Functional Diversity of Non-Histone Chromosomal Protein HmgB1
Abstract
1. Introduction
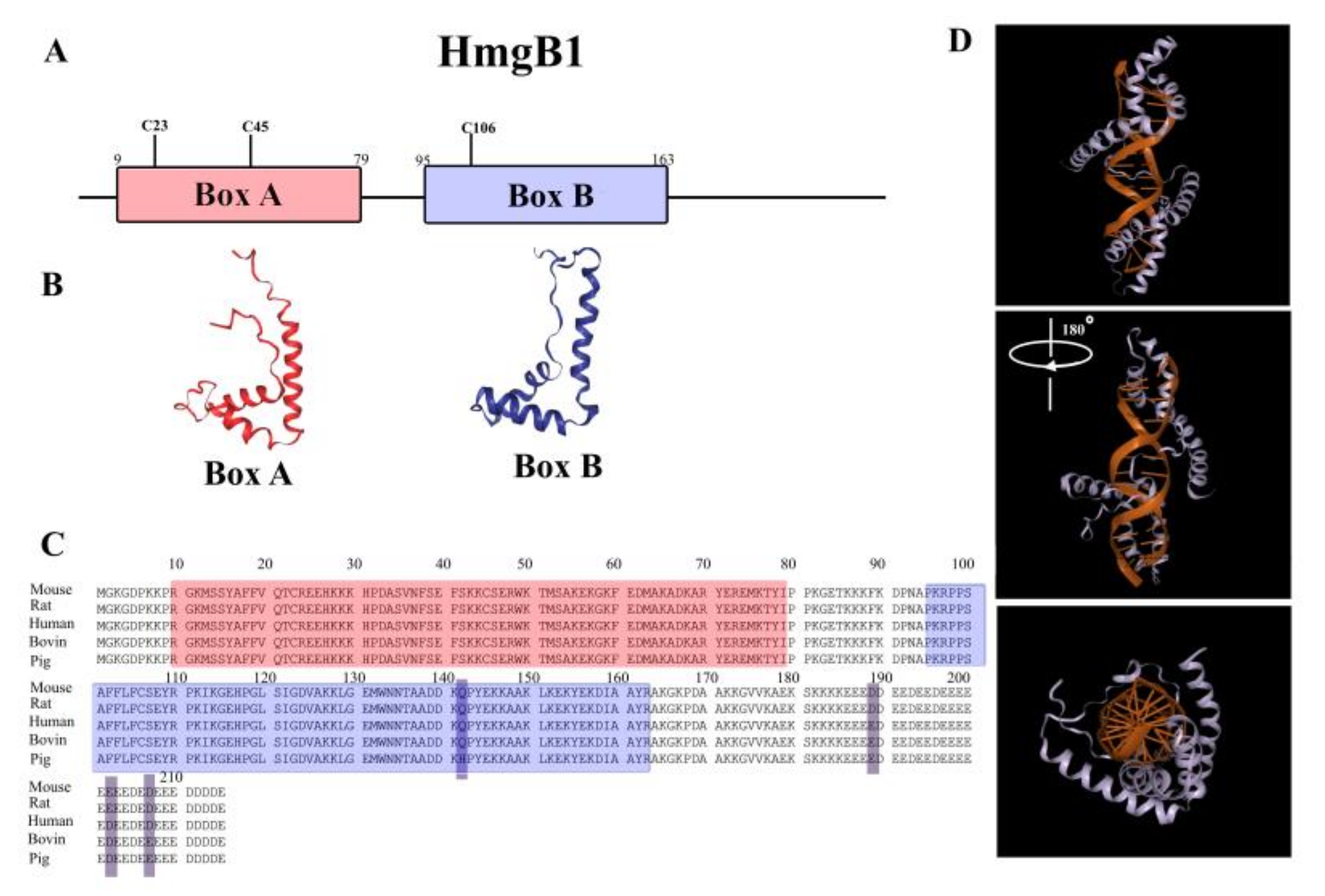
2. Structure of HmgB1 Protein
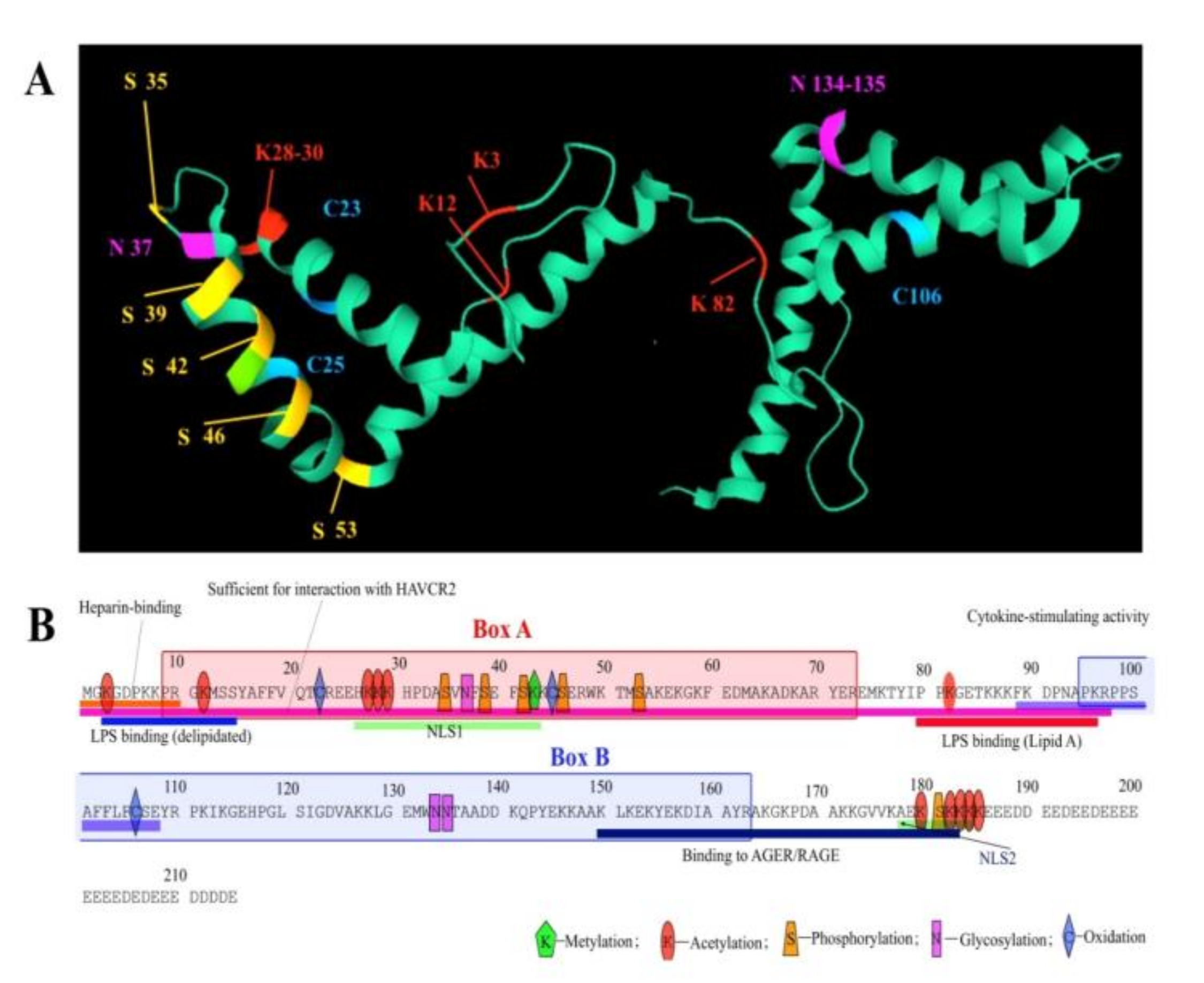
3. Post-Translational Modifications and Redox State of HmgB1 Protein
3.1. Phosphorylation
3.2. Acetylation
3.3. Methylation
3.4. Redox State
3.5. Glycosylation
3.6. ADP-Ribosylation
4. Functions of HmgB1 Protein in Cell Nucleus
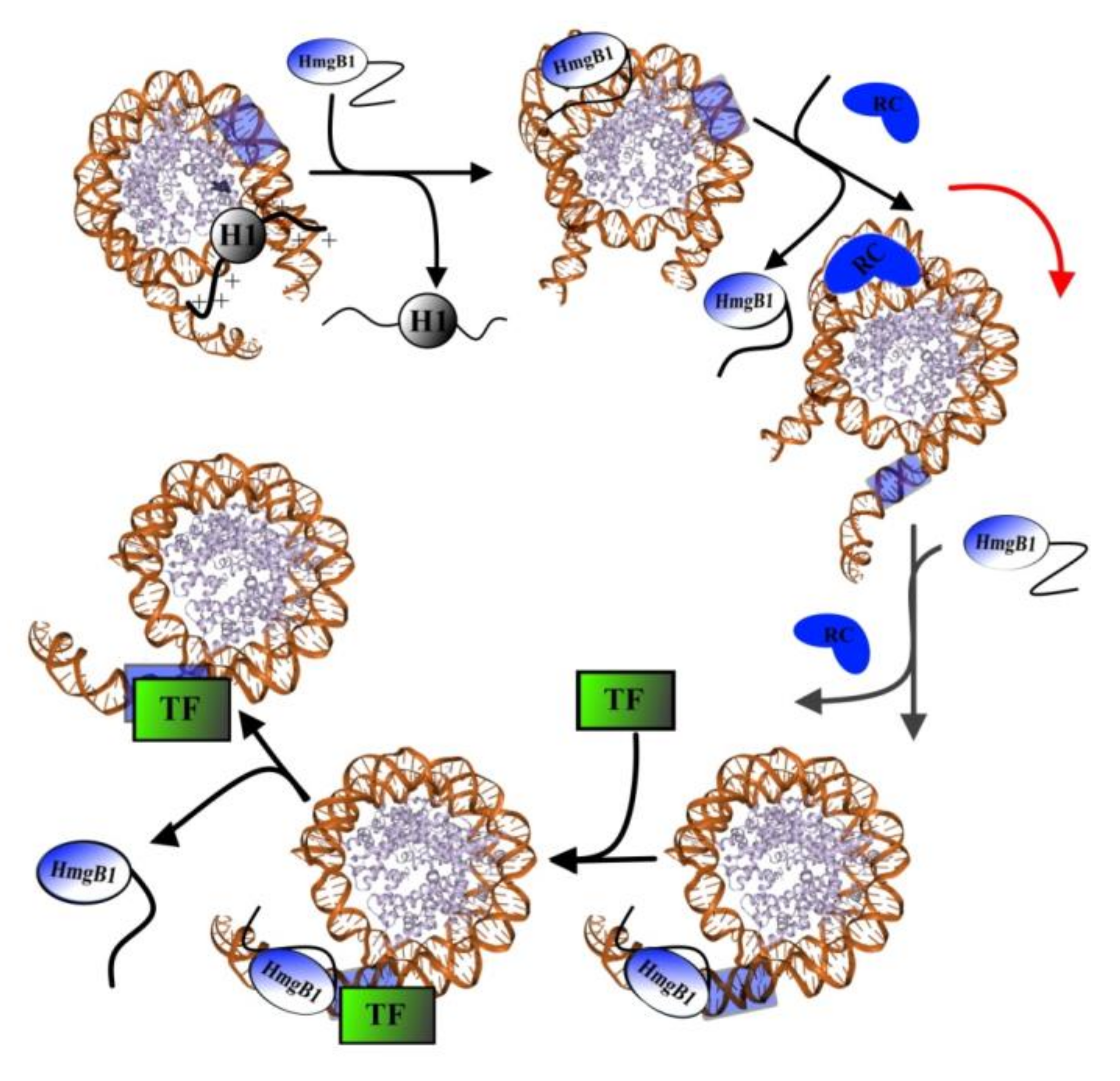
4.1. Interaction with DNA
4.2. Interaction of HmgB1 with Other Proteins
4.3. Effect of Expression Levels of HmgB1 and HmgB2 on Mouse and Human Stem Cell Chromatin, Mouse Embryogenesis, and Postnatal Development
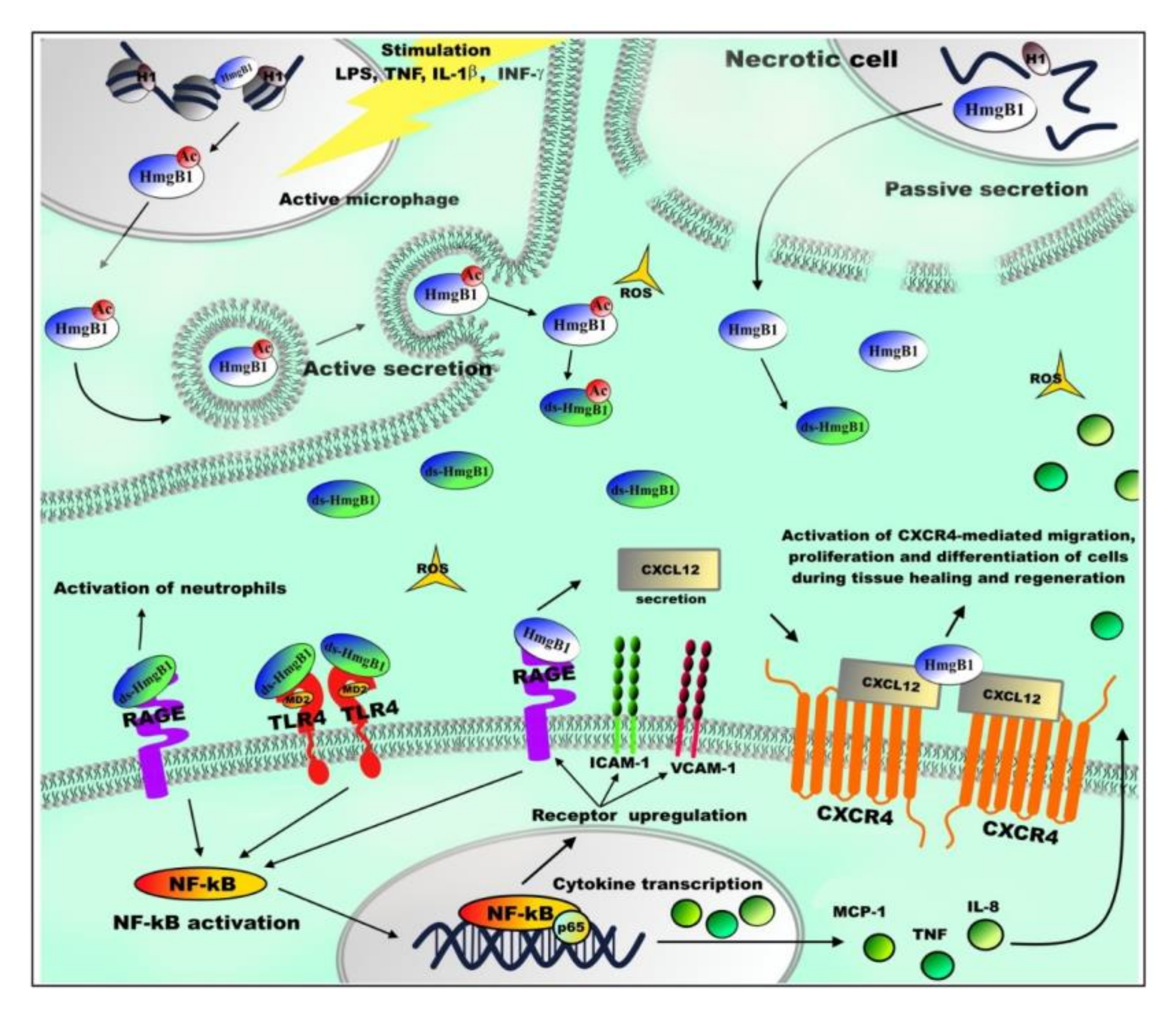
5. Extranuclear Functions of HmgB1
5.1. HmgB1 and Bioenergetics of Eukaryotic Cells
5.2. HMGB1 and Cellular Senescence
6. HMGB1 and Structural Organization of Genome
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Woodcock, C.L.; Ghosh, R.P. Chromatin Higher-order Structure and Dynamics. Gold Spring Harb. Perspect. Biol. 2010, a000296. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New insights into nucleosome and chromatin structure: An ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Chikhirzhina, E.; Chikhirzhina, G.; Polyanichko, A. Chromatin structure: The role of “linker” Proteins. Biomed. Spectr. Imaging 2014, 3, 345–358. [Google Scholar] [CrossRef]
- White, A.E.; Hieb, A.R.; Luger, K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci. Rep. 2016, 6, 19122. [Google Scholar] [CrossRef] [PubMed]
- Chikhirzhina, E.; Starkova, T.; Polyanichko, A. The Role of Linker Histones in Chromatin Structural Organization. 2. The interactions with DNA and nuclear proteins. Biophysics 2020, 65, 202–212. [Google Scholar] [CrossRef]
- Crane-Robinson, C. Linker histones: History and current perspectives. Biochim. Biophys. Acta 2016, 1859, 431–435. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chikhirzhina, E.; Starkova, T.; Polyanichko, A. The Role of Linker Histones in Chromatin Structural Organization. 1. H1 Family Histones. Biophysics 2018, 63, 858–865. [Google Scholar] [CrossRef]
- Nicolas, R.H.; Goodwin, G.H. Isolation and Analysis. In Book The HMG Chromosomal Proteins; Johns, E.W., Ed.; Academic Press Inc.: London, UK, 1982; pp. 43–45, 59–60. ISBN 9780323158749. [Google Scholar] [CrossRef]
- Bustin, M.; Reeves, R. High-mobility-group chromosomal proteins: Architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996, 54, 35–100. [Google Scholar] [CrossRef]
- Stros, M.; Launholt, D.; Grasser, K.D. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007, 64, 2590–2606. [Google Scholar] [CrossRef]
- Stros, M. HMGB proteins: Interactions with DNA and chromatin. Biochim. Biophys. Acta 2010, 1799, 101–113. [Google Scholar] [CrossRef]
- Stros, M.; Polanska, E.; Kucirek, M.; Pospisilova, S. Histone H1 differentially inhibits DNA bending by reduced and oxidized HMGB1 protein. PLoS ONE 2015, 10, e0138774. [Google Scholar] [CrossRef]
- Reeves, R. High mobility group (HMG) proteins, modulators of chromatin structure and DNA repair in mammalian cells. Dna Repair 2015, 36, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Bustin, M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 1999, 19, 5237–5246. [Google Scholar] [CrossRef]
- Ozturk, N.; Singh, I.; Mehta, A.; Braun, T.; Barreto, G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Biol. 2014, 2, 5. [Google Scholar] [CrossRef]
- Bustin, M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef]
- Postnikov, Y.V.; Trieschmann, L.; Rickers, A.; Bustin, M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J. Mol. Biol. 1995, 252, 423–432. [Google Scholar] [CrossRef]
- Phair, R.D.; Scaffidi, P.; Elbi, C.; Vecerova, J.; Dey, A.; Ozato, K.; Brown, D.T.; Hager, G.; Bustin, M.; Misteli, T. Global nature of dynamic protein-chromatin interactions In Vivo: Three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004, 24, 6393–6402. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; van Ingen, H.; Zhou, B.R.; Feng, H.; Bustin, M.; Kay, L.E.; Bai, Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. USA 2011, 108, 12283–12288. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.J.; Cutter, A.R.; Fang, H.; Postnikov, Y.V.; Bustin, M.; Hayes, J.J. HMGN1 and 2 remodel core and linker histone tail domains within chromatin. Nucleic Acids Res. 2017, 45, 9917–9930. [Google Scholar] [CrossRef]
- Chiefari, E.; Foti, D.P.; Sgarra, R.; Pegoraro, S.; Arcidiacono, B.; Brunetti, F.S.; Greco, M.; Manfioletti, G.; Brunetti, A. Transcriptional Regulation of Glucose Metabolism: The Emerging Role of the HMGA1 Chromatin Factor. Front. Endocrin. (Lausanne) 2018, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Read, C.M.; Cary, P.D.; Crane-Robinson, C.; Driscoll, P.C.; Norman, D.G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993, 21, 3427–3436. [Google Scholar] [CrossRef]
- Polyanichko, A.M.; Chikhirzhin, E.V.; Skvortsov, A.N.; Kostyleva, E.I.; Colson, P.; Houssier, C.; Vorob’ev, V.I. The HMG1 Ta(i)le. J. Biomol. Struct. Dyn. 2002, 19, 1053–1062. [Google Scholar] [CrossRef]
- Chikhirzhina, E.V.; Polyanichko, A.M.; Skvortsov, A.N.; Kostyleva, E.I.; Houssier, C.; Vorobyev, V.I. HMG1 Domains: The Victims of the Circumstances. Mol. Biol. (Mosk.) 2002, 36, 412–418. [Google Scholar] [CrossRef]
- Chikhirzhina, E.V.; Starkova, T.Y.; Kostyleva, E.I.; Chikhirzhina, G.I.; Vorobyev, V.I.; Polyanichko, A.M. Interaction of DNA with Sperm-Specific Histones of the H1 Family. Cell Tissue Biol. 2011, 5, 536–542. [Google Scholar] [CrossRef]
- Chikhirzhina, E.; Starkova, T.; Polyanichko, A. The structural organization of the nuclear protein HMGB1 and its effect on the formation of the ordered supramolecular complexes. Biophysics 2000, in press. [Google Scholar]
- Chikhirzhina, E.V.; Polyanichko, A.M.; Starkova, T.Y. Extranuclear functions of nonhistone protein HMGB1. Tsitologiya 2020, 62, 1–10. [Google Scholar] [CrossRef]
- Chikhirzhina, E.; Polyanichko, A.; Leonenko, Z.; Wieser, H.; Vorob’ev, V. C-terminal domain of nonhistone protein HMGB1 as a modulator of HMGB1-DNA structural interactions. Spectroscopy 2010, 24, 361–366. [Google Scholar] [CrossRef]
- Polyanichko, A.; Chikhirzhina, E. Interaction between DNA and chromosomal proteins HMGB1 and H1 studied by IR/VCD spectroscopy. J. Mol. Struct. 2013, 1044, 167–172. [Google Scholar] [CrossRef]
