Aerobic Cytotoxicity of Aromatic N-Oxides: The Role of NAD(P)H:Quinone Oxidoreductase (NQO1)
Abstract
:1. Introduction
2. Results
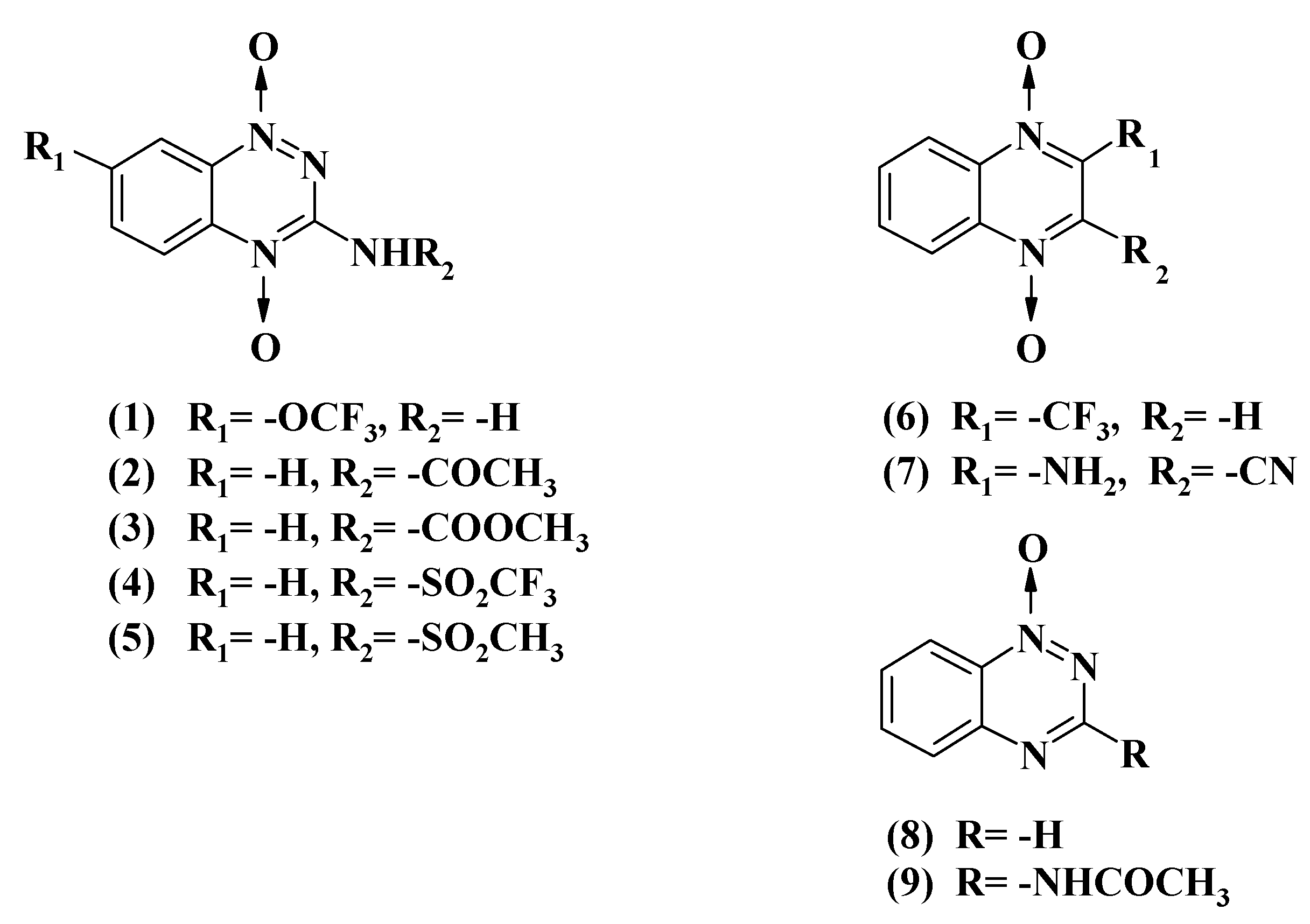
2.1. Reactions of Aromatic N-Oxides with Flavoenzymes Dehydrogenases-Electrontransferases
| No. | Compound | E17 (V) a | kcat/Km (M−1·s−1) | log kcat/Km (avge) | |
|---|---|---|---|---|---|
| P-450R | PfFNR | ||||
| 1 | 7-CF3O-tirapazamine | 4.6 ± 0.4 × 104 | 3.8 ± 0.4 × 104 | 4.62 | |
| 2 | 3-CH3CONH-1,2,4-benzotriazine-1,4-dioxide | 7.0 ± 0.5 × 104 | 6.2 ± 0.5 × 104 | 4.82 | |
| 3 | 3-CH3OCONH-1,2,4-benzotriazine-1,4-dioxide | 8.0 ± 0.9 × 104 | 4.8 ± 0.4 × 104 | 4.78 | |
| 4 | 3-CF3SO2NH-1,2,4-benzotriazine-1,4-dioxide | 2.5 ± 0.3 × 104 | 2.6 ± 0.3 × 104 | 4.41 | |
| 5 | 3-CH3SO2NH-1,2,4-benzotriazine-1,4-dioxide | 2.7 ± 0.3 × 103 | 7.9 ± 0.5 × 103 | 3.67 | |
| 6 | 2-CF3-quinoxaline-1,4-dioxide | (−0.465) b | 2.7 ± 0.3 × 104 | 8.9 ± 0.7 × 103 | 4.19 |
| 7 | 2-NH2-3-CN-quinoxaline-1,4-dioxide | 4.7 ± 0.4 × 103 | 1.8 ± 0.2 × 104 | 3.96 | |
| 8 | 1,2,4-Benzotriazine-1-oxide | (−0.431) b | 1.7 ± 0.2 × 104 | 4.3 ± 0.3 × 103 | 3.94 |
| 9 | 3-CH3CONH-1,2,4-benzotriazine-1-oxide | 8.7 ± 0.9 × 103 | 1.6 ± 0.1 × 103 | 3.58 | |
| ArN→O with available E17 values | |||||
| 10 | 1,2,4-Benzotriazine-1,4-dioxide | −0.318 | 4.3 ± 0.4 × 105 c | 2.5 ± 0.3 × 104 | 5.00 |
| 11 | 7-CF3-tirapazamine | −0.345 | 8.7 ± 0.7 × 104 c | 5.2 ± 0.4 × 104 d | 4.83 |
| 12 | 7-Cl-tirapazamine | −0.400 | 6.9 ± 0.7 × 104 c | 3.7 ± 0.4 × 104 d | 4.71 |
| 13 | 7-F-tirapazamine | −0.400 | 3.4 ± 0.3 × 104 c | 2.7 ± 0.2 × 104 d | 4.48 |
| 14 | Tirapazamine | −0.455 | 1.1 ± 0.1 × 104 c | 4.4 ± 0.5 × 103 d | 3.84 |
| 15 | 7-CH3-tirapazamine | −0.474 | 8.6 ± 0.7 × 103 c | 5.0 ± 0.6 × 103 d | 3.82 |
| 16 | 7-C2H5O-tirapazamine | −0.494 | 4.5 ± 0.5 × 103 c | 4.5 ± 0.5 × 103 d | 3.65 |
| 17 | 3-Amino-1,2,4-benzotriazine-1-oxide | −0.568 | 2.8 ± 0.2 × 103 c | 3.2 ± 0.2 × 103 d | 3.48 |
| 18 | Quinoxaline-1,4-dioxide | −0.575 | 3.3 ± 0.2 × 103 c | 8.2 ± 0.9 × 102 d | 3.22 |
2.2. NQO1-Catalyzed Reduction of Aromatic N-Oxides
2.3. Cytotoxicity of Aromatic N-Oxides
3. Discussion
4. Materials and Methods
4.1. Enzymes and Chemicals
4.2. Enzymatic Assays
4.3. Cytotoxicity Assays
4.4. Statistical Analysis and Calculations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ArN→O | Heteroaromatic N-oxide |
| BCNU | 1,3-bis(2-chloroethyl)-1-nitrosourea |
| cL50 | Concentration for 50% cell survival |
| DESF | Desferrioxamine |
| DIC | Dicoumarol |
