Genetically Encoded Biosensor-Based Screening for Directed Bacteriophage T4 Lysozyme Evolution
Abstract
:1. Introduction
2. Results and Discussion
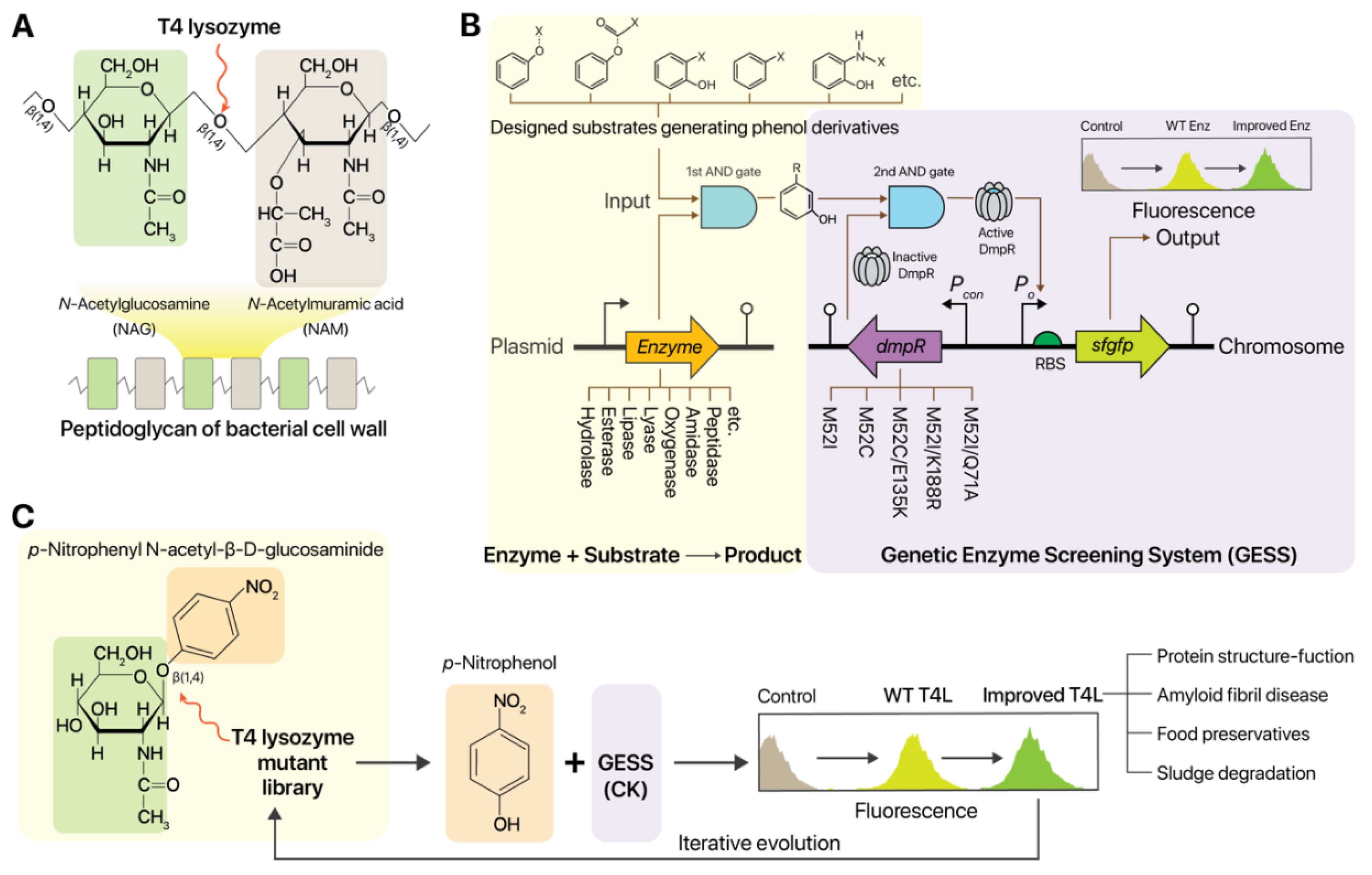
2.1. Design of T4L Screening Using Genetic Circuits
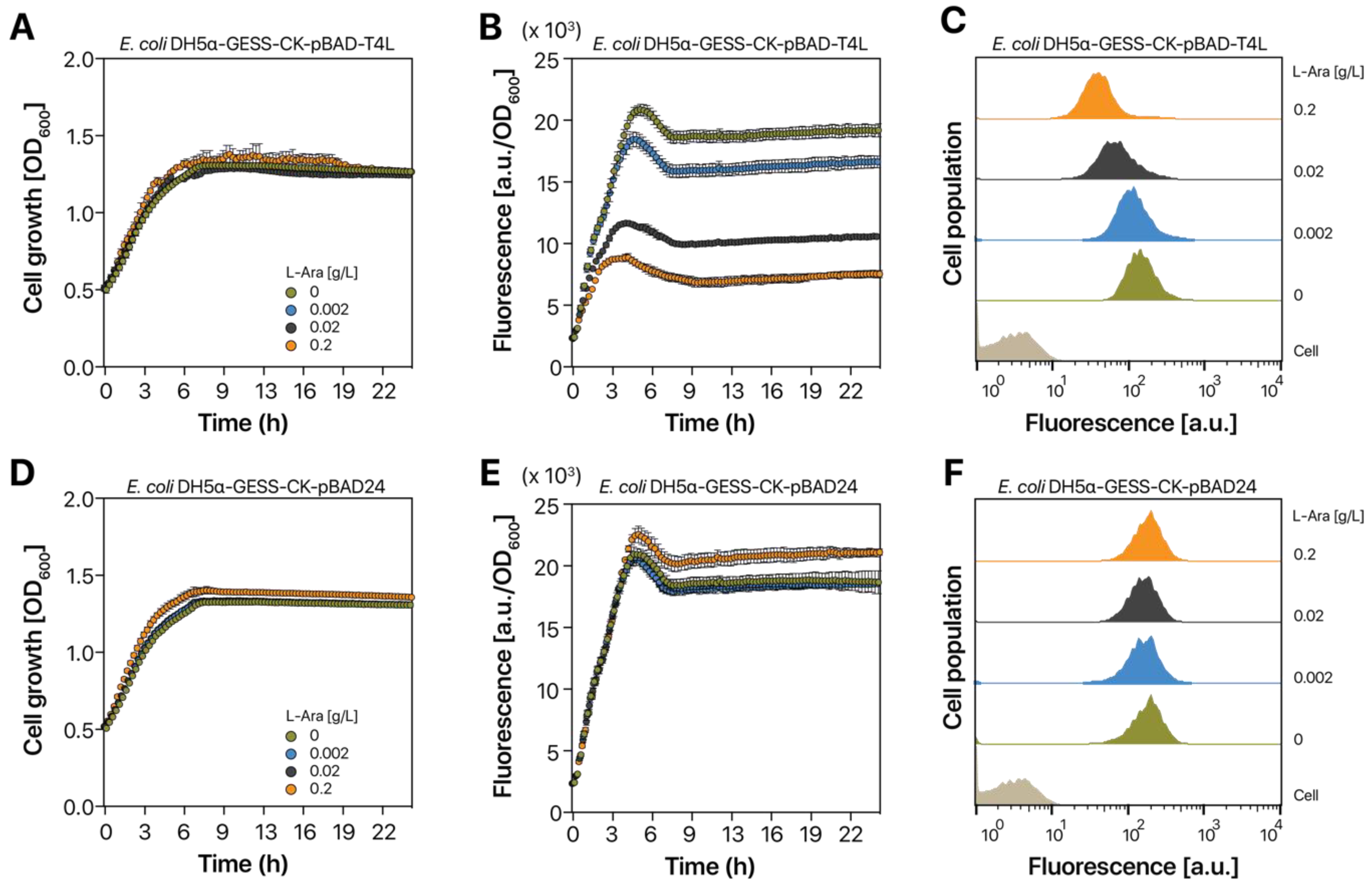
2.2. Tunable T4 Lysozyme Expression
2.3. T4L Expression in E. coli DH5α-GESS-CK Cells
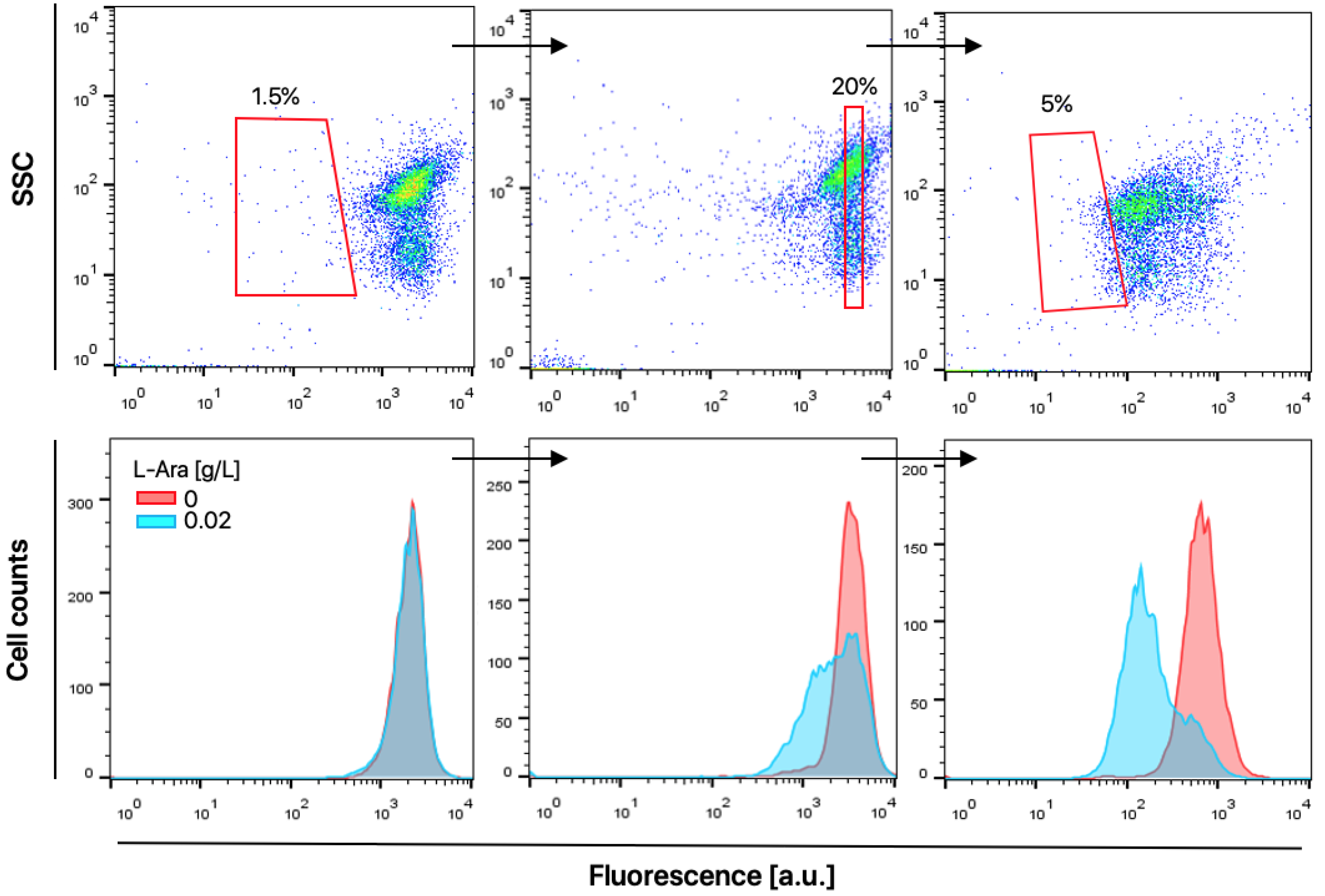
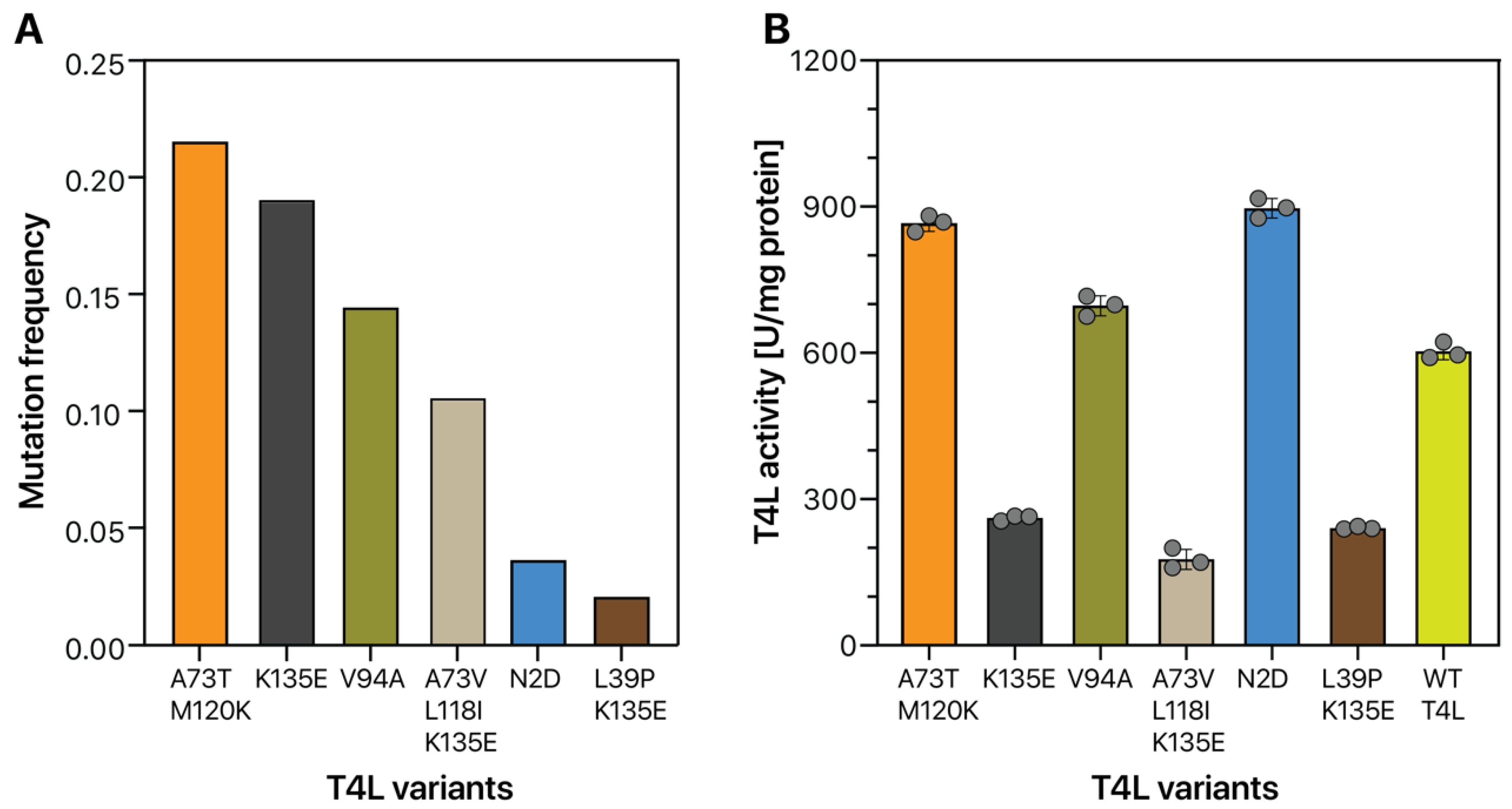
2.4. HTS of a T4L Mutant Library
3. Conclusions
4. Materials and Methods
4.1. Plasmids and Bacterial Strains
4.2. Protein Expression and Purification
4.3. Monitoring of Cell Growth and Fluorescence
4.4. Flow Cytometry Analysis
4.5. Random Mutagenesis Library Construction
4.6. HTS of T4 Lysozyme Mutant Libraries by FACS
4.7. Lysozyme Assay
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vocadlo, D.J.; Davies, G.J.; Laine, R.; Withers, S.G. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature 2001, 412, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, R.; Ravi, V.K.; Kumar, S.; Kumar, M.V.S.; Chandra, N. Lysozyme: A model protein for amyloid research. In Advances in Protein Chemistry and Structural Biology; Donev, R., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 84, pp. 63–111. [Google Scholar]
- Proctor, V.A.; Cunningham, F.E.; Fung, D.Y.C. The chemistry of lysozyme and its use as a food preservative and a pharmaceutical. CRC Crit. Rev. Food Sci. Nutr. 1988, 26, 359–395. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.O.; Holley, R.A. Interactive inhibition of meat spoilage and pathogenic bacteria by lysozyme, nisin and EDTA in the presence of nitrite and sodium chloride at 24 °C. Int. J. Food Microbiol. 2003, 80, 251–259. [Google Scholar] [CrossRef]