- Polyanichko, A.M.; Vorob’ev, V.I.; Chikhirzhina, E.V. Structure of DNA complexes with chromosomal protein HMGB1 and histone H1 in the presence of manganese ions, 2. Vibrational circular dichroism spectroscopy. Mol. Biol. 2013, 47, 299–306. [Google Scholar] [CrossRef]
- Chikhirzhina, E.V.; Starkova, T.J.; Polyanichko, A.M. Interaction between chromosomal protein HMGB1 and DNA studied by DNA-melting analysis. J. Spectr. 2014, 2014, 387138. [Google Scholar] [CrossRef]
- Muller, S.; Scaffidi, P.; Degryse, B.; Bonaldi, T.; Ronfani, L.; Agresti, A.; Beltrame, M.; Bianchi, M.E. New EMBO members’ review: The double life of HMGB1 chromatin protein: Architectural factor and extracellular signal. Embo J. 2001, 20, 4337–4340. [Google Scholar] [CrossRef] [PubMed]
- Atcha, F.A.; Syed, A.; Wu, B.; Hoverter, N.P.; Yokoyama, N.N.; Ting, J.H.; Munguia, J.E.; Mangalam, H.J.; Marsh, J.L.; Waterman, M.L. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol. Cell. Biol. 2007, 27, 8352–8363. [Google Scholar] [CrossRef] [PubMed]
- Pevny, L.H.; Nicolis, S.K. Sox2 roles in neural stem cells. Int. J. Biochem. Cell. Biol. 2010, 42, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.; Harley, V.R. Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. Int. J. Biochem. Cell. Biol. 2010, 42, 400–410. [Google Scholar] [CrossRef]
- Oppel, F.; Müller, N.; Schackert, G.; Hendruschk, S.; Martin, D.; Geiger, K.D.; Temme, A. SOX2-RNAi attenuates S-phase entry and induces RhoA-dependent switch to protease-independent amoeboid migration in human glioma cells. Mol. Cancer 2011, 10, 137. [Google Scholar] [CrossRef]
- Leis, O.; Eguiara, A.; Lopez-Arribillaga, E.; Alberdi, M.J.; Hernandez-Garcia, S.; Elorriaga, K.; Pandiella, A.; Rezola, R.; Martin, A.G. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31, 1354–1365. [Google Scholar] [CrossRef]
- Chi, T. BAF-centred view of the immune system. Nat. Rev. Immunol. 2004, 4, 965–977. [Google Scholar] [CrossRef]
- Lai, D.; Wan, M.; Wu, J.; Preston-Hurlburt, P.; Kushwaha, R.; Grundström, T.; Imbalzano, A.N.; Chi, T. Induction of TLR4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling. Proc. Natl. Acad. Sci. USA 2009, 106, 1169–1174. [Google Scholar] [CrossRef]
- Catena, R.; Escoffier, E.; Caron, C.; Khochbin, S.; Martianov, I.; Davidson, I. HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of longating spermatids. Biol. Reprod. 2009, 80, 358–366. [Google Scholar] [CrossRef]
- Park, S.; Lippard, S.J. Binding Interaction of HMGB4 with Cisplatin-Modified DNA. Biochemistry 2012, 51, 6728–6737. [Google Scholar] [CrossRef]
- Sullivan, G.J.; McStay, B. Dimerization and HMG box domains 1–3 present in Xenopus UBF are sufficient for its role in transcriptional enhancement. Nucleic Acids Res. 1998, 26, 3555–3561. [Google Scholar] [CrossRef] [PubMed]
- Decoville, M.; Giacomello, E.; Leng, M.; Locker, D. DSP1, an HMG-like protein, is involved in the regulation of homeotic genes. Genetics 2001, 157, 237–244. [Google Scholar] [PubMed]
- Panday, A.; Grove, A. The high mobility group protein HMO1 functions as a linker histone in yeast. Epigenetics Chromatin 2016, 9, 13. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Wilson, J.J.; Lippard, S.J. Monofunctional and higher-valent platinum anticancer agents. Inorg. Chem. 2013, 52, 12234–12249. [Google Scholar] [CrossRef]
- Totsingan, F.; Bell, A.J., Jr. Interaction of HMG proteins and H1 with hybrid PNA-DNA junctions. Protein Sci. 2013, 22, 1552–1562. [Google Scholar] [CrossRef]
- Thomas, J.O.; Stott, K. H1 and HMGB1: Modulators of chromatin structure. Biochem. Soc. Trans. 2012, 40, 341–346. [Google Scholar] [CrossRef]
- Begum, N.; Pash, J.M.; Bhorjee, J.S. Expression and synthesis of high mobility group chromosomal proteins in different rat skeletal cell lines during myogenesis. J. Biol. Chem. 1990, 265, 11936–11941. [Google Scholar]
- Muller, S.; Ronfani, L.; Bianchi, M.E. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J. Intern. Med. 2004, 255, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Campana, L.; Bosurgi, L.; Rovere-Querini, P. HMGB1: A two-headed signal regulating tumor progression and immunity. Curr. Opin. Immunol. 2008, 20, 518–523. [Google Scholar] [CrossRef]
- Mosevitsky, M.I.; Novitskaya, V.A.; Iogannsen, M.G.; Zabezhinsky, M.A. Tissue specificity of nucleocytoplasmic distibution of HMG1 and HMG2 proteins and their probable functions. Eur. J. Biochem. 1989, 185, 303–310. [Google Scholar] [CrossRef]
- Raucci, A.; Di Maggio, S.; Scavello, F.; D’Ambrosio, A.; Bianchi, M.E.; Capogrossi, M.C. The Janus face of HMGB1 in heart disease: A necessary update. Cell. Mol. Life Sci. 2019, 76, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Knapp, S.; Muller, S.; Digilio, G.; Bonaldi, T.; Bianchi, M.E.; Musco, G. The Long Acidic Tail of High Mobility Group Box 1 (HMGB1) Protein Forms an Extended and Flexible Structure That Interacts with Specific Residues within and between the HMG Boxes. Biochemistry 2004, 43, 11992–11997. [Google Scholar] [CrossRef]
- Stott, K.; Watson, M.; Howe, F.S.; Grossmann, J.G.; Thomas, J.O. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J. Mol. Biol. 2010, 403, 706–722. [Google Scholar] [CrossRef]
- Carrozza, M.J.; Deluca, N. The High Mobility Group Protein 1 Is a Coactivator of Herpes Simplex Virus ICP4 In Vitro. J. Virol. 1998, 72, 6752–6757. [Google Scholar] [CrossRef] [PubMed]
- Zetterstrom, C.K.; Bergman, T.; Rynnel-Dagoo, B.; Erlandsson, H.H.; Soder, O.; Andersson, U.; Boman, H.G. High mobility group box chromosomal protein 1 (HMGB1) is an antibacterial factor produced by the human adenoid. Pediatr. Res. 2002, 52, 148–154. [Google Scholar] [CrossRef]
- Gong, W.; Li, Y.; Chao, F.; Huang, G.; He, F. Amino acid residues 201-205 in C-terminal acidic tail region plays a crucial role in antibacterial activity of HMGB1. J. Biomed. Sci. 2009, 16, 83. [Google Scholar] [CrossRef]
- Broadhurst, R.W.; Hardman, C.H.; Thomas, J.O.; Laue, E.D. Backbone dynamics of the A-domain of HMG1 as studied by 15N NMR spectroscopy. Biochemistry 1995, 34, 16608–16617. [Google Scholar] [CrossRef]
- Hardman, C.H.; Broadhurst, R.W.; Raine, A.R.; Grasser, K.D.; Thomas, J.O.; Laue, E.D. Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry 1995, 34, 16596–16607. [Google Scholar] [CrossRef]
- Teo, S.H.; Grasser, K.D.; Thomas, J.O. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur. J. Biochem. 1995, 230, 943–950. [Google Scholar] [CrossRef]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef]