| DPPD | N,N′-diphenyl-p-phenylene diamine |
| E17 | Single-electron reduction midpoint potential at pH 7.0 |
| GI50 | Concentration for 50% inhibition of maximal cell proliferation |
| kcat | Catalytic constant |
| kcat/Km | Bimolecular rate constant |
| kcat/Km (avge) | Geometric average of kcat/Km in P-450R- and PfFNR-catalyzed reactions |
| kcat/Km (NQO1) | kcat/Km in NQO1-catalyzed reaction |
| log D | Octanol/water distribution coefficient at pH 7.0 |
| NQO1 | NAD(P)H:quinone oxidoreductase |
| P-450R | NADPH:cytochrome P-450 reductase |
| PfFNR | Plasmodium falciparum ferredoxin:NADP+ oxidoreductase |
| SOD | Superoxide dismutase |
| TPZ | Tirapazamine |
References
- Wardman, P.; Dennis, M.F.; Everett, S.A.; Patel, K.B.; Stratford, M.R.L.; Tracy, M. Radicals from one-electron reduction of nitro compounds, aromatic N-oxides and quinones: The kinetic basis for hypoxia-selective, bioreductive drugs. Biochem. Soc. Symp. 1995, 61, 171–194. [Google Scholar] [PubMed] [Green Version]
- Shen, X.; Gates, K.S. Enzyme-activated generation of reactive oxygen species from heterocyclic N-oxides under aerobic and anaerobic conditions and its relevance to hypoxia-selective prodrugs. Chem. Res. Toxicol. 2019, 32, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Shah, Z.; Mahnuba, R.; Turcotte, B. The anticancer drug tirapazamine has antimicrobial activity against Escherichia coli, Staphylococcus aureus and Clostridium difficile. FEMS Microbiol. Lett. 2013, 347, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil, A.; Pabon, A.; Galiano, S.; Burguete, A.; Perez-Silanes, S.; Deharo, E.; Monge, A.; Aldana, I. Synthesis, biological evaluation and structure-activity relationships of new quinoxaline derivatives as anti-Plasmodium falciparum agents. Molecules 2014, 19, 2166–2180. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.; Li, B.; Wang, C.; Zhang, H.; Liang, G.; Weng, Z.; Hao, H.; Wang, X.; Liu, Z.; Dai, M.; et al. Systematic and molecular basis of the antibacterial action of quinoxaline 1,4-di-N-oxides against Escherichia coli. PLoS ONE 2015, 10, e0136450. [Google Scholar] [CrossRef] [Green Version]
- Perez-Silanes, S.; Torrers, E.; Arbillaga, L.; Varela, J.; Cerecetto, H.; Gonzalez, M.; Azqueta, A.; Moreno-Viguri, E. Synthesis and biological evaluation of quinoxaline di-N-oxide derivatives with in vitro trypanocidal activity. Bioorg. Med. Chem. Lett. 2016, 26, 903–906. [Google Scholar] [CrossRef]