- Moak, M.; Molineux, I.J. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 2004, 51, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Lu, H.P. Placing single-molecule T4 lysozyme enzymes on a bacterial cell surface: Toward probing single-molecule enzymatic reaction in living cells. Biophys. J. 2004, 87, 656–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, X.-D.; He, J.-G.; Qiu, W.; Tang, J.; Liu, T.-T. Microbial community related to lysozyme digestion process for boosting waste activated sludge biodegradability. Bioresour. Technol. 2015, 175, 112–119. [Google Scholar] [CrossRef]
- Wawrzynczyk, J.; Recktenwald, M.; Norrlöw, O.; Dey, E.S. The function of cation-binding agents in the enzymatic treatment of municipal sludge. Water Res. 2008, 42, 1555–1562. [Google Scholar] [CrossRef]
- Teo, C.W.; Wong, P.C.Y. Enzyme augmentation of an anaerobic membrane bioreactor treating sewage containing organic particulates. Water Res. 2014, 48, 335–344. [Google Scholar] [CrossRef]
- Khanal, S.K.; Grewell, D.; Sung, S.; van Leeuwen, J. Ultrasound applications in wastewater sludge pretreatment: A review. Crit. Rev. Environ. Sci. Technol. 2007, 37, 277–313. [Google Scholar] [CrossRef]
- Kameswari, K.S.B.; Kalyanaraman, C.; Thanasekaran, K. Evaluation of various pre-treatment processes on tannery sludge for enhancement of soluble chemical oxygen demand. Clean Technol. Environ. Policy 2014, 16, 369–376. [Google Scholar] [CrossRef]
- Yang, Q.; Luo, K.; Li, X.-M.; Wang, D.-B.; Zheng, W.; Zeng, G.-M.; Liu, J.-J. Enhanced efficiency of biological excess sludge hydrolysis under anaerobic digestion by additional enzymes. Bioresour. Technol. 2010, 101, 2924–2930. [Google Scholar] [CrossRef] [PubMed]
