Extracellular Matrix-Specific Platelet Activation Leads to a Differential Translational Response and Protein De Novo Synthesis in Human Platelets
Abstract
:1. Introduction
2. Results
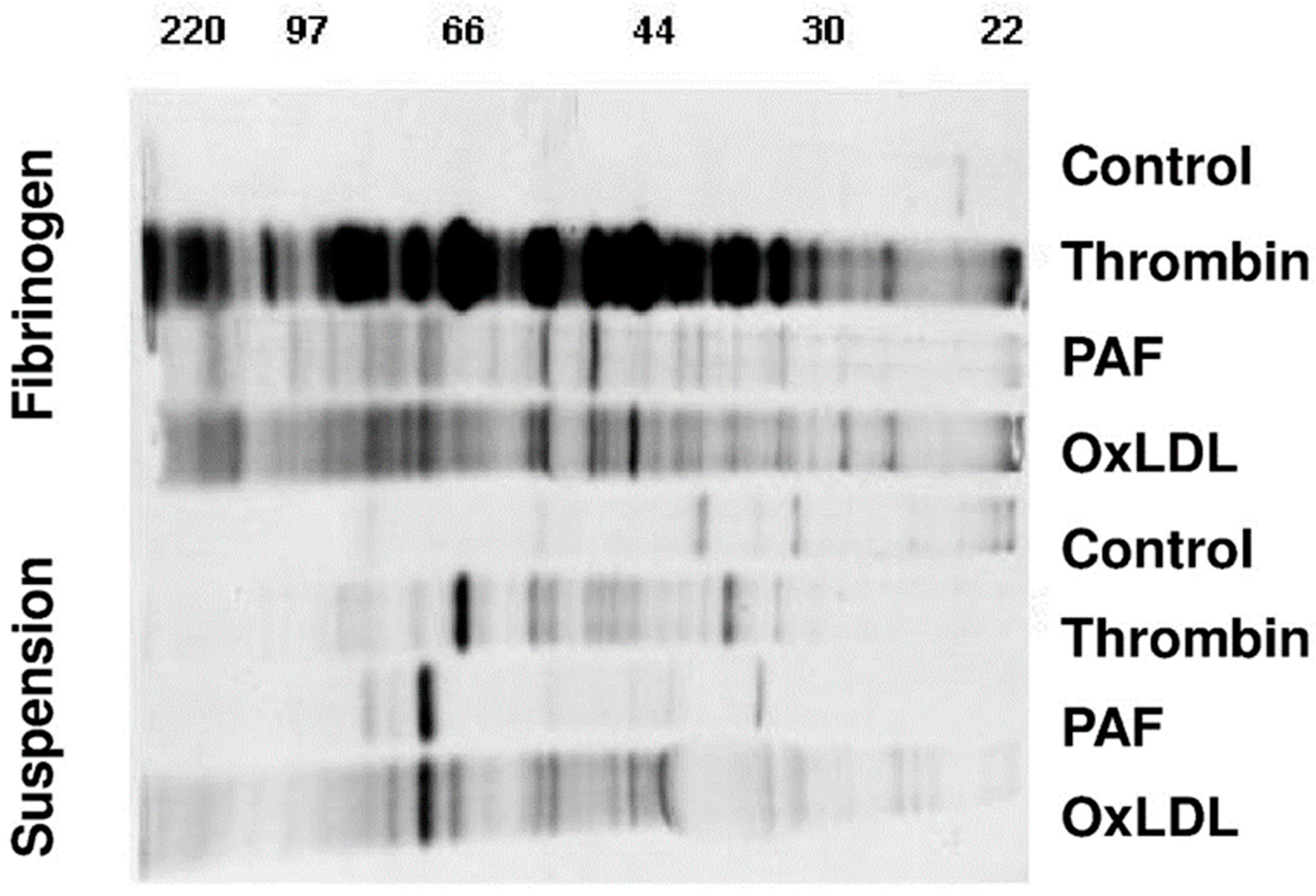
2.1. De Novo Protein Synthesis of Activated Platelets Is Markedly Increased by Fibrinogen Binding
2.2. Adherent Platelet Morphology Is Specific to the Extracellular Matrix Stimulus
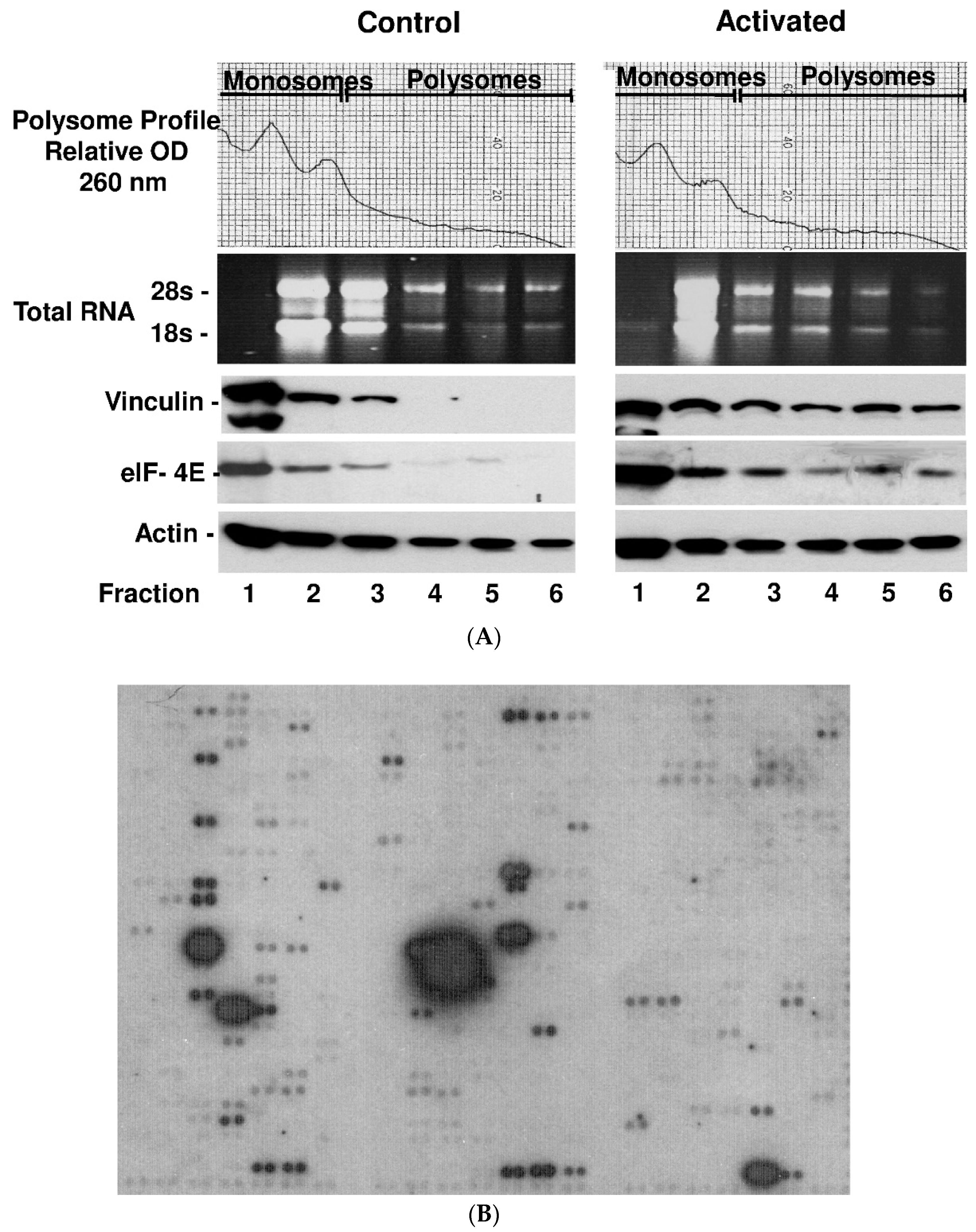
2.3. Platelet Activation Results in Polysomal Accumulation and Assembly of the Translational Machinery (Consisting of eIF4E, mRNAs and Vinculin)
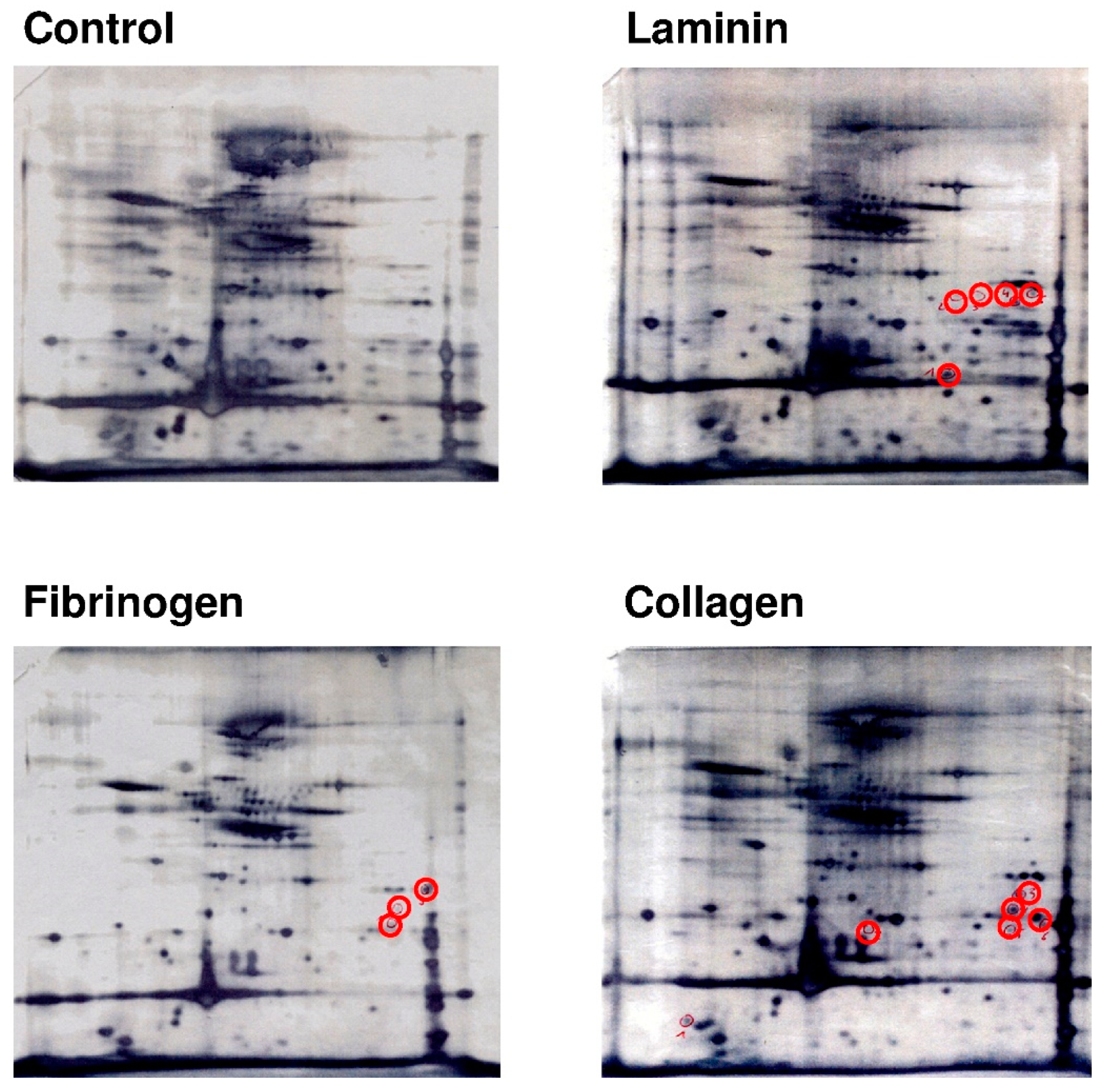
2.4. Platelets Show a Differential and Stimulus-Dependent Protein Expression Profile after Exposure to Extracellular Matrix Proteins Laminin or Collagen and to Fibrinogen
2.5. Platelets That Are Adherent to Laminin Synthesize Proteins That Are Essential for Platelet Migration
3. Discussion
4. Material and Methods
4.1. Platelet Isolation
4.2. Metabolic Radiolabeling of Human Platelet Proteins
4.3. Two-Dimensional Electrophoresis
4.4. Immunoblotting
4.5. Confocal Microscopy
4.6. Polysome Analysis
4.7. cDNA Arrays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Denis, M.M.; Tolley, N.D.; Bunting, M.; Schwertz, H.; Jiang, H.; Lindemann, S.; Yost, C.C.; Rubner, F.J.; Albertine, K.H.; Swoboda, K.J.; et al. Escaping the Nuclear Confines: Signal-Dependent Pre-mRNA Splicing in Anucleate Platelets. Cell 2005, 122, 379–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Májek, P.; Reicheltová, Z.; Štikarová, J.; Suttnar, J.; Sobotková, A.; Dyr, J.E. Proteome changes in platelets activated by arachidonic acid, collagen, and thrombin. Proteome Sci. 2010, 8, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nassa, G.; Giurato, G.; Cimmino, G.; Rizzo, F.; Ravo, M.; Salvati, A.; Nyman, T.A.; Zhu, Y.; Vesterlund, M.; Lehtiö, J.; et al. Splicing of platelet resident pre-mRNAs upon activation by physiological stimuli results in functionally relevant proteome modifications. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Czapiga, M.; Gao, J.