- Shen, X.; Rajapakse, A.; Galazzi, F.; Junnotula, V.; Fuchs-Knotts, T.; Glaser, R.; Gates, K.S. Isotopic labeling experiments that elucidate the mechanism of DNA strand cleavage by the hypoxia-selective antitumor agent 1,2,4-benzotriazine 1,4-di-N-oxide. Chem. Res. Toxicol. 2013, 22, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Yadav, P.; Marshall, A.J.; Reynisson, J.; Denny, W.A.; Hay, M.P.; Anderson, R.F. Fragmentation of the quinoxaline N-oxide bond to the .OH radical upon one-electron bioreduction. Chem. Commun. 2014, 50, 13729–13731. [Google Scholar] [CrossRef]
- Shinde, S.S.; Maroz, A.; Hay, M.P.; Patterson, A.V.; Denny, V.A.; Anderson, R.F. Characterization of radicals formed following enzymatic reduction of 3-substituted analogues of the hypoxia-selective cytotoxin 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine). J. Am. Chem. Soc. 2010, 132, 2591–2599. [Google Scholar] [CrossRef]
- Anderson, R.F.; Yadav, P.; Shinde, S.S.; Hong, C.R.; Pullen, S.M.; Reynisson, J.; Wilson, W.R.; Hay, M.P. Radical chemistry and cytotoxicity of bioreductive 3-substituted quinoxaline di-N-oxides. Chem. Res. Toxicol. 2016, 29, 1310–1324. [Google Scholar] [CrossRef]
- Fuchs, T.; Chowdhury, G.; Fuchs, C.L.; Gates, K.S. 3-Amino-1,2,4-benzotriazine 4-oxide: Characterization of a new metabolite arising from bioreductive processing of the antitumour agent 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine). J. Org. Chem. 2001, 66, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Zarranz, B.; Jaso, A.; Aldana, I.; Monge, A. Synthesis and anticancer activity evaluation of new 2-alkylcarbonyl and 2-benzoyl-1-trifluoromethyl-quinoxaline-1,4-di-N-oxide derivatives. Bioorg. Med. Chem. 2004, 12, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xia, Q.; Shangguan, S.; Liu, X.; Hu, Y.; Sheng, R. Synthesis and biological evaluation of 3-aryl-quinoxaline-2-carbonitrile 1,4-di-N-oxide derivatives as hypoxic selective anti-tumour agents. Molecules 2012, 17, 9683–9696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, G.; Sarkar, U.; Pullen, S.; Wilson, W.R.; Rajapakse, A.; Fuchs-Knotts, T.; Dates, K.S. DNA strand cleavage by the phenazine di-N-oxide natural product myxin under both aerobic and anaerobic conditions. Chem. Res. Toxicol. 2012, 25, 197–206. [Google Scholar] [CrossRef]
- Gu, Y.; Chang, T.T.-A.; Wang, J.; Jaiswal, J.K.; Edwards, D.; Downes, N.J.; Liyanage, H.D.S.; Lynch, C.H.R.; Pruijn, F.B.; Hickey, A.J.R.; et al. Reductive metabolism influences the toxicity and pharmacokinetics of the hypoxia-targeted benzotriazine-dioxide anticancer agent SN30000 in mice. Front. Pharmacol. 2017, 8, 531. [Google Scholar] [CrossRef] [Green Version]
