E3 Ubiquitin Ligase APC/CCdh1 Negatively Regulates FAH Protein Stability by Promoting Its Polyubiquitination
Abstract
:1. Introduction
2. Results
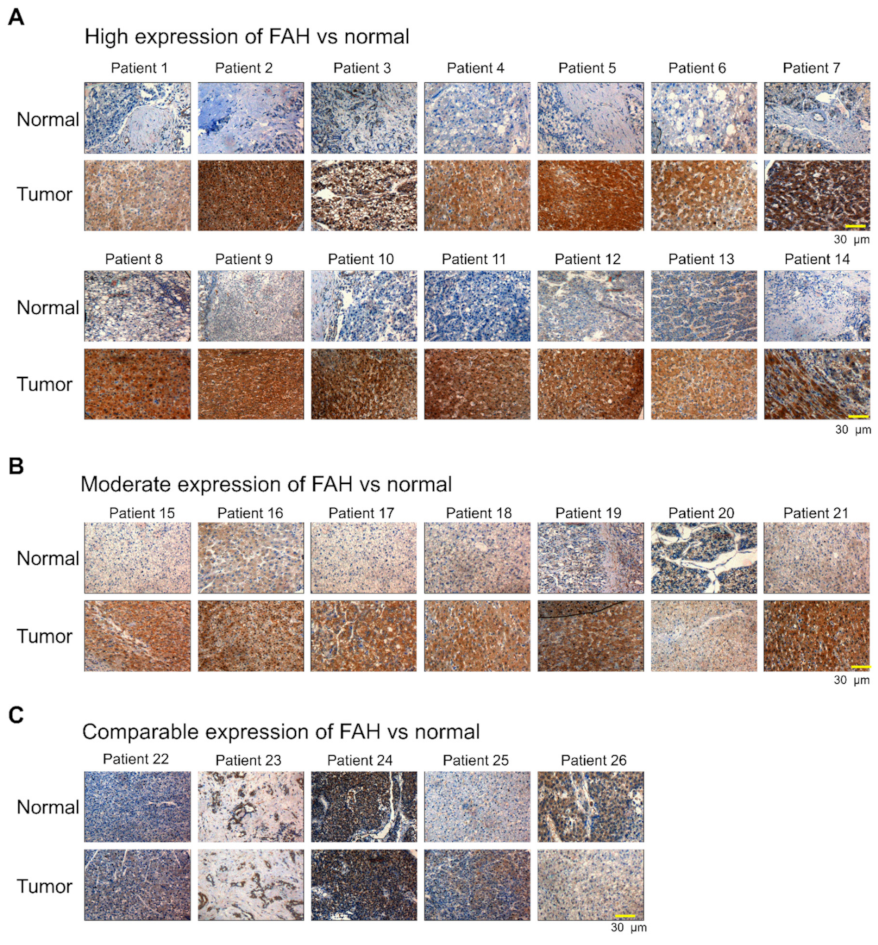
2.1. FAH Protein Expression in Human HCC Tumor Tissues
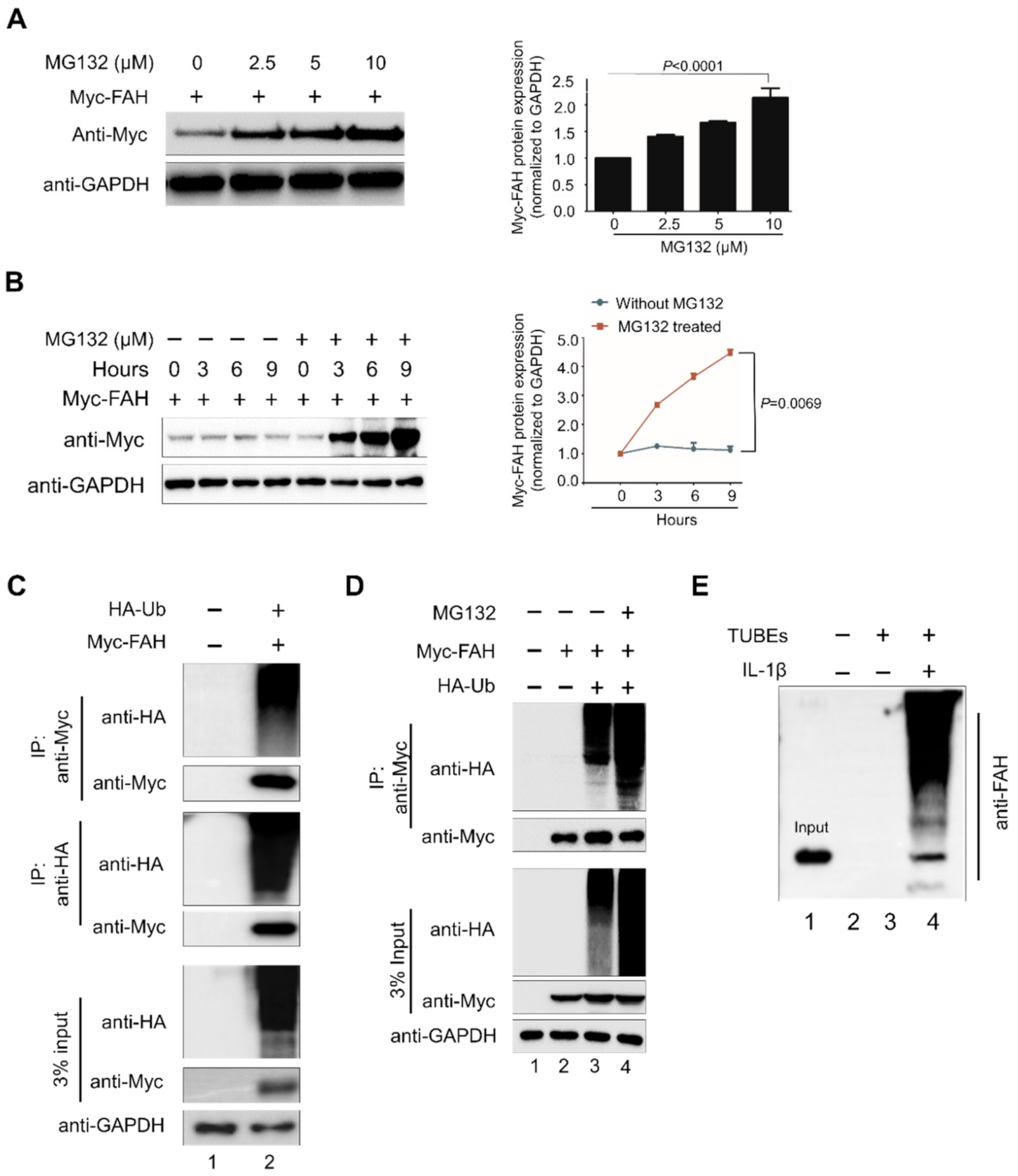
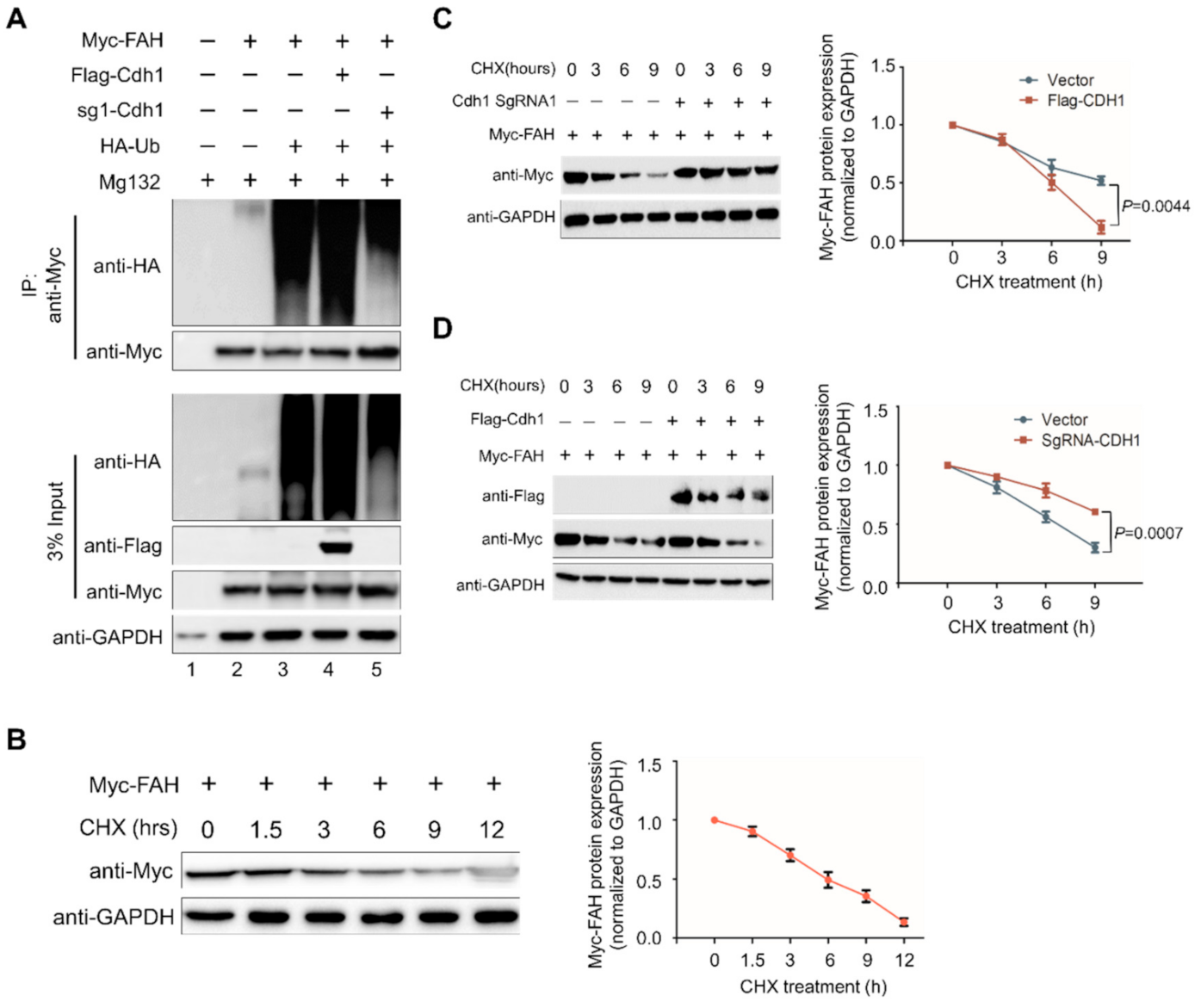
2.2. FAH Undergoes Proteolysis through the 26S Proteasomal Pathway
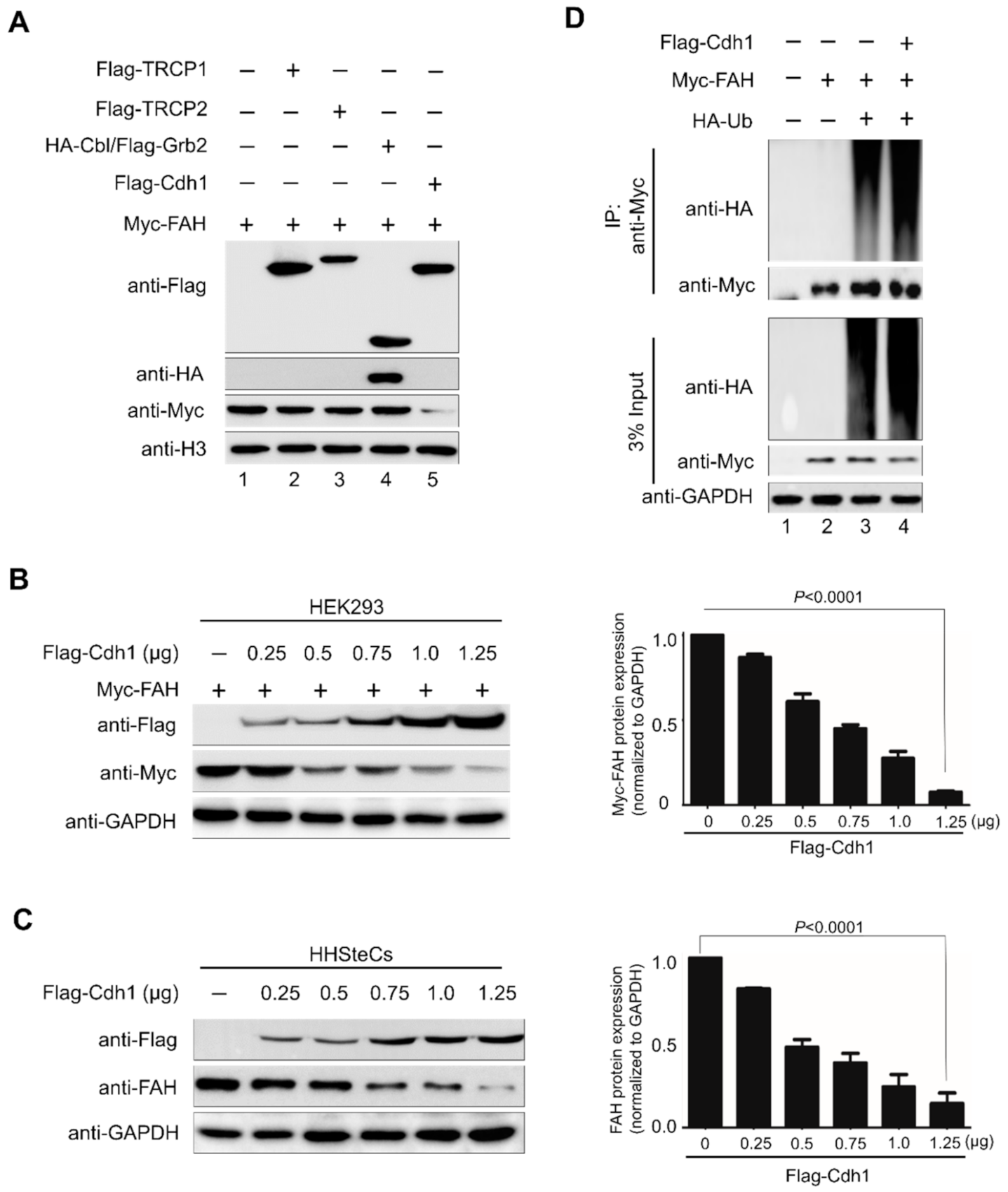
2.3. E3 Ligase APC/CCdh1 Destabilizes FAH Protein
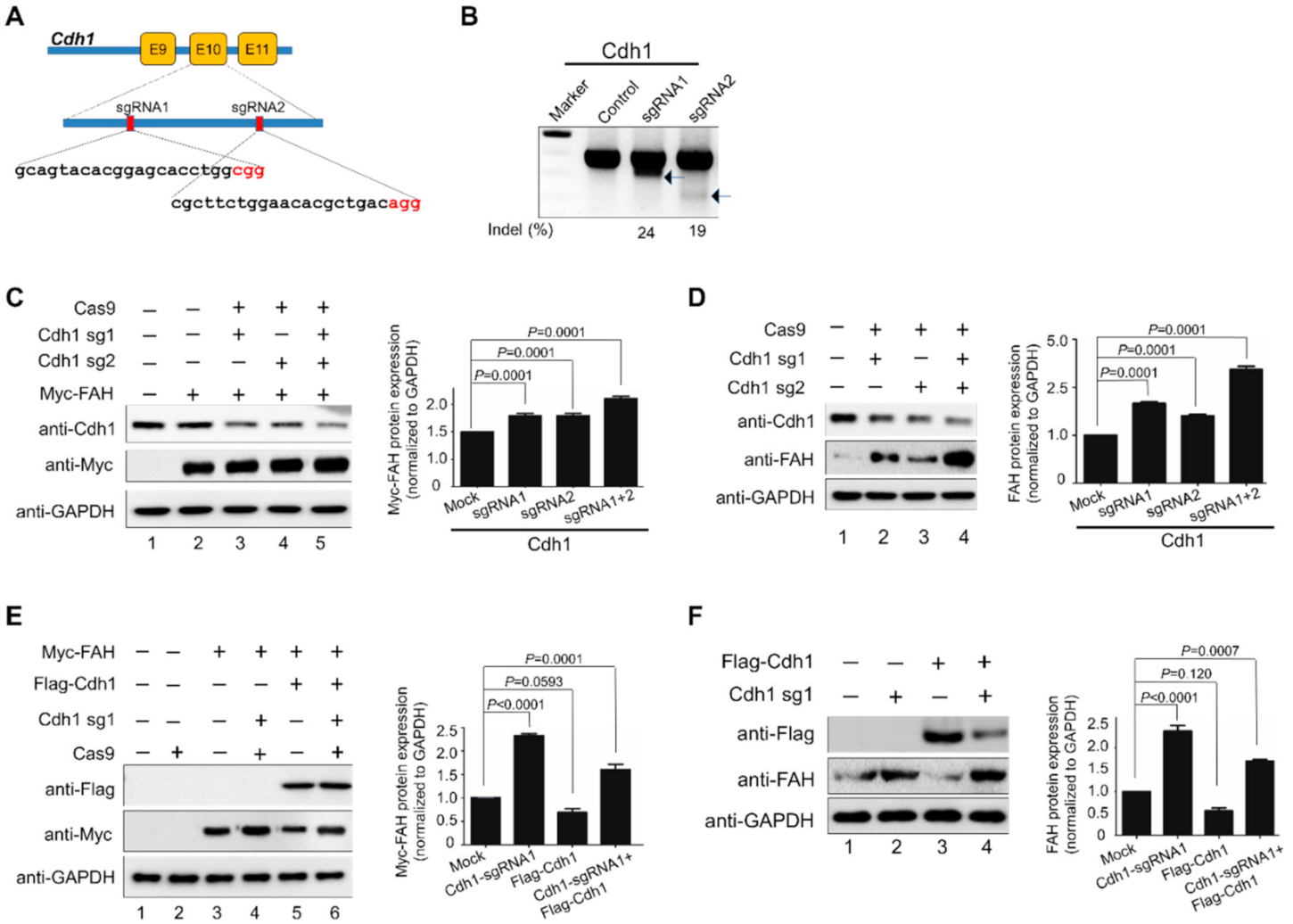
2.4. APC/CCdh1 Acts as a Negative Regulator of FAH Protein Stability
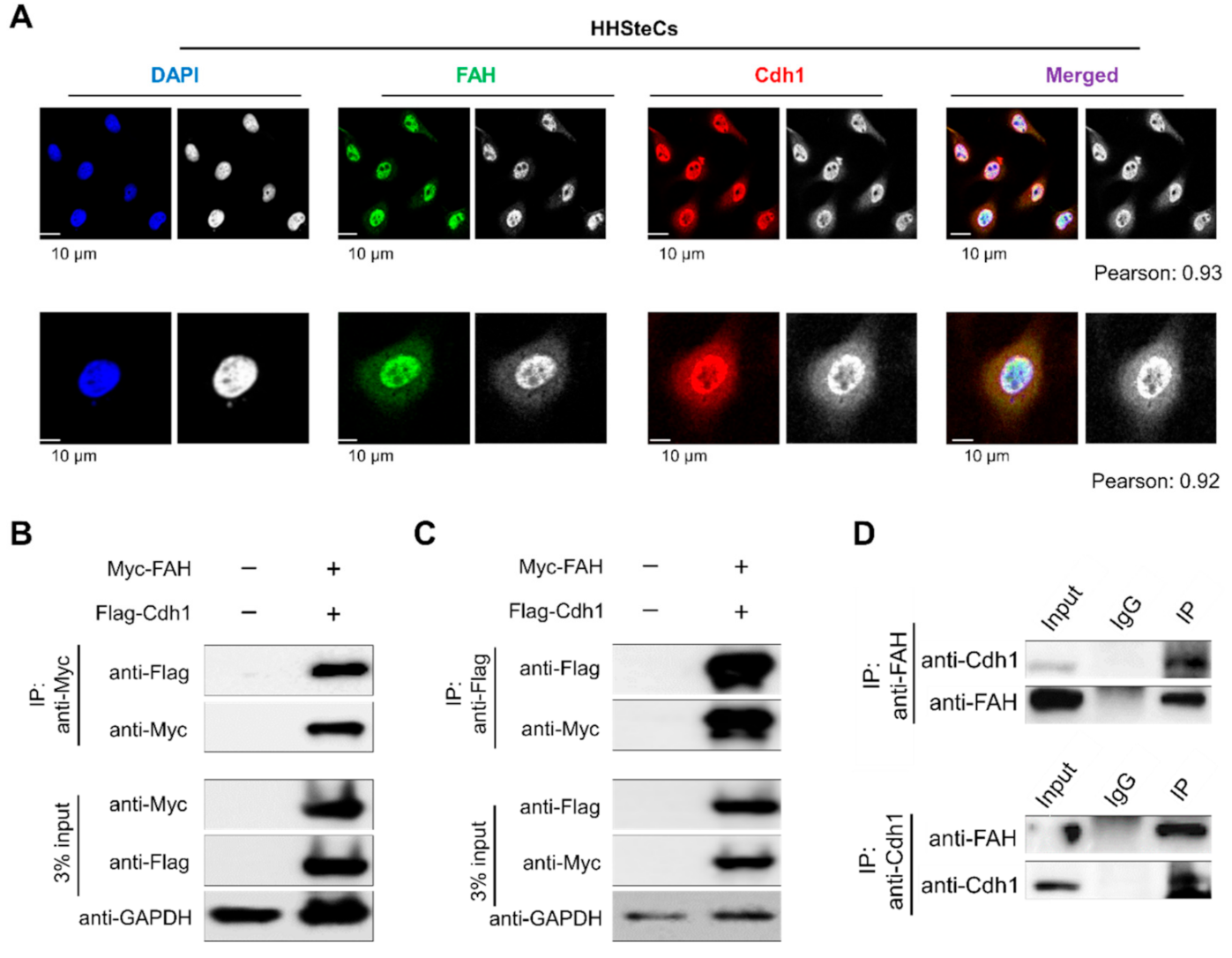
2.5. APC/CCdh1 Co-Localizes and Interacts with FAH
2.6. APC/CCdh1 Promotes FAH Polyubiquitination
2.7. APC/CCdh1 Regulates FAH Protein Half-Life
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Antibodies and Reagents
4.3. Cas9 and sgRNA Constructs
4.4. Cell Culture and Transfection
4.5. Real-Time PCR
4.6. T7 Endonuclease I (T7E1) Assay
4.7. Immunoprecipitation and Immunoblotting
4.8. TUBEs Ubiquitination Assay
4.9. Immunofluorescence Microscopy
4.10. Expression and Survival Analysis Based on The Cancer Genome Atlas Data
4.11. Immunohistochemistry
4.12. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Parthasarathy, A.; Cross, P.J.; Dobson, R.C.J.; Adams, L.E.; Savka, M.A.; Hudson, A.O. A Three-Ring Circus: Metabolism of the Three Proteogenic Aromatic Amino Acids and Their Role in the Health of Plants and Animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Agsteribbe, E.; Van Faassen, H.; Hartog, M.V.; Reversma, T.; Taanman, J.