-L.; Kirk, A.; Lekstrom-Himes, J. Human platelets exhibit chemotaxis using functional N-formyl peptide receptors. Exp. Hematol. 2005, 33, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Nagy, J.A.; Pyne, K.; Dvorak, H.F.; Dvorak, A.M. Platelets exit venules by a transcellular pathway at sites of F-met peptide-induced acute inflammation in guinea pigs. Int. Arch. Allergy Immunol. 1998, 116, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Gaertner, F.; Ahmad, Z.; Rosenberger, G.; Fan, S.; Nicolai, L.; Busch, B.; Yavuz, G.; Luckner, M.; Ishikawa-Ankerhold, H.; Hennel, R.; et al. Migrating Platelets are Mechano-scavengers that Collect and Bundle Bacteria. Cell 2017, 171, 1368–1382. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, B.F.; Borst, O.; Gehring, E.M.; Schoenberger, T.; Urban, B.; Ninci, E.; Seizer, P.; Schmidt, C.; Bigalke, B.; Koch, M.; et al. PI3 kinase-dependent stimulation of platelet migration by stromal cell-derived factor 1 (SDF-1). J. Mol. Med. 2010, 88, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, B.F.; Schmidt, C.; Urban, B.; Bigalke, B.; Schwanitz, L.; Koch, M.; Seizer, P.; Schaller, M.; Gawaz, M.; Lindemann, S. High shear flow induces migration of adherent human platelets. Platelets 2011, 22, 415–421. [Google Scholar] [CrossRef]
- Pitchford, S.C.; Momi, S.; Baglioni, S.; Casali, L.; Giannini, S.; Rossi, R.; Page, C.P.; Gresele, P. Allergen Induces the Migration of Platelets to Lung Tissue in Allergic Asthma. Am. J. Respir. Crit. Care Med. 2008, 177, 604–612. [Google Scholar] [CrossRef]
- Schmidt, E.M.; Kraemer, B.F.; Borst, O.; Münzer, P.; Schönberger, T.; Schmidt, C.; Leibrock, C.; Towhid, S.T.; Seizer, P.; Kuhl, D.; et al. SGK1 Sensitivity of Platelet Migration. Cell. Physiol. Biochem. 2012, 30, 259–268. [Google Scholar] [CrossRef]