- Breydo, L.; Redington, J.M.; Uversky, V.N. Effects of Intrinsic and Extrinsic Factors on Aggregation of Physiologically Important Intrinsically Disordered Proteins. Int. Rev. Cell Mol. Biol. 2017, 329, 145–185. [Google Scholar] [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder, biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef]
- Watson, M.; Stott, K.; Thomas, J.O. Mapping Intramolecular Interaction between Domains in HMGB1 using Tail-truncation Approach. J. Mol. Biol. 2007, 374, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, Y. High mobility group proteins and their post-translational modifications. Biochim. Biophys. Acta 2008, 1784, 1159–1166. [Google Scholar] [CrossRef]
- Richard, S.A.; Jiang, Y.; Xiang, L.H.; Zhou, S.; Wang, J.; Su, Z.; Xu, H. Post-translational modifications of high mobility group box 1 and cancer. Am. J. Transl. Res. 2017, 9, 5181–5196, 1943-8141/AJTR0068988. [Google Scholar]
- Ugrinova, I.; Pasheva, E.A.; Armengaud, J.; Pashev, I.G. In Vivo acetylation of HMG1 protein enhances its binding affinity to distorted DNA structures. Biochemistry 2001, 40, 14655–14660. [Google Scholar] [CrossRef]
- Ugrinova, I.; Pashev, I.G.; Pasheva, E.A. Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry 2009, 48, 6502–6507. [Google Scholar] [CrossRef]
- Ugrinova, I.; Zlateva, S.; Pashev, I.G.; Pashev, E.A. Native HMGB1 protein inhibits repair of cisplatin-damaged nucleosomes in vitro. Int. J. Biochem. Cell Biol. 2009, 41, 1556–1562. [Google Scholar] [CrossRef]
- Ugrinova, I.; Pashev, I.G.; Pasheva, E.A. Post-synthetic acetylation of HMGB1 protein modulates its interactions with supercoiled DNA. Mol. Biol. Rep. 2009, 6, 1399–1404. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, M.; Wang, W.; Tang, J. The HMGB1 acidic tail regulates HMGB1 DNA binding specificity by a unique mechanism. Biochem. Biophys. Res. Commun. 2007, 360, 14–19. [Google Scholar] [CrossRef]
- Cato, L.; Stott, K.; Watson, M.; Thomas, J.O. The Interaction of HMGB1 and Linker Histones Occurs Through their Acidic and Basic Tails. J. Mol. Biol. 2008, 384, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Pasheva, E.; Sarov, M.; Bidjekov, K.; Ugrinova, I.; Sarg, B.; Lindner, H.; Pashev, I.G. In Vitro Acetylation of HMGB-1 and -2 Proteins by CBP: The Role of the Acidic Tail. Biochemistry 2004, 43, 2935–2940. [Google Scholar] [CrossRef] [PubMed]
- Bonaldi, T.; Talamo, F.; Scaffidi, P.; Ferrera, D.; Porto, A.; Bachi, A.; Rubartelli, A.; Agresti, A.; Bianchi, M.E. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. Embo J. 2003, 22, 5551–5560. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.H.; Shin, J.S.; Immunol, J. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 2006, 177, 7889–7897. [Google Scholar] [CrossRef] [PubMed]
- Ito, I.; Fukazawa, J.; Yoshida, M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J. Biol. Chem. 2007, 282, 16336–16344. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.J.; Youn, J.H.; Ji, Y.; Lee, S.E.; Lim, K.J.; Choi, J.E.; Shin, J.-S. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium dependent mechanism. J. Immunol. 2009, 182, 5800–5809. [Google Scholar] [CrossRef] [PubMed]
- Venereau, E.; Casalgrandi, M.; Schiraldi, M.; Antoine, D.J.; Cattaneo, A.; De Marchis, F.; Liu, J.; Antonelli, A.; Preti, A.; Raeli, L.; et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 2012, 209, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lundback, P.; Ottosson, L.; Erlandsson-Harris, H.; Venereau, E.; Bianchi, M.E.; Al-Abed, Y.; Andersson, U.; Tracey, K.J.; Antoine, D.J. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med. 2012, 18, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Tsung, A.; Tohme, S.; Billiar, T.R. High-mobility group box-1 in sterile inflammation. J. Intern. Med. 2014, 276, 425–443. [Google Scholar] [CrossRef]
- Boffa, L.C.; Sterner, R.; Vidali, G.; Allfrey, V.G. Post-synthetic modifications of nuclear proteins high mobility group proteins are methylated. Biochem. Biophys. Res. Commun. 1979, 89, 1322–1327. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Zeh, H.J., 3rd; Lotze, M.T. High-mobility group box 1, oxidative stress, and disease. Antioxid. Redox Signal. 2011, 14, 1315–1335. [Google Scholar] [CrossRef]
- Polanska, E.; Pospisilova, S.; Stros, M. Binding of histone H1 to DNA is differentially modulated by redox state of HMGB1. PLoS ONE 2014, 9, e89070. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lippard, S.J. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry 2011, 50, 2567–2574. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kwak, M.S.; Park, J.B.; Lee, S.A.; Choi, J.E.; Cho, H.S.; Shin, J.S. N-linked glycosylation plays a crucial role in the secretion of HMGB1. J. Cell Sci. 2016, 129, 29–38. [Google Scholar] [CrossRef]
- Ditsworth, D.; Zong, W.X.; Thompson, C.B. Activation of poly (ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J. Biol. Chem. 2007, 282, 17845–17854. [Google Scholar] [CrossRef]
- Zong, W.X.; Ditsworth, D.; Bauer, D.E.; Wang, Z.Q.; Thompson, C.B. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004, 18, 1272–1282. [Google Scholar] [CrossRef]
- Davis, K.; Banerjee, S.; Friggeri, A.; Bell, C.; Abraham, E.; Zerfaoui, M. Poly (ADP-ribosyl) ation of high mobility group box 1 (HMGB1) protein enhances inhibition of efferocytosis. Mol. Med. 2012, 18, 359–369. [Google Scholar] [CrossRef]
- Huang, H.; Nace, G.W.; McDonald, K.A.; Tai, S.; Klune, J.R.; Rosborough, B.R.; Ding, Q.; Loughran, P.; Zhu, X.; Beer-Stolz, D.; et al. Hepatocyte specific HMGB1 deletion worsens the injury in liver ischemia/reperfusion: A role for intracellular HMGB1 in cellular protection. Hepatology 2014, 59, 1984–1997. [Google Scholar] [CrossRef]
- Joshi, S.R.; Sarpong, Y.C.; Peterson, R.C.; Scovell, W.M. Nucleosome dynamics: HMGB1 relaxes canonical nucleosome structure to facilitate estrogen receptor binding. Nucleic Acids Res. 2012, 40, 10161–10171. [Google Scholar] [CrossRef][Green Version]
- Polyanichko, A.M.; Davydenko, S.G.; Chikhirzhina, E.V.; Vorob’ev, V.I. Interaction of superhelical DNA with the nonhistone protein HMG1. Tsitologiya 2000, 42, 787–793. [Google Scholar]
- Rodionova, T.Y.; Chikhirzhina, E.V.; Vorob’yov, V.I.; Polyanichko, A.M. Changes in the secondary structure of HMGB1 protein bonded to DNA. J. Struct. Chem. 2010, 50, 976–981. [Google Scholar] [CrossRef]