-W.; Pannekoek, H.; Evers, R.; Welling, G.; Berger, R. Nucleotide sequence of cDNA encoding human fumarylacetoacetase. Nucleic Acids Res. 1990, 18, 1887. [Google Scholar] [CrossRef] [Green Version]
- Phaneuf, D.; Labelle, Y.; Bérubé, D.; Arden, K.; Cavenee, W.; Gagné, R.; Tanguay, R.M. Cloning and expression of the cDNA encoding human fumarylacetoacetate hydrolase, the enzyme deficient in hereditary tyrosinemia: Assignment of the gene to chromosome 15. Am. J. Hum. Genet. 1991, 48, 525–535. [Google Scholar]
- Labelle, Y.; Phaneuf, D.; Leclerc, B.; Tanguay, R.M. Characterization of the human fumarylacetoacetate hydrolase gene and identification of a missense mutation abolishing enzymatic activity. Hum. Mol. Genet. 1993, 2, 941–946. [Google Scholar] [CrossRef]
- Bateman, R.L.; Ashworth, J.; Witte, J.F.; Baker, L.-J.; Bhanumoorthy, P.; Timm, D.E.; Hurley, T.D.; Grompe, M.; Mcclard, R.W. Slow-onset inhibition of fumarylacetoacetate hydrolase by phosphinate mimics of the tetrahedral intermediate: Kinetics, crystal structure and pharmacokinetics. Biochem. J. 2007, 402, 251–260. [Google Scholar] [CrossRef]
- Bateman, R.L.; Bhanumoorthy, P.; Witte, J.F.; Mcclard, R.W.; Grompe, M.; Timm, D.E. Mechanistic Inferences from the Crystal Structure of Fumarylacetoacetate Hydrolase with a Bound Phosphorus-based Inhibitor. J. Biol. Chem. 2001, 276, 15284–15291. [Google Scholar] [CrossRef] [Green Version]