- Schmidt, E.-M.; Münzer, P.; Borst, O.; Kraemer, B.F.; Schmid, E.; Urban, B.; Lindemann, S.; Ruth, P.; Gawaz, M.; Lang, F. Ion channels in the regulation of platelet migration. Biochem. Biophys. Res. Commun. 2011, 415, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Fogh, B.S.; Multhaupt, H.A.; Couchman, J.R. Protein Kinase C, Focal Adhesions and the Regulation of Cell Migration. J. Histochem. Cytochem. 2013, 62, 172–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, S.J. Regulating cell migration: Calpains make the cut. J. Cell Sci. 2005, 118, 3829–3838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, G.; Mingle, L.; van de Water, L.; Liu, G. Control of cell migration through mRNA localization and local translation. Wiley Interdiscip. Rev. RNA 2014, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Lindemann, S.; Tolley, N.D.; Eyre, J.R.; Kraiss, L.W.; Mahoney, T.M.; Weyrich, A.S. Integrins Regulate the Intracellular Distribution of Eukaryotic Initiation Factor 4E in Platelets. J. Biol. Chem. 2001, 276, 33947–33951. [Google Scholar] [CrossRef] [Green Version]
- Weyrich, A.S.; Dixon, D.A.; Pabla, R.; Elstad, M.R.; McIntyre, T.M.; Prescott, S.M.; Zimmerman, G.A. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc. Natl. Acad. Sci. USA 1998, 95, 5556–5561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pabla, R.; Weyrich, A.S.; Dixon, D.A.; Bray, P.F.; McIntyre, T.M.; Prescott, S.M.; Zimmerman, G.A. Integrin-dependent Control of Translation: Engagement of Integrin αIIbβ3 Regulates Synthesis of Proteins in Activated Human Platelets. J. Cell Biol. 1999, 144, 175–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy-Toledano, S.; Gallet, C.; Nadal, F.; Bryckaert, M.; Maclouf, J.; Rosa, J.P. Phosphorylation and Dephosphorylation Mechanisms in Platelet Function: A Tightly Regulated Balance. Thromb. Haemost. 1997, 78, 226–233. [Google Scholar] [CrossRef]
- Kraemer, B.F.; Lindemann, S.; Weyrich, A.S. Protein degradation systems in platelets. Thromb. Haemost. 2013, 110, 920–924. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Li, W.; Willard, B.; Silverstein, R.L.; McIntyre, T.M. Proteasome Proteolysis Supports Stimulated Platelet Function and Thrombosis. Arter. Thromb. Vasc. Biol. 2014, 34, 160–168. [Google Scholar] [CrossRef] [Green Version]