- Reeves, R. Nuclear functions of the HMG proteins. Biochim. Biophys. Acta 2010, 1799, 3–14. [Google Scholar] [CrossRef]
- Jung, Y.; Lippard, S.J. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry 2003, 42, 2664–2671. [Google Scholar] [CrossRef]
- Muller, S.; Bianchi, M.E.; Knapp, S. Thermodynamics of HMGB1 Interaction with Duplex DNA. Biochemistry 2001, 40, 10254–10261. [Google Scholar] [CrossRef] [PubMed]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef] [PubMed]
- Polyanichko, A.M.; Leonenko, Z.V.; Cramb, D.; Wieser, H.; Vorob’ev, V.I.; Chikhirzhina, E.V. Visualization of DNA complexes with HMGB1 and its C-truncated form HMGB1(A+B). Biophysics 2008, 53, 202–206. [Google Scholar] [CrossRef]
- Stott, K.; Tang, G.S.; Lee, K.B.; Thomas, J.O. Structure of a complex of tandem HMG boxes and DNA. J. Mol. Biol. 2006, 360, 90–104. [Google Scholar] [CrossRef]
- Kohlstaedt, L.A.; Cole, R.D. Specific interaction between H1 histone and high mobility protein HMG1. Biochemistry 1994, 3, 570–575. [Google Scholar] [CrossRef]
- Kohlstaedt, L.A.; Sung, E.C.; Fujishige, A.; Cole, R.D. Non-histone chromosomal protein HMG1 modulates the histone H1-induced condensation of DNA. J. Biol. Chem. 1987, 262, 524–526. [Google Scholar]
- Fonin, A.V.; Stepanenko, O.V.; Turoverov, K.K.; Vorob’ev, V.I. Interaction between non-histone chromatin protein HMGB1 and linker histone H1. Tsitologiia 2010, 52, 946–949. [Google Scholar] [CrossRef]
- McBryant, S.J.; Lu, X.; Hansen, J.C. Multifunctionality of the linker histones: An emerging role for protein-protein interactions. Cell Res. 2010, 20, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.-Q.; Liu, L.-P.; Hess, D.; Rietdorf, J.; Sun, F.-L. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006, 20, 1959–1973. [Google Scholar] [CrossRef] [PubMed]
- Postnikov, Y.V.; Bustin, M. Functional interplay between histone H1 and HMG proteins in chromatin. Biochim. Biophys. Acta 2016, 1859, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V. Histone structure and the organization of the nucleosome. Ann. Rev. Biophys. Biomol. Struct. 1997, 26, 83. [Google Scholar] [CrossRef] [PubMed]
- Polyanichko, A.M.; Chikhirzhina, E.V.; Andrushchenko, V.V.; Vorob’ev, V.I.; Wieser, H. The effect of manganese(II) on the structure of DNA/HMGB1/H1 complexes: Electronic and vibrational circular dichroism studies. Biopolymers 2006, 83, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Polyanichko, A.; Chikhirzhina, E. Interaction between nonhistone protein HMGB1 and linker histone H1 facilitates the formation of structurally ordered DNA-protein complexes. Spectroscopy 2012, 27, 393–398. [Google Scholar] [CrossRef]
- Polyanichko, A.; Chikhirzhina, E. Supramolecular organization of the complexes of DNA with chromosomal proteins HMGB1 and H1. Adv. Biomed. Spectrosc. 2013, 7, 185–190. [Google Scholar] [CrossRef]
- Polyanichko, A.; Wieser, H. Fourier transform infrared/vibrational circular dichroism spectroscopy as an informative tool for the investigation of large supramolecular complexes of biological macromolecules. Biopolymers 2005, 78, 329–339. [Google Scholar] [CrossRef]
- Grosshedl, R.; Giese, K.; Pagel, J. HMG domain proteins: Architectural elements in assembly nucleoprotein structures. Trends Genet. 1994, 10, 94–100. [Google Scholar] [CrossRef]
- Ito, H.; Fujita, K.; Tagawa, K.; Chen, X.; Homma, H.; Sasabe, T.; Shimizu, J.; Shimizu, S.; Tamura, T.; Muramatsu, S.; et al. HMGB1 facilitates repair of mitochondrial DNA damage and extends the lifespan of mutant ataxin-1 knock-in mice. Embo Mol. Med. 2014, 7, 78–101. [Google Scholar] [CrossRef]
- Varga-Weisz, P.; van Holde, K.; Zlatanova, J. Competition between linker histones and HMG1 for binding to four-way junction DNA: Implications for transcription. Biochem. Biophys. Res. Commun. 1994, 203, 1904–1911. [Google Scholar] [CrossRef] [PubMed]
- Krynetski, E.Y.; Krynetskaia, N.F.; Bianchi, M.E.; Evans, W.E. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70, ERp60, andglyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res. 2003, 63, 100–106. [Google Scholar] [PubMed]
- Yuan, F.; Gu, L.; Guo, S.; Wang, C.; Li, G.M. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J. Biol. Chem. 2004, 279, 20935–20940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yuan, F.; Presnell, S.R.; Tian, K.; Gao, Y.; Tomkinson, A.E.; Gu, L.; Li, G.M. Reconstitution of 5′-directed human mismatch repair in purified system. Cell 2005, 122, 693–705. [Google Scholar] [CrossRef]
- Prasad, R.; Liu, Y.; Deterding, L.J.; Poltoratsky, V.P.; Kedar, P.S.; Horton, J.K.; Kanno, S.; Asagoshi, K.; Hou, E.W.; Khodyreva, S.N.; et al. HMGB1 is a cofactor in mammalian base excision repair. Mol. Cell 2007, 27, 829–841. [Google Scholar] [CrossRef]
- Malina, J.; Kasparkova, J.; Natile, G.; Brabec, V. Recognition of major DNA adducts of en antiomeric cisplatin analogs by HMG box proteins and nucleotide excision repair of these adducts. Chem. Biol. 2002, 9, 629–638. [Google Scholar] [CrossRef]
- Liu, Y.; Prasad, R.; Wilson, S.H. HMGB1: Roles in base excision repair and related function. Biochim. Biophys. Acta 2010, 1799, 119–130. [Google Scholar] [CrossRef]
- Sawchuk, D.J.; Mansilla-Soto, J.; Alarcon, C.; Singha, N.C.; Langen, H.; Bianchi, M.E.; Lees-Miller, S.P.; Nussenzweig, M.C.; Cortes, P. Ku70/Ku80 and DNA-dependent protein kinase catalytic subunit modulate RAG-mediated cleavage: Implications for the enforcement of the 12/23 rule. J. Biol. Chem. 2004, 279, 29821–29831. [Google Scholar] [CrossRef]
- Van Gent, D.C.; Hiom, K.; Paull, T.T.; Gellert, M. Stimulation of V(D)J cleavage by high mobility group proteins. Embo J. 1997, 16, 2665–2670. [Google Scholar] [CrossRef]
- Stros, M.; Cherny, D.; Jovin, T.M. HMG1 protein stimulates DNA end joining by promoting association of DNA molecules via their ends. Eur. J. Biochem. 2000, 267, 4088–4097. [Google Scholar] [CrossRef]
- Little, A.J.; Corbett, E.; Ortega, F.; Schatz, D.G. Cooperative recruitment of HMGB1 during V(D)Jrecombination through interactions with RAG1 and DNA. Nucleic Acids Res. 2013, 41, 3289–3301. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, E.R.; Lippard, S.J. Stopped-flow fluorescence studies of HMG-domain protein binding to cisplatin-modified DNA. Biochemistry 2000, 39, 8426–8438. [Google Scholar] [CrossRef] [PubMed]