- Ran, T.; Gao, Y.; Marsh, M.; Zhu, W.; Wang, M.; Mao, X.; Xu, L.; Xu, D.; Wang, W. Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for the FAH family. Biochem. J. 2012, 449, 51–60. [Google Scholar] [CrossRef]
- Jorquera, R.; Tanguay, R.M. The Mutagenicity of the Tyrosine Metabolite, Fumarylacetoacetate, Is Enhanced by Glutathione Depletion. Biochem. Biophys. Res. Commun. 1997, 232, 42–48. [Google Scholar] [CrossRef]
- Mohamed, S.; Kambal, M.; Jurayyan, N.; Alnemri, A.R.M.; Babiker, A.M.; Hasanato, R.; Al-Jarallah, A.S. Tyrosinemia type 1: A rare and forgotten cause of reversible hypertrophic cardiomyopathy in infancy. BMC Res. Notes 2013, 6, 362. [Google Scholar] [CrossRef] [Green Version]
- Kvittingen, E. Hereditary tyrosinemia type I—An overview. Scand. J. Clin. Lab. Invest. Suppl. 1986, 184, 27–34. [Google Scholar]
- Macias, I.; Laín, A.; Bernardo-Seisdedos, G.; Gil, D.; Gonzalez, E.; Falcon-Perez, J.M.; Millet, O. Hereditary tyrosinemia type I–associated mutations in fumarylacetoacetate hydrolase reduce the enzyme stability and increase its aggregation rate. J. Biol. Chem. 2019, 294, 13051–13060. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Siepka, S.M.; Cox, K.H.; Kumar, V.; De Groot, M.; Chelliah, Y.; Chen, J.; Tu, B.; Takahashi, J.S. Tissue-specific FAH deficiency alters sleep–wake patterns and results in chronic tyrosinemia in mice. Proc. Natl. Acad. Sci. USA 2019, 116, 22229–22236. [Google Scholar] [CrossRef]