- Kuglarz, M.; Karakashev, D.; Angelidaki, I. Microwave and thermal pretreatment as methods for increasing the biogas potential of secondary sludge from municipal wastewater treatment plants. Bioresour. Technol. 2013, 134, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Cassini, S.T.; Andrade, M.C.E.; Abreu, T.A.; Keller, R.; Gonçalves, R.F. Alkaline and acid hydrolytic processes in aerobic and anaerobic sludges: Effect on total EPS and fractions. Water Sci. Technol. 2006, 53, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, R.T.; Zhang, R.; Teter, S.; McGarvey, J.A. The effect of enzyme addition on anaerobic digestion of Jose Tall Wheat Grass. Bioresour. Technol. 2009, 100, 4564–4571. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Shi, Z.; Chen, S.-Y.; Luo, L. Feasibility of using lysozyme to reduce excess sludge in activated sludge process. J. Cent. South Univ. 2013, 20, 2472–2477. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [Green Version]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407. [Google Scholar] [CrossRef] [Green Version]
- Loessner, M.J. Bacteriophage endolysins—Current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef]
- Donovan, D.; Becker, S.; Dong, S.; Baker, J.; Foster-Frey, J.; Pritchard, D.J.B.I. Peptidoglycan hydrolase enzyme fusions for treating multi-drug resistant pathogens. Biotechnol. Int. 2009, 21, 6. [Google Scholar]
- Hayano, T.; Hirose, M.; Kikuchi, M. Protein disulfide isomerase mutant lacking its isomerase activity accelerates protein folding in the cell. FEBS Lett. 1995, 377, 505–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, T.; Tanaka, R.; Suetsugu, M.; Ishibashi, M.; Tokunaga, H.; Kikuchi, M.; Tokunaga, M. Efficient secretion of human lysozyme from the yeast, Kluyveromyces lactis. Biotechnol. Lett. 2004, 26, 1803–1808. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.T.; Tang, C.D.; Wu, M.C.; Liu, G.L.; Shi, H.L.; Li, J.F. Cloning and functional expression of a human lysozyme gene (hly) from human leukocytes in Pichia pastoris. Mol. Med. Rep. 2012, 6, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Busso, D.; Peleg, Y.; Heidebrecht, T.; Romier, C.; Jacobovitch, Y.; Dantes, A.; Salim, L.; Troesch, E.; Schuetz, A.; Heinemann, U.; et al. Expression of protein complexes using multiple Escherichia coli protein co-expression systems: A benchmarking study. J. Struct. Biol. 2011, 175, 159–170. [Google Scholar] [CrossRef]
- Fischer, B.; Perry, B.; Phillips, G.; Sumner, I.; Goodenough, P. Physiological consequence of expression of soluble and active hen egg white lysozyme in Escherichia coli. Appl. Microbiol. Biotechnol. 1993, 39, 537–540. [Google Scholar] [CrossRef]
- Časaitė, V.; Bružytė, S.; Bukauskas, V.; Šetkus, A.; Morozova-Roche, L.A.; Meškys, R. Expression and purification of active recombinant equine lysozyme in Escherichia coli. Protein Eng. Des. Sel. 2009, 22, 649–654. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.-L.; Rha, E.; Lee, S.J.; Kim, H.; Kwon, K.; Jeong, Y.-S.; Rhee, Y.H.; Song, J.J.; Kim, H.-S.; Lee, S.-G. Toward a generalized and high-throughput enzyme screening system based on artificial genetic circuits. ACS Synth. Biol. 2014, 3, 163–171. [Google Scholar] [CrossRef]