- Murray, V. Nucleosomes and cisplatin. Chem. Biol. 2010, 17, 1271–1272. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Todd, R.C.; Lippard, S.J. Inhibition of transcription by platinum antitumor compounds. Metallomics 2009, 1, 280–291. [Google Scholar] [CrossRef]
- Todd, R.C.; Lippard, S.J. Consequences of cisplatin binding on nucleosome structure and dynamics. Chem. Biol. 2010, 17, 1334–1343. [Google Scholar] [CrossRef]
- Reeves, R.; Adair, J.E. Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Repair. 2005, 4, 926–938. [Google Scholar] [CrossRef]
- He, Q.; Liang, C.H.; Lippard, S.J. Steroid hormones induce HMG1 overexpression and sensitize breast cancer cells to cisplatin and carboplatin. Proc. Natl. Acad. Sci. USA 2000, 97, 5768–5772. [Google Scholar] [CrossRef]
- Zamble, D.B.; Mikata, Y.; Eng, C.H.; Sandman, K.E.; Lippard, S.J. Testis-specific HMG-domain protein alters the responses of cells to cisplatin. J. Inorg. Biochem. 2002, 91, 451–462. [Google Scholar] [CrossRef]
- Bang, S.; Kaur, S.; Kurokawa, M. Regulation of the p53 Family Proteins by the Ubiquitin Proteasomal Pathway. Int. J. Mol. Sci. 2020, 21, 261. [Google Scholar] [CrossRef]
- Rowell, P.; Simpson, K.L.; Stott, K.; Watson, M.; Thomas, J.O. HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure 2012, 20, 2014–2024. [Google Scholar] [CrossRef]
- McKinney, K.; Prives, C. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol. Cell. Biol. 2002, 22, 6797–6808. [Google Scholar] [CrossRef] [PubMed]
- Stros, M.; Kucirek, M.; Sani, S.A.; Polanska, E. HMGB1-mediated DNA bending: Distinct roles in increasing p53 binding to DNA and the transactivation of p53-responsive gene promoters. Biochim. Biophys. Acta Gen. Regul. Mech. 2018, 1861, 200–210. [Google Scholar] [CrossRef]
- Imamura, T.; Izumi, H.; Nagatani, G.; Ise, T.; Nomoto, M.; Iwamoto, Y.; Kohno, K. Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein. J. Biol. Chem. 2001, 276, 7534–7540. [Google Scholar] [CrossRef]
- Livesey, K.M.; Kang, R.; Zeh, H.J., 3rd; Lotze, M.T.; Tang, D. Direct molecular interactions between HMGB1 and TP53in colorectal cancer. Autophagy 2012, 8, 846–848. [Google Scholar] [CrossRef]
- Livesey, K.M.; Kang, R.; Vernon, P.; Buchser, W.; Loughran, P.; Watkins, S.C.; Zhang, L.; Manfredi, J.J.; Zeh, H.J., 3rd; Li, L.; et al. p53/HMGB1 complexes regulate autophagy andapoptosis. Cancer Res. 2012, 72, 1996–2005. [Google Scholar] [CrossRef]
- Bagherpoor, A.J.; Dolezalova, D.; Barta, T.; Kučírek, M.; Sani, S.A.; Ešner, M.; Bosakova, M.K.; Vinarský, V.; Peskova, L.; Hampl, A.; et al. Properties of Human Embryonic Stem Cells and their Differentiated Derivatives Depend on Non-histone DNA-Binding HMGB1 and HMGB2 Proteins. Stem Cells Dev. 2017, 27, 328–340. [Google Scholar] [CrossRef]
- Huang, Z.; Zhong, Z.; Zhang, L.; Wang, X.; Xu, R.; Zhu, L.; Wang, Z.; Hu, S.; Zhao, X. Down-regulation of HMGB1 expression by shRNA constructs inhibits the bioactivity of urothelial carcinoma cell lines via the NF-κB pathway. Sci. Rep. 2015, 5, 12807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, C.; Hou, R. Knockdown of HMGB1 improves apoptosis and suppresses proliferation and invasion of glioma cells. Chin. J. Cancer Res. 2014, 26, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Shie, M.Y.; Ding, S.J. Integrin binding and MAPK signal pathways in primary cell responses to surface chemistry of calcium silicate cements. Biomaterials 2013, 34, 6589–6606. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.; Rudnicki, M. Oct4 Interaction with Hmgb2 Regulates Akt Signaling and Pluripotency. Stem Cells. 2013, 31, 1107–1120. [Google Scholar] [CrossRef]
- Schnerch, A.; Cerdan, C.; Bhatia, M. Distinguishing between mouse and human pluripotent stem cell regulation: The best laid plans of mice and men. Stem Cells 2010, 28, 419–430. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.-L.; et al. Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Polanská, E.; Dobšáková, Z.; Dvořáčková, M.; Fajkus, J.; Štros, M. HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma 2012, 121, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Rouhiainen, A.; Li, Z.; Guo, S.; Rauvala, H. Regulation of Neurogenesis in Mouse Brain by HMGB1. Cells 2020, 9, 1714. [Google Scholar] [CrossRef] [PubMed]
- Damalas, A.; Ben-Ze’ev, A.; Simcha, I.; Shtutman, M.; Leal, J.F.; Zhurinsky, J.; Geiger, B.; Oren, M. Excess beta-catenin promotes accumulation of transcriptionally active p53. Embo J. 1999, 18, 3054–3063. [Google Scholar] [CrossRef] [PubMed]
- Patapoutian, A.; Reichardt, L.F. Roles of Wnt proteins in neural development and maintenance. Curr. Opin. Neurobiol. 2000, 10, 392–399. [Google Scholar] [CrossRef]
- Calogero, S.; Grassi, F.; Aguzzi, A.; Voigtländer, T.; Ferrier, P.; Ferrari, S.; Bianchi, M.E. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycemia in newborn mice. Nat. Gen. 1999, 22, 276–280. [Google Scholar] [CrossRef]
- Ronfani, L.; Ferraguti, M.; Croci, L.; Ovitt, C.E.; Schöler, H.R.; Consalez, G.G.; Bianchi, M.E. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 2001, 128, 1265–1273. [Google Scholar]
- Sato, M.; Miyata, K.; Tian, Z.; Kadomatsu, T.; Ujihara, Y.; Morinaga, J.; Horiguchi, H.; Endo, M.; Zhao, J.; Zhu, S.; et al. Loss of Endogenous HMGB2 Promotes Cardiac Dysfunction and Pressure Overload-Induced Heart Failure in Mice. Circ. J. 2019, 83, 368–378. [Google Scholar] [CrossRef]
- Kuznik, B.I.; Khavinson, V.K.; Linkova, N.S.; Sall, T.S. Alarmin1 (HMGB1) and Age-Related Pathologies. Epygenetic Regulatory Mechanisms. Usp. Fiziol. Nauk 2017, 48, 40–55. [Google Scholar]
- Brien, M.-E.; Baker, B.; Duval, C.; Gaudreault, V.; Jones, R.L.; Girard, S. Alarmins at the maternal-fetal interface: Involvement of inflammation in placental dysfunction and pregnancy complications. Can. Pfysiol. Pharmacor. 2019, 96, 206–212. [Google Scholar] [CrossRef]
- Zhang, X.; Wheeler, D.; Tang, Y.; Guo, L.; Shapiro, R.A.; Ribar, T.J.; Means, A.R.; Billiar, T.R.; Angus, D.C.; Rosengart, M.R. Calcium/calmodulin dependent protein kinase (CaMK) IV mediates nucleocytoplasmic shuttling and release of HMGB1 during lipopolysaccharide stimulation of macrophages. J. Immunol. 2008, 181, 5015–5023. [Google Scholar] [CrossRef]