- Kaushal, K.; Antao, A.M.; Kim, K.-S.; Ramakrishna, S. Deubiquitinating enzymes in cancer stem cells: Functions and targeted inhibition for cancer therapy. Drug Discov. Today 2018, 23, 1974–1982. [Google Scholar] [CrossRef]
- Sarodaya, N.; Suresh, B.; Kim, K.-S.; Ramakrishna, S. Protein Degradation and the Pathologic Basis of Phenylketonuria and Hereditary Tyrosinemia. Int. J. Mol. Sci. 2020, 21, 4996. [Google Scholar] [CrossRef]
- Antao, A.M.; Tyagi, A.; Kim, K.-S.; Ramakrishna, S. Advances in Deubiquitinating Enzyme Inhibition and Applications in Cancer Therapeutics. Cancers 2020, 12, 1579. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The Ubiquitin System. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Pines, J. Cubism and the cell cycle: The many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 2011, 12, 427–438. [Google Scholar] [CrossRef]
- Peters, J.-M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006, 7, 644–656. [Google Scholar] [CrossRef]
- Reis, A.; Levasseur, M.; Chang, H.-Y.; Elliott, D.J.; Jones, K.T. The CRY box: A second APC cdh1 -dependent degron in mammalian cdc20. EMBO Rep. 2006, 7, 1040–1045. [Google Scholar] [CrossRef] [Green Version]
- Lau, A.W.; Inuzuka, H.; Fukushima, H.; Wan, L.; Liu, P.; Gao, D.; Sun, Y.; Wei, W. Regulation of APCCdh1 E3 ligase activity by the Fbw7/cyclin E signaling axis contributes to the tumor suppressor function of Fbw7. Cell Res. 2013, 23, 947–961. [Google Scholar] [CrossRef] [Green Version]
- Lukas, C.; Sørensen, C.S.; Kramer, E.R.; Santoni-Rugiu, E.; Lindeneg, C.; Peters, J.-M.; Bartek, J.; Lukas, J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nat. Cell Biol. 1999, 401, 815–818. [Google Scholar] [CrossRef]