- Yeom, S.-J.; Kim, M.; Kwon, K.K.; Fu, Y.; Rha, E.; Park, S.-H.; Lee, H.; Kim, H.; Lee, D.-H.; Kim, D.-M.; et al. A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts. Nat. Commun. 2018, 9, 5053. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Baek, J.I.; Kim, S.J.; Kwon, K.K.; Rha, E.; Yeom, S.J.; Kim, H.; Lee, D.H.; Kim, D.M.; Lee, S.G. Sensitive and rapid phenotyping of microbes with soluble methane monooxygenase using a droplet-based assay. Front. Bioeng. Biotechnol. 2020, 8, 358. [Google Scholar] [CrossRef]
- Kwon, K.K.; Yeom, S.J.; Choi, S.L.; Rha, E.; Lee, H.; Kim, H.; Lee, D.H.; Lee, S.G. Acclimation of bacterial cell state for high-throughput enzyme engineering using a DmpR-dependent transcriptional activation system. Sci. Rep. 2020, 10, 6091. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Sung, B.H.; Oh, S.H.; Kwon, K.K.; Lee, H.; Kim, H.; Lee, D.H.; Yeom, S.J.; Lee, S.G. C1 compound biosensors: Design, functional study, and applications. Int. J. Mol. Sci. 2019, 20, 2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.K.; Kim, S.H.; Subhadra, B.; Woo, S.G.; Rha, E.; Kim, S.W.; Kim, H.; Lee, D.H.; Lee, S.G. A genetically encoded biosensor for monitoring isoprene production in engineered Escherichia coli. ACS Synth. Biol. 2018, 7, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.K.; Yeom, S.J.; Lee, D.H.; Jeong, K.J.; Lee, S.G. Development of a novel cellulase biosensor that detects crystalline cellulose hydrolysis using a transcriptional regulator. Biochem. Biophys. Res. Commun. 2018, 495, 1328–1334. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.K.; Lee, D.H.; Kim, S.J.; Choi, S.L.; Rha, E.; Yeom, S.J.; Subhadra, B.; Lee, J.; Jeong, K.J.; Lee, S.G. Evolution of enzymes with new specificity by high-throughput screening using DmpR-based genetic circuits and multiple flow cytometry rounds. Sci. Rep. 2018, 8, 2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patten, P.A.; Sonoda, T.; Davis, M.M. Directed evolution studies with combinatorial libraries of T4 lysozyme mutants. Mol. Divers. 1996, 1, 97–108. [Google Scholar] [CrossRef]