- Grundler, K.; Rotter, R.; Tilley, S.; Pircher, J.; Czermak, T.; Yakac, M.; Gaitzsch, E.; Massberg, S.; Krötz, F.; Sohn, H.Y.; et al. The proteasome regulates collagen-induced platelet aggregation via nuclear-factor-kappa-B (NFĸB) activation. Thromb. Res. 2016, 148, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, B.F.; Lamkemeyer, T.; Franz-Wachtel, M.; Lindemann, S. The Integrin Activating Protein Kindlin-3 is Cleaved in Human Platelets during ST-Elevation Myocardial Infarction. Int. J. Mol. Sci. 2019, 20, 6154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraemer, B.F.; Mannell, H.; Lamkemeyer, T.; Franz-Wachtel, M.; Lindemann, S. Heat-Shock Protein 27 (HSPB1) is Upregulated and Phosphorylated in Human Platelets during ST-Elevation Myocardial Infarction. Int. J. Mol. Sci. 2019, 20, 5968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, P.F.; McKenzie, S.E.; Edelstein, L.C.; Nagalla, S.; Delgrosso, K.; Ertel, A.; Kupper, J.; Jing, Y.; Londin, E.; Loher, P.; et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genom. 2013, 14, 1. [Google Scholar] [CrossRef] [Green Version]
- Schwertz, H.; Tolley, N.D.; Foulks, J.M.; Denis, M.M.; Risenmay, B.W.; Buerke, M.; Tilley, R.E.; Rondina, M.T.; Harris, E.M.; Kraiss, L.W.; et al. Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J. Exp. Med. 2006, 203, 2433–2440. [Google Scholar] [CrossRef] [Green Version]
- Weyrich, A.S.; Schwertz, H.; Kraiss, L.W.; Zimmerman, G.A. Protein synthesis by platelets: Historical and new perspectives. J. Thromb. Haemost. 2009, 7, 241–246. [Google Scholar] [CrossRef] [Green Version]
- Gnatenko, D.V.; Dunn, J.J.; Schwedes, J.; Bahou, W.F. Transcript Profiling of Human Platelets Using Microarray and Serial Analysis of Gene Expression (SAGE). In DNA and RNA Profiling in Human Blood; Bugert, P., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 496, pp. 245–272. [Google Scholar]
- Nassa, G.; de Filippo, M.R.; Giurato, G.; Ravo, M.; Rizzo, F.; Conte, S.; Pellegrino, G.; Cirillo, P.; Calabrò, P.; Öhman, T.; et al. Activating stimuli induce platelet microRNA modulation and proteome reorganisation. Thromb. Haemost. 2015, 114, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.K.; Rubenstein, P.A.; DeMali, K.A. Vinculin Nucleates Actin Polymerization and Modifies Actin Filament Structure. J. Biol. Chem. 2009, 284, 30463–30473. [Google Scholar] [CrossRef] [Green Version]