- Rabadi, M.M.; Xavier, S.; Vasko, R.; Kaur, K.; Goligorksy, M.S.; Ratliff, B.B. High-mobility group box 1 is a novel deacetylation target of Sirtuin1. Kidney Int. 2015, 87, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Tang, D. PKR-dependent inflammatory signals. Sci. Signal. 2012, 5(247), 47. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.W.; Jiang, W.; Reich, C.F., 3rd; Pisetsky, D.S. The extracellular release of HMGB1 during apoptotic cell death. Am. J. Physiol. Cell Physiol. 2006, 291, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Ju, Z.; Ragab, A.A.; Lundbäck, P.; Long, W.; Valdes-Ferrer, S.I.; He, M.; Pribis, J.P.; Li, J.; et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J. Exp. Med. 2015, 212, 5–14. [Google Scholar] [CrossRef]
- Verrijdt, G.; Haelens, A.; Schoenmakers, E.; Rombauts, W.; Claessens, F. Comparative analysis of the influence of the high-mobility group box 1 protein on DNA binding and transcriptional activation by the androgen, glucocorticoid, progesterone and mineralocorticoid receptors. Biochem. J. 2002, 361, 97–103. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Livesey, K.M.; Zeh, H.J., 3rd; Lotze, M.T. High Mobility Group Box 1 (HMGB1) Activates an Autophagic Response to Oxidative Stress. Antioxid. Redox Signal. 2011, 15, 2185–2195. [Google Scholar] [CrossRef]
- Zandarashvili, L.; Sahu, D.; Lee, K.; Lee, Y.S.; Singh, P.; Rajarathnam, K.; Iwahara, J. Real-time kinetics of high-mobility group box 1 (HMGB1) oxidation in extracellular fluids studied by in situ protein NMR spectroscopy. J. Biol. Chem. 2013, 288, 11621–11627. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-M.; Kim, B.J.; Toro, L.N.; Skoultchi, A.I. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl. Acad. Sci. USA 2013, 110, 1708–1713. [Google Scholar] [CrossRef]
- Stark, K.; Philippi, V.; Stockhausen, S.; Busse, J.; Antonelli, A.; Miller, M.; Schubert, I.; Hoseinpour, P.; Chandraratne, S.; von Brühl, M.L.; et al. Disulfide HMGB1 derived from platelets coordinates venous thrombosis in mice. Blood 2016, 128, 2435–2449. [Google Scholar] [CrossRef]
- Tirone, M.; Tran, N.L.; Ceriotti, C.; Gorzanelli, A.; Canepari, M.; Bottinelli, R.; Raucci, A.; Di Maggio, S.; Santiago, C.; Mellado, M.; et al. High mobility group box 1 orchestrates tissue regeneration via CXCR4. J. Exp. Med. 2018, 215, 303–318. [Google Scholar] [CrossRef]
- Venereau, E.; Ceriotti, C.; Bianchi, M.E. DAMPs from cell death to new life. Front. Immunol. 2015, 6, 442. [Google Scholar] [CrossRef]
- Ferrara, M.; Chialli, G.; Ferreira, L.M.; Ruggieri, E.; Careccia, G.; Preti, A.; Piccirillo, R.; Bianchi, M.E.; Sitia, G.; Venereau, E. Oxidation of HMGB1 Is a Dynamically Regulated Process in Physiological and Pathological Conditions. Front. Immunol. 2020, 11, 1122. [Google Scholar] [CrossRef]
- Tian, J.; Avalos, A.M.; Mao, S.Y.; Chen, B.; Senthil, K.; Wu, H.; Parroche, P.; Drabic, S.; Golenbock, D.; Sirois, C.; et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007, 8, 487–496. [Google Scholar] [CrossRef]
- Orlova, V.V.; Choi, E.Y.; Xie, C.; Chavakis, E.; Bierhaus, A.; Ihanus, E.; Ballantyne, C.M.; Gahmberg, C.G.; Bianchi, M.E.; Nawroth, P.P.; et al. A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. Embo J. 2007, 26, 1129–1139. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. Eaten alive: A history of macroautophagy. Nat. Cell Biol. 2010, 12, 814–822. [Google Scholar] [CrossRef]
- Kang, R.; Livesey, K.M.; Zeh, H.J., III; Lotze, M.T.; Tang, D. Metabolic regulation by HMGB1-mediated autophagy and mitophagy. Autophagy 2011, 7, 1256–1258. [Google Scholar] [CrossRef]
- Lee, G.; Santo, A.I.E.; Zwingenberger, S.; Cai, L.; Vogl, T.; Feldmann, M.; Horwood, N.J.; Chan, J.K.; Nanchahalm, J. Fully reduced HMGB1 accelerates the regeneration of multiple tissues by transitioning stem cells to GAlert. Proc. Natl. Acad. Sci. USA 2018, 115, E4463–E4472. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; Blood, D.C.; del Toro, G.; Canet, A.; Lee, D.C.; Qu, W.; Tanji, N.; Lu, Y.; Lalla, E.; Fu, C.; et al. Blockade of RAGE amphoterin signalling suppresses tumour growth and metastases. Nature 2000, 405, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, H.J.; Fages, C.; Kuja-Panula, J.; Ridley, A.J.; Rauvala, H. Receptor for advanced glycation end products-binding COOH-terminal motif of amphoterin inhibits invasive migration and metastasis. Cancer Res. 2002, 62, 4805–4811. [Google Scholar] [PubMed]
- Hanahan, D.; Weinberg, R.A. HMGB1 in Cancer: Good, Bad, or Both? Clin. Cancer Res. 2013, 19, 4046–4057. [Google Scholar] [CrossRef]
- Zhong, H.; Li, X.; Zhou, S.; Jiang, P.; Liu, X.; Ouyang, M.; Nie, Y.; Chen, X.; Zhang, L.; Liu, Y.; et al. Interplay between RAGE and TLR4 Regulates HMGB1-Induced Inflammation by Promoting Cell Surface Expression of RAGE and TLR4. J. Immunol. 2020, 205, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Lanzardo, S.; Arigoni, M.; Antonazzo, R.; Radaelli, E.; Cantarella, D.; Calogero, R.A.; Cavallo, F. The noninflammatory role of high mobility group box 1/Toll-like receptor 2 axis in the self-renewal of mammary cancer stem cells. Faseb J. 2013, 27, 4731–4744. [Google Scholar] [CrossRef]
- Qiu, Y.; Yang, J.; Wang, W.; Zhao, W.; Peng, F.; Xiang, Y.; Chen, G.; Chen, T.; Chai, C.; Zheng, S.; et al. HMGB1-promoted and TLR2/4-dependent NK cell maturation and activation take part in rotavirus-induced murine biliary atresia. PLoS Pathog. 2014, 10, e1004011. [Google Scholar] [CrossRef]
- Chen, G.Y.; Tang, J.; Zheng, P.; Liu, Y. CD24 and Siglec-10 selectively repress tissue damageinduced immune responses. Science 2009, 323, 1722–1725. [Google Scholar] [CrossRef]
- Maugeri, N.; Rovere-Querini, P.; Baldini, M.; Baldissera, E.; Sabbadini, M.G.; Bianchi, M.E.; Manfredi, A.A. Oxidative stress elicits platelet/leukocyte inflammatory interactions via HMGB1: A candidate for microvessel injury in systemic sclerosis. Antioxid. Redox Signal. 2014, 20, 1060–1074. [Google Scholar] [CrossRef]
- Davalos, A.R.; Kawahara, M.; Malhotra, G.K.; Schaum, N.; Huang, J.; Ved, U.; Beausejour, C.M.; Coppe, J.P.; Rodier, F.; Campisi, J. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol. 2013, 201, 613–629. [Google Scholar] [CrossRef]
- Lohani, N.; Rajeswari, M.R. Dichotomous life of DNA binding High Mobility Group Box 1 protein in human health and disease. Curr. Protein Pept. Sci. 2016, 17, 762–775. [Google Scholar] [CrossRef]
- Bianchi, M.E.; Crippa, M.P.; Manfredi, A.A.; Mezzapelle, R.; Rovere Querini, P.; Venereau, E. High-mobility group box 1 protein orchestrates responses to tissue damage via inflammation, innate and adaptive immunity, and tissue repair. Immunol. Rev. 2017, 280, 74–82. [Google Scholar] [CrossRef]