- Yoshida, Y.; Saeki, Y.; Murakami, A.; Kawawaki, J.; Tsuchiya, H.; Yoshihara, H.; Shindo, M.; Tanaka, K. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc. Natl. Acad. Sci. USA 2015, 112, 4630–4635. [Google Scholar] [CrossRef] [Green Version]
- Medes, G. A new error of tyrosine metabolism: Tyrosinosis. The intermediary metabolism of tyrosine and phenylalanine. Biochem. J. 1932, 26, 917–940. [Google Scholar] [CrossRef] [Green Version]
- Lindblad, B.; Lindstedt, S.; Steen, G. On the enzymic defects in hereditary tyrosinemia. Proc. Natl. Acad. Sci. USA 1977, 74, 4641–4645. [Google Scholar] [CrossRef] [Green Version]
- Fällström, S.-P.; Lindblad, B.; Lindstedt, S.; Steen, G. Hereditary tyrosinemia–Fumary lacetoacetase deficiency. Pediatr. Res. 1979, 13, 78. [Google Scholar] [CrossRef]
- Berger, R.; Smit, O.; Vries, S.-D.; Duran, M.; Ketting, D.; Wadman, S. Deficiency of fumarylacetoacetase in a patient with hereditary tyrosinemia. Clin. Chim. Acta 1981, 114, 37–44. [Google Scholar] [CrossRef]
- Cassiman, D.; Zeevaert, R.; Kvittingen, E.-A.; Jaeken, J. A novel mutation causing mild, atypical fumarylacetoacetase deficiency (Tyrosinemia type I): A case report. Orphanet J. Rare Dis. 2009, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Malik, S.H.; NiMhurchadha, S.; Jackson, C.; Eliasson, L.; Weinman, J.; Roche, S.; Walter, J.; Zschocke, J.; Baumgartner, M.; Gibson, K.M.; et al. Treatment Adherence in Type 1 Hereditary