- Chanpeng, P.; Svasti, S.; Paiboonsukwong, K.; Smith, D.R.; Leecharoenkiat, K. Platelet proteome reveals specific proteins associated with platelet activation and the hypercoagulable state in beta-thalassmia/HbE patients. Sci. Rep. 2019, 9, 6059. [Google Scholar] [CrossRef] [Green Version]
- Heijnen, H.; van der Sluijs, P. Platelet secretory behaviour: As diverse as the granules … or not? J. Thromb. Haemost. 2015, 13, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Manne, B.K.; Xiang, S.C.; Rondina, M.T. Platelet secretion in inflammatory and infectious diseases. Platelets 2017, 28, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Rijkers, M.; Eshof, B.L.V.D.; van der Meer, P.F.; van Alphen, F.P.J.; de Korte, D.; Leebeek, F.W.G.; Meijer, A.B.; Voorberg, J.; Jansen, A.J.G. Monitoring storage induced changes in the platelet proteome employing label free quantitative mass spectrometry. Sci. Rep. 2017, 7, 11045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, R.; Hao, J.; Han, W.; Ni, Y.; Huang, X.; Hu, Q. Gelsolin regulates proliferation, apoptosis, migration and invasion in human oral carcinoma cells. Oncol. Lett. 2015, 9, 2129–2134. [Google Scholar] [CrossRef] [PubMed]
- Jannie, K.M.; Ellerbroek, S.M.; Zhou, D.W.; Chen, S.; Crompton, D.J.; García, A.J.; DeMali, K.A. Vinculin-dependent actin bundling regulates cell migration and traction forces. Biochem. J. 2015, 465, 383–393. [Google Scholar] [CrossRef] [Green Version]
- Pick, R.; Begandt, D.; Stocker, T.J.; Salvermoser, M.; Thome, S.; Böttcher, R.T.; Montañez, E.; Harrison, U.; Forné, I.; Khandoga, A.G.; et al. Coronin 1A, a novel player in integrin biology, controls neutrophil trafficking in innate immunity. Blood 2017, 130, 847–858. [Google Scholar] [CrossRef]