- Kim, H.; Seong, W.; Rha, E.; Lee, H.; Kim, S.K.; Kwon, K.K.; Park, K.-H.; Lee, D.-H.; Lee, S.-G. Machine learning linked evolutionary biosensor array for highly sensitive and specific molecular identification. Biosens. Bioelectron. 2020, 170, 112670. [Google Scholar] [CrossRef]
- Siegele, D.A.; Hu, J.C. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 1997, 94, 8168–8172. [Google Scholar] [CrossRef] [Green Version]
- Duarte, J.M.; Barbier, I.; Schaerli, Y. Bacterial microcolonies in gel beads for high-throughput screening of libraries in synthetic biology. ACS Synth. Biol. 2017, 6, 1988–1995. [Google Scholar] [CrossRef] [Green Version]
- Guzman, L.-M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.Y.; Park, H.J.; Yoo, Y.J. Flexibility analysis of activity-enhanced mutants of bacteriophage T4 lysozyme. J. Mol. Catal. B Enzym. 2014, 106, 95–99. [Google Scholar] [CrossRef]
- Hardy, L.W.; Poteete, A.R. Reexamination of the role of Asp20 in catalysis by bacteriophage T4 lysozyme. Biochemistry 1991, 30, 9457–9463. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Woo, S.-G.; Lee, J.; Lee, D.-H.; Hwang, S. Evaluation of feasibility of using the bacteriophage t4 lysozyme to improve the hydrolysis and biochemical methane potential of secondary sludge. Energies 2019, 12, 3644. [Google Scholar] [CrossRef] [Green Version]


| Strains or Plasmids | Description | Source |
|---|---|---|
| E. coli DH5α | F-, Φ80lacZ·ΔM15·f(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk,- mk+) phoA supE44 thi-1 gyrA96 relA1 | Thermo Fisher Scientific |