Tyrosinaemia (HT1): A Mixed-Method Investigation into the Beliefs, Attitudes and Behaviour of Adolescent Patients, Their Families and Their Health-Care Team. JIMD Rep. 2014, 18, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Zuo, D.; Esubjeck, J.; Ewang, X.-Y. Unfolding the Role of Large Heat Shock Proteins: New Insights and Therapeutic Implications. Front. Immunol. 2016, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- Kevei, É.; Pokrzywa, W.; Hoppe, T. Repair or destruction-an intimate liaison between ubiquitin ligases and molecular chaperones in proteostasis. FEBS Lett. 2017, 591, 2616–2635. [Google Scholar] [CrossRef]
- Taipale, M.; Tucker, G.; Peng, J.; Krykbaeva, I.; Lin, Z.-Y.; Larsen, B.; Choi, H.; Berger, B.; Gingras, A.-C.; Lindquist, S. A Quantitative Chaperone Interaction Network Reveals the Architecture of Cellular Protein Homeostasis Pathways. Cell 2014, 158, 434–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, A.K.; Albertini, E.; Holzknecht, M.; Cappuccio, E.; Dorigatti, I.; Krahbichler, A.; Damisch, E.; Gstach, H.; Jansen-Dürr, P. Regulation of cellular senescence by eukaryotic members of the FAH superfamily–A role in calcium homeostasis? Mech. Ageing Dev. 2020, 190, 111284. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [Green Version]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Dimova, N.V.; Hathaway, N.A.; Lee, B.-H.; Kirkpatrick, D.S.; Berkowitz, M.L.; Gygi, S.P.; Finley, D.; King, R.W. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat. Cell Biol. 2012, 14, 168–176. [Google Scholar] [CrossRef] [Green Version]