- Yasinska, I.M.; Silva, I.G.; Sakhnevych, S.S.; Ruegg, L.; Hussain, R.; Siligardi, G.; Fiedler, W.; Wellbrock, J.; Bardelli, M.; Varani, L.; et al. High mobility group box 1 (HMGB1) acts as an “alarmin” to promote acute myeloid leukaemia progression. Oncoimmunology 2018, 7, e1438109. [Google Scholar] [CrossRef]
- Lin, F.; Xue, D.; Xie, T.; Pan, Z. HMGB1 promotes cellular chemokine synthesis and potentiates mesenchymal stromal cell migration via Rap1 activation. Mol. Med. Rep. 2016, 14, 1283–1289. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chan, D.C. Mitochondria: Dynamic organelles in disease, aging, and development. Cell 2006, 125, 1241–1252. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Livesey, K.M.; Kroemer, G.; Billiar, T.R.; Van Houten, B.; Zeh III, H.J.; Lotze, M.T. High-Mobility Group Box 1 Is Essential for Mitochondrial Quality Control. Cell Metab. 2011, 13, 701–711. [Google Scholar] [CrossRef]
- Garrido, C. Size matters: Of the small HSP27 and its large oligomers. Cell Death Differ. 2002, 9, 483–485. [Google Scholar] [CrossRef]
- Arrigo, A.P. The cellular ‘‘networking’’ of mammalian Hsp27 and its functions in the control of protein folding, redox state and apoptosis. Adv. Exp. Med. Biol. 2007, 594, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Marino, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Kim, I.; Rodriguez-Enriquez, S.; Lemasters, J.J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007, 462, 245–253. [Google Scholar] [CrossRef]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, J.N.; Gingras-Breton, G.; Tanguay, R.M.; Landry, J. Induction of Chinese hamster HSP27 gene expression in mouse cells confers resistance to heat shock. HSP27 stabilization of the microfilament organization. J. Biol. Chem. 1993, 268, 3420–3429. [Google Scholar] [PubMed]
- Lavoie, J.N.; Hickey, E.; Weber, L.A.; Landry, J. Modulation of actin microfilament dynamics and fluid phase pinocytosis by phosphorylation of heat shock protein 27. J. Biol. Chem. 1993, 268, 24210–24214. [Google Scholar] [PubMed]
- Boldogh, I.R.; Pon, L.A. Interactions of mitochondria with the actin cytoskeleton. Biochim. Biophys. Acta 2006, 1763, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, S.; Houle, F.; Landry, J.; Huot, J. p38 MAP kinase activation by vascular endothelial growth factor mediates actin reorganization and cell migration in human endothelial cells. Oncogene 1997, 15, 2169–2177. [Google Scholar] [CrossRef]
- Green, D.R. Apoptotic pathways: Ten minutes to dead. Cell 2005, 121, 671–674. [Google Scholar] [CrossRef]
- Rodier, F.; Campisi, J. Four faces of cellular senescence. J. Cell Biol. 2011, 192, 547–556. [Google Scholar] [CrossRef]
- Sofiadis, K.; Nikolic, M.; Kargapolova, Y.; Josipovic, N.; Zirkel, A.; Papadakis, A.; Papadionysiou, I.; Loughran, G.; Keanes, J.; Michel, A.; et al. HMGB1 coordinates SASP-related chromatin folding and RNA homeostasis on the path to senescence. bioRxiv 2020. [Google Scholar] [CrossRef]
- Giavara, S.; Kosmidou, E.; Hande, M.P.; Bianchi, M.E.; Morgan, A.; d’Adda di Fagagna, F.; Jackson, S.P. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Curr. Biol. 2005, 15, 68–72. [Google Scholar] [CrossRef]
- Gazzar, M.E.; Yoza, B.K.; Chen, X.; Garcia, B.A.; Young, N.L.; McCall, C.E. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol. Cell. Biol. 2009, 29, 1959–1971. [Google Scholar] [CrossRef] [PubMed]
- Celona, B.; Weiner, A.; Di Felice, F.; Mancuso, F.M.; Cesarini, E.; Rossi, R.L.; Gregory, L.; Baban, D.; Rossetti, G.; Grianti, P.; et al. Substantial histone reduction modulates genome wide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011, 9, e1001086. [Google Scholar] [CrossRef]
- De Toma, I.; Rossetti, G.; Zambrano, S.; Bianchi, M.E.; Agresti, A. Nucleosome loss facilitates the chemotactic response of macrophages. J. Intern. Med. 2014, 276, 454–469. [Google Scholar] [CrossRef]
- Nikopoulou, C.; Panagopoulos, G.; Sianidis, G.; Psarra, E.; Ford, E.; Thanos, D. The transcription factor ThPOK orchestrates stochastic interchromosomal interactions required for IFNB1 virus-inducible gene expression. Mol. Cell 2018, 71, 352–361. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Zirkel, A.; Nikolic, M.; Sofiadis, K.; Mallm, J.P.; Brackley, C.A.; Gothe, H.; Drechsel, O.; Becker, C.; Altmüller, J.; Josipovic, N.; et al. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell 2018, 70, 730–744. [Google Scholar] [CrossRef]
- Criscione, S.W.; De Cecco, M.; Siranosian, B.; Zhang, Y.; Kreiling, J.A.; Sedivy, J.M.; Neretti, N. Reorganization of chromosome architecture in replicative cellular senescence. Sci. Adv. 2016, 2, e1500882. [Google Scholar] [CrossRef]
- Sexton, T.; Cavalli, G. The role of chromosome domains in shaping the functional genome. Cell 2015, 160, 1049–1059. [Google Scholar] [CrossRef]
- Fraser, J.; Ferrai, C.; Chiariello, A.M.; Schueler, M.; Rito, T.; Laudanno, G.; Barbieri, M.; Moore, B.L.; Kraemer, D.C.; Aitken, S.; et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 2015, 11, 852. [Google Scholar] [CrossRef]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef]
- Wang, W.K.; Lu, Q.H.; Zhang, J.N.; Wang, B.; Liu, X.J.; An, F.S.; Qin, W.D.; Chen, X.Y.; Dong, W.Q.; Zhang, C.; et al. HMGB1 mediates hyperglycaemia-induced cardiomyocyte apoptosis via ERK/Ets-1 signalling pathway. J. Cell. Mol. Med. 2014, 18, 2311–2320. [Google Scholar] [CrossRef]
- Tsung, A.; Sahai, R.; Tanaka, H.; Nakao, A.; Fink, M.P.; Lotze, M.T.; Yang, H.; Li, J.; Tracey, K.J.; Geller, D.A.; et al. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemiareperfusion. J. Exp. Med. 2005, 201, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Andrassy, M.; Volz, H.C.; Igwe, J.C.; Funke, B.; Eichberger, S.N.; Kaya, Z.; Buss, S.; Autschbach, F.; Pleger, S.T.; Lukic, I.K.; et al. High-mobility group box-1 in ischemia-reperfusion injury of the heart. Circulation 2008, 117, 3216–3226. [Google Scholar] [CrossRef] [PubMed]
- Mollica, L.; De Marchis, F.; Spitaleri, A.; Dallacosta, C.; Pennacchini, D.; Zamai, M.; Agresti, A.; Trisciuoglio, L.; Musco, G.; Bianchi, M.E. Glycyrrhizin Binds to High-Mobility Group Box 1 Protein and Inhibits Its Cytokine Activities. Chem. Biol. 2007, 14, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.; Zhang, M.; Zhang, Y.; Xu, H.; Wang, Y.; An, G.; Wang, Y.; Li, L. Glycyrrhizin protects rat heart against ischemia-reperfusion injury through blockade of HMGB1-dependent phospho-JNK/Bax pathway. Acta Pharmacol. Sin. 2012, 33, 1477–1487. [Google Scholar] [CrossRef]