- Zhao, Y.; Malinin, N.L.; Meller, J.; Ma, Y.; West, X.Z.; Bledzka, K.; Qin, J.; Podrez, E.A.; Byzova, T.V. Regulation of Cell Adhesion and Migration by Kindlin-3 Cleavage by Calpain. J. Biol. Chem. 2012, 287, 40012–40020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, R.; Wang, Y.; Cuddy, K.; Sun, C.; Magalhaes, J.; Grynpas, M.; Glogauer, M. Filamin A Regulates Monocyte Migration Through Rho Small GTPases During Osteoclastogenesis. J. Bone Miner. Res. 2009, 25, 1077–1091. [Google Scholar] [CrossRef] [PubMed]
- Weyrich, A.S.; Elstad, M.R.; McEver, R.P.; McIntyre, T.M.; Moore, K.L.; Morrissey, J.H.; Prescott, S.M.; Zimmerman, G.A. Activated platelets signal chemokine synthesis by human monocytes. J. Clin. Investig. 1996, 97, 1525–1534. [Google Scholar] [CrossRef] [Green Version]
- Hála, M.; Cole, R.; Synek, L.; Drdová, E.; Pečenková, T.; Nordheim, A.; Lamkemeyer, T.; Madlung, J.; Hochholdinger, F.; Fowler, J.E.; et al. An Exocyst Complex Functions in Plant Cell Growth in Arabidopsis and Tobacco. Plant Cell 2008, 20, 1330–1345. [Google Scholar] [CrossRef] [Green Version]
- Tiffert, Y.; Franz-Wachtel, M.; Fladerer, C.; Nordheim, A.; Reuther, J.; Wohlleben, W.; Mast, Y. Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 2011, 89, 1149–1159. [Google Scholar] [CrossRef] [PubMed]
- Kaspar, R.L.; Gehrke, L. Peripheral blood mononuclear cells stimulated with C5a or lipopolysaccharide to synthesize equivalent levels of IL-1 beta mRNA show unequal IL-1 beta protein accumulation but similar polyribosome profiles. J. Immunol. 1994, 153, 277–286. [Google Scholar] [PubMed]


| Protein Spot # | Protein Identified |
|---|---|
| spot #1055 | Tubulin beta-1 chain |
| spot #1090 | Isoform 2 of gelsolin |
| spot #1091 | Isoform 2 of gelsolin |
| spot #1242 | FERMT3 Isoform 2 of Fermitin family homolog 3 |
| spot #1245 | FERMT3 Isoform 2 of Fermitin family homolog |
| Transaldolase (TALDO1) | |
| spot #1462 | CAPZB cDNA FLJ60094, highly similar to F-actin |
| capping protein subunit beta | |
| spot #1532 | FGA Isoform 1 of Fibrinogen alpha chain |
| spot #1799 | CAP1 Isoform 1 of Adenylyl cyclase-associated protein1 |
| HK1 Isoform 3 of Hexokinase-1 | |
| spot #478 | VCL Isoform 1 of Vinculin |
| TLN1 Talin-1 | |