- Shao, R.; Liu, J.; Yan, G.; Zhang, J.; Han, Y.; Guo, J.; Xu, Z.; Yuan, Z.; Liu, J.; Malumbres, M.; et al. Cdh1 regulates craniofacial development via APC-dependent ubiquitination and activation of Goosecoid. Cell Res. 2016, 26, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Meyer, H.-J.; Rape, M. Processive ubiquitin chain formation by the anaphase-promoting complex. Semin. Cell Dev. Biol. 2011, 22, 544–550. [Google Scholar] [CrossRef] [Green Version]
- Maestre, C.; Delgado-Esteban, M.; Gomez-Sanchez, J.C.; Bolaños, J.P.; Almeida, A. Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. EMBO J. 2008, 27, 2736–2745. [Google Scholar] [CrossRef] [Green Version]
- Kirstein, M.M.; Schweitzer, N.; Schmidt, S.; Klöpper, A.; I Ringe, K.; Lehmann, U.; Manns, M.P.; Wedemeyer, H.; Vogel, A. Long-lasting tumour response to sorafenib therapy in advanced hepatocellular carcinoma. Acta Gastro-Enterol. Belg. 2014, 77, 386–388. [Google Scholar]
- Jorquera, R.; Tanguay, R.M. Fumarylacetoacetate, the metabolite accumulating in hereditary tyrosinemia, activates the ERK pathway and induces mitotic abnormalities and genomic instability. Hum. Mol. Genet. 2001, 10, 1741–1752. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, S.; Dad, A.-B.K.; Beloor, J.; Gopalappa, R.; Lee, S.-K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, J.; Parmryd, I. Quantifying colocalization by correlation: The Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytom. Part A 2010, 77, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Mishra, M.K.; Eltoum, I.