- Bangert, A.; Andrassy, M.; Müller, A.; Bockstahler, M.; Fischer, A.; Volz, C.H.; Leib, C.; Göser, S.; Korkmaz-Icöz, S.; Zittrich, S.; et al. Critical role of RAGE and HMGB1 in inflammatory heart disease. Proc. Natl. Acad. Sci. USA 2016, 113, E155–E164. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, Z.; Barnie, P.A.; Su, Z. Dual faced HMGB1 plays multiple roles in cardiomyocyte senescence and cardiac inflammatory injury. Cytokine Gr. Factor Rev. 2019, 47, 74–82. [Google Scholar] [CrossRef]
- Wei, J.; Alfajaro, M.M.; DeWeirdt, P.C.; Hanna, R.E.; Lu-Culligan, W.J.; Cai, W.L.; Strine, M.S.; Zhang, S.-M.; Graziano, V.R.; Schmitz, C.O.; et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 2020. [Google Scholar] [CrossRef]
| Localization of Protein | Partner | Functions |
|---|---|---|
| Nuclear HmgB1 | Linker histone H1 | C-terminal sequence of HmgB1 binds N-terminal region of H1, disrupting its interactions with DNA and leading to the displacement of H1 [29,30] to facilitate the sliding of nucleosome along the DNA, which is essential for the binding of transcription factors [12,72,83]. |
| Transcription factors | The interaction of transcription factors with DNA might occur via the formation of an intermediate complex TF1–DNA–HmgB1, whose formation induces attachment of other regulatory chromatin proteins [11]. | |
| Hsp70, MSH2, MLH1 | HmgB1 works during the initial stages of recognition/damage of the DNA and interacts with MMR proteins, [113,114,115]. | |
| Enzymes which take part in BER and NER | Base excision repair (BER) and nucleotide excision repair (NER) are also associated with the interaction between HmgB1 and various enzymes [67,68,69,70,116,117,118]. | |
| DNA-dependent protein kinase RAG1 and RAG2 proteins T4 DNA ligase | Repair of double-strand breaks (DSBR) involves DNA-dependent protein kinase (DNA-PKcs) [119]: in vitro HmgB1 protein stimulates activity of DNA-Pkcs [119], RAG1, and RAG2 proteins [120] and increases the activity of T4 DNA ligase [121]. The complex between RAG1, RAG2, and HmgB1 proteins induces the formation of hairpin on DNA [122]. | |
| Tumor suppressor p53 protein | Interaction of p53 with the A-domain of HmgB1 is regulated by C-terminal tail of HmgB1 [131]. HmgB1/p53 complex regulates apoptosis and autophagy [135,136]. | |
| Nuclear factor (NF)-κB Steroid hormone receptors Glucocorticoid receptors | HmgB1 regulates the transcriptional activity of these proteins [133,160]. | |
| Nuclear export protein CRM1 | N-glycosylation of HmgB1 is important for binding with CRM1 [85]. | |
| LPS (lipopolysaccharide) | N-glycosylation of HmgB1 is mediated by phorbol 12-myristate 13-acetate (PMA), trichostatin A (TSA) and lipopolysaccharide (LPS) and can lead to the secretion of the protein into the extracellular space as a result of decreasing HmgB1-DNA binding affinity and increasing association with nuclear export protein CRM1 [85]. | |
| cPKC (calcium/phospholipid-dependent protein kinase C) | In vertebrates, phosphorylation of HmgB1 involves calcium/phospholipid-dependent protein kinase C (cPKC) by the PI3K-PKC signaling pathway [77]. | |
| Phosphatase inhibitors (TNF-α or okadaic acid) | HmgB1 can be phosphorylated in mouse macrophage cells RAW264.7 and human monocytes after their treatment with these phosphatase inhibitors, leading to HmgB1 translocation to cytoplasm with possible subsequent secretion into the extracellular space [75]. | |
| PARP1 (poly-ADP-ribose polymerase 1) | PARP1 promotes repair of damaged bases and single-stranded DNA breaks by modulating the structure of chromatin and binding DNA repair factors [89]. Thus, there is a cross-link between ADP-ribosylation of HmgB1 and PARP1 in regulating cell death. | |
| Extranuclear HmgB1 | RAGE (receptor for advanced glycation end products) | ADP-ribosylation affects the binding of HmgB1 to RAGE [88]. fr-HmgB1 can bind to RAGE and promote autophagy by inhibiting mTOR kinase and promoting Beclin1–Ptdlns3KC3 complex formation [171]. RAGE binding to fr-HmgB1 stimulates production of CXCL12. ds-HmgB1–RAGE interaction leads to activation of neutrophils and the formation of their extracellular traps [164,165]. |
| Toll-like receptor TLR9 | HmgB1 interacts with TLR9, increasing cytokine production [168]. | |
| Toll-like receptors TLR2 and TLR4 | ds-HmgB1/TLR4-MD2 complex stimulates release of inflammatory and angiogenic factors by activation of transcription nuclear factor NF-κB. The interaction of HmgB1 with TLR2 or TLR4 regulates inflammation process when lungs or liver are damaged due to epilepsy, heart disease, or cancer [52]. | |
| Toll-like receptor TLR5 | The HmgB1–TLR5 complex activates the NF-κB signaling pathway through the adapter protein MyD88, involved in signal transmission from toll-like receptors, which leads to increased synthesis of pro-inflammatory cytokines [152]. | |
| TIM-3 (T cell immunoglobulin mucin-3) | The interaction of HmgB1 with TIM-3 inhibits the activity of dendritic cells [152]. | |
| Rap1 (Ras-associated protein-1) | HmgB1 activates signal pathway of Rap1 [185]. | |
| TLR9 | HmgB1 interacts with TLR9 of endoplasmic reticulum and Golgi complex [152]. | |
| Illexin IL-6 | The oxidized form of the HMGB1 protein stimulates the secretion of pro-inflammatory cytokines, including IL-6 by activation of the TLR-4 receptor [152]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chikhirzhina, E.; Starkova, T.; Beljajev, A.; Polyanichko, A.; Tomilin, A. Functional Diversity of Non-Histone Chromosomal Protein HmgB1. Int. J. Mol. Sci. 2020, 21, 7948. https://doi.org/10.3390/ijms21217948
Chikhirzhina E, Starkova T, Beljajev A, Polyanichko A, Tomilin A. Functional Diversity of Non-Histone Chromosomal Protein HmgB1. International Journal of Molecular Sciences. 2020; 21(21):7948. https://doi.org/10.3390/ijms21217948
Chicago/Turabian StyleChikhirzhina, Elena, Tatyana Starkova, Anton Beljajev, Alexander Polyanichko, and Alexey Tomilin. 2020. "Functional Diversity of Non-Histone Chromosomal Protein HmgB1" International Journal of Molecular Sciences 21, no. 21: 7948. https://doi.org/10.3390/ijms21217948
APA StyleChikhirzhina, E., Starkova, T., Beljajev, A., Polyanichko, A., & Tomilin, A. (2020). Functional Diversity of Non-Histone Chromosomal Protein HmgB1. International Journal of Molecular Sciences, 21(21), 7948. https://doi.org/10.3390/ijms21217948