| ACTN1 Alpha-actinin-1 | |
| FGG Isoform Gamma-B of Fibrinogen gamma chain | |
| ACTN4 Alpha-actinin-4 | |
| spot #479 | VCL Isoform 1 of Vinculin |
| TLN1 Talin-1 | |
| ACTN1 Alpha-actinin-1 | |
| FGG Isoform Gamma-B of Fibrinogen gamma chain | |
| ACTN4 Alpha-actinin-4 | |
| spot #562 | TLN1 Talin-1 |
| FLNA Isoform 2 of Filamin-A | |
| TLN2 Talin-2 | |
| spot #585 | VCL Isoform 1 of Vinculin |
| TLN1 Talin-1 | |
| GSN Isoform 2 of Gelsolin | |
| spot #588 | VCL Isoform 1 of Vinculin |
| GSN Isoform 2 of Gelsolin | |
| spot #6083 | VCL Isoform 1 of Vinculin |
| spot #961 | CORO1C Coronin-1C |
| spot #997 | CORO1A Coronin-1A |
| ENO1 Isoform alpha-enolase of alpha-enolase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kraemer, B.F.; Geimer, M.; Franz-Wachtel, M.; Lamkemeyer, T.; Mannell, H.; Lindemann, S. Extracellular Matrix-Specific Platelet Activation Leads to a Differential Translational Response and Protein De Novo Synthesis in Human Platelets. Int. J. Mol. Sci. 2020, 21, 8155. https://doi.org/10.3390/ijms21218155
Kraemer BF, Geimer M, Franz-Wachtel M, Lamkemeyer T, Mannell H, Lindemann S. Extracellular Matrix-Specific Platelet Activation Leads to a Differential Translational Response and Protein De Novo Synthesis in Human Platelets. International Journal of Molecular Sciences. 2020; 21(21):8155. https://doi.org/10.3390/ijms21218155
Chicago/Turabian StyleKraemer, Bjoern F., Marc Geimer, Mirita Franz-Wachtel, Tobias Lamkemeyer, Hanna Mannell, and Stephan Lindemann. 2020. "Extracellular Matrix-Specific Platelet Activation Leads to a Differential Translational Response and Protein De Novo Synthesis in Human Platelets" International Journal of Molecular Sciences 21, no. 21: 8155. https://doi.org/10.3390/ijms21218155
APA StyleKraemer, B. F., Geimer, M., Franz-Wachtel, M., Lamkemeyer, T., Mannell, H., & Lindemann, S. (2020). Extracellular Matrix-Specific Platelet Activation Leads to a Differential Translational Response and Protein De Novo Synthesis in Human Platelets. International Journal of Molecular Sciences, 21(21), 8155. https://doi.org/10.3390/ijms21218155




