossRef] [PubMed]

| Rab GTPases | Human Osteoclasts | Rodent Osteoclasts | Autophagy (Non-Osteoclastic Cells) | Bone Resorption | |
|---|---|---|---|---|---|
| Gene Expression | Gene Expression | Protein | |||
| 1 | X (b) | X | X | ||
| 2 | X (b) | ||||
| 3 (a–d) | X (c) | X (a, b/c, d) | X (d) | ||
| 4 (a, b) | X (a) | X (b) | |||
| 5 | X (a, b) | X (c) | X | X | X (c) |
| 6 | X | ||||
| 7 | X | X | X | X | X |
| 8 | X (b) | X | |||
| 9 (a, b) | X (a) | X | X | ||
| 10 | X | X | |||
| 11 (a, b) | X (a) | X (b) | X (b) | X | X (b) |
| 12 | X | X | |||
| 13 | X | -- | |||
| 14 | X | X | |||
| 15, 17, 19 | -- | ||||
| 18 | X | X | X | ||
| 20 | X | ||||
| 21 | X | X | |||
| 22a | X | ||||
| 23 | X | X | |||
| 24 | X | X | |||
| 25, 26 | -- | ||||
| 27 (a, b) | X (a) | X (a) | X (a) | X (a) | |
| 28, 29 | -- | ||||
| 30 | X | ||||
| 31 | X | ||||
| 32 | X | X | |||
| 33 (a, b) | X (a, b) | X (b) | |||
| 34 | X | ||||
| 35 | X | ||||
| 36, 37 | -- | X | |||
| 38 | X | X | -- | ||
| 39a/b, 40 | -- | ||||
| 44 | X | X | X 1 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, M.; Roux, S. Rab GTPases in Osteoclastic Bone Resorption and Autophagy. Int. J. Mol. Sci. 2020, 21, 7655. https://doi.org/10.3390/ijms21207655
Roy M, Roux S. Rab GTPases in Osteoclastic Bone Resorption and Autophagy. International Journal of Molecular Sciences. 2020; 21(20):7655. https://doi.org/10.3390/ijms21207655
Chicago/Turabian StyleRoy, Michèle, and Sophie Roux. 2020. "Rab GTPases in Osteoclastic Bone Resorption and Autophagy" International Journal of Molecular Sciences 21, no. 20: 7655. https://doi.org/10.3390/ijms21207655
APA StyleRoy, M., & Roux, S. (2020). Rab GTPases in Osteoclastic Bone Resorption and Autophagy. International Journal of Molecular Sciences, 21(20), 7655. https://doi.org/10.3390/ijms21207655