-E.A.; Bae, S.; Singh, R. CCR5/CCL5 axis interaction promotes migratory and invasiveness of pancreatic cancer cells. Sci. Rep. 2018, 8, 1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaushal, K.; Woo, S.H.; Tyagi, A.; Kim, D.H.; Suresh, B.; Kim, K.-S.; Ramakrishna, S. E3 Ubiquitin Ligase APC/CCdh1 Negatively Regulates FAH Protein Stability by Promoting Its Polyubiquitination. Int. J. Mol. Sci. 2020, 21, 8719. https://doi.org/10.3390/ijms21228719
Kaushal K, Woo SH, Tyagi A, Kim DH, Suresh B, Kim K-S, Ramakrishna S. E3 Ubiquitin Ligase APC/CCdh1 Negatively Regulates FAH Protein Stability by Promoting Its Polyubiquitination. International Journal of Molecular Sciences. 2020; 21(22):8719. https://doi.org/10.3390/ijms21228719
Chicago/Turabian StyleKaushal, Kamini, Sang Hyeon Woo, Apoorvi Tyagi, Dong Ha Kim, Bharathi Suresh, Kye-Seong Kim, and Suresh Ramakrishna. 2020. "E3 Ubiquitin Ligase APC/CCdh1 Negatively Regulates FAH Protein Stability by Promoting Its Polyubiquitination" International Journal of Molecular Sciences 21, no. 22: 8719. https://doi.org/10.3390/ijms21228719
APA StyleKaushal, K., Woo, S. H., Tyagi, A., Kim, D. H., Suresh, B., Kim, K. -S., & Ramakrishna, S. (2020). E3 Ubiquitin Ligase APC/CCdh1 Negatively Regulates FAH Protein Stability by Promoting Its Polyubiquitination. International Journal of Molecular Sciences, 21(22), 8719. https://doi.org/10.3390/ijms21228719