| E. coli LMG194 | F- ΔlacX74 gal E thi rpsL ΔphoA (Pvu II) Δara714 leu::Tn10 | Invitrogen |
| E. coli DH5α GESS-CK | DH5α derivatives with CK mutant dmpR GESS integrated into the chromosomal bglA locus of E. coli DH5α | [37] |
| pBAD/Myc-His/lacZ | Expression vector, PBAD:lacZ, pBR322 ori, AmpR | Invitrogen |
| pBAD24 | Expression vector, PBAD, pBR322 ori, AmpR | Invitrogen |
| pBAD-sfGFP | E. coli codon-optimized superfolder GFP expressing plasmid, PBAD, pBR322 ori, AmpR | Addgene (Plasmid #85482) |
| pBAD-T4L | T4 lysozyme-expressing plasmid with an N-terminal His6 tag, AmpR | [43] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woo, S.-G.; Kim, S.K.; Oh, B.-R.; Lee, S.-G.; Lee, D.-H. Genetically Encoded Biosensor-Based Screening for Directed Bacteriophage T4 Lysozyme Evolution. Int. J. Mol. Sci. 2020, 21, 8668. https://doi.org/10.3390/ijms21228668
Woo S-G, Kim SK, Oh B-R, Lee S-G, Lee D-H. Genetically Encoded Biosensor-Based Screening for Directed Bacteriophage T4 Lysozyme Evolution. International Journal of Molecular Sciences. 2020; 21(22):8668. https://doi.org/10.3390/ijms21228668
Chicago/Turabian StyleWoo, Seung-Gyun, Seong Keun Kim, Baek-Rock Oh, Seung-Goo Lee, and Dae-Hee Lee. 2020. "Genetically Encoded Biosensor-Based Screening for Directed Bacteriophage T4 Lysozyme Evolution" International Journal of Molecular Sciences 21, no. 22: 8668. https://doi.org/10.3390/ijms21228668
APA StyleWoo, S.-G., Kim, S. K., Oh, B.-R., Lee, S.-G., & Lee, D.-H. (2020). Genetically Encoded Biosensor-Based Screening for Directed Bacteriophage T4 Lysozyme Evolution. International Journal of Molecular Sciences, 21(22), 8668. https://doi.org/10.3390/ijms21228668