- Hunter, F.W.; Young, R.J.; Shalev, Z.; Vellanki, R.N.; Wang, J.; Gu, Y.; Joshi, N.; Sreebhavan, S.; Weinreb, J.; Goldstein, D.P.; et al. Identification of P450 oxidoreductase as a major determinant of sensitivity to hypoxia-activated prodrugs. Cancer Res. 2015, 75, 4211–4223. [Google Scholar] [CrossRef] [Green Version]
- Delahussaye, Y.M.; Evans, J.W.; Brown, J.M. Metabolism of tirapazamine by multiple reductases in the nucleus. Biochem. Pharmacol. 2001, 62, 1201–1209. [Google Scholar]
- Ross, D.; Siegel, D. NAD(P)H:quinone oxidoreductase I (NQO1, DT-diaphorase), functions and pharmacogenetics. Meth. Enzymol. 2004, 382B, 115–144. [Google Scholar]
- Anusevičius, Ž.; Šarlauskas, J.; Čėnas, N. Two-electron reduction of quinones by rat liver NAD(P)H: Quinone oxidoreductase: Quantitative structure-activity relationships. Arch. Biochem. Biophys. 2002, 404, 254–262. [Google Scholar] [CrossRef]
- Misevičienė, L.; Anusevičius, Ž.; Šarlauskas, J.; Čėnas, N. Reduction of nitroaromatic compounds by NAD(P)H:quinone oxidoreductase (NQO1): The role of electron-accepting potency and structural parameters in the substrate specificity. Acta Biochim. Pol. 2006, 53, 569–576. [Google Scholar] [CrossRef]
- Di Francesco, A.; Ward, T.; Butler, J. Diaziridinylbenzoquinones. Meth. Enzymol. 2004, 382B, 174–193. [Google Scholar]
- Nemeikaitė-Čėnienė, A.; Šarlauskas, J.; Jonušienė, V.; Marozienė, A.; Misevičienė, L.; Yantsevich, A.V.; Čėnas, N. Kinetics of flavoenzyme-catalyzed reduction of tirapazamine derivatives: Implications for their prooxidant cytotoxicity. Int. J. Mol. Sci. 2019, 20, 4602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plumb, J.A.; Gerritsen, M.; Workman, P. DT-diaphorase protects cells from the hypoxic cytotoxicity of indoloquinone EO9. Br. J. Cancer 1994, 70, 1136–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elwell, J.H.; Siim, B.G.; Evans, J.W.; Brown, J.M. Adaptation of human tumour cells to tirapazamine under aerobic conditions. Implications of increased antioxidant enzyme activity to mechanism of aerobic toxicity. Biochem. Pharmacol. 1997, 54, 249–257. [Google Scholar] [CrossRef]
- Sharp, S.Y.; Kelland, L.R.; Valenti, M.R.; Brunton, L.A.; Hobbs, S.; Workman, P. Establishment of an isogenic human colon tumor model for NQO1 gene expression: Application to investigate the role of DT-diaphorase in bioreductive drug activation in vitro and in vivo. Mol. Pharmacol. 2000, 58, 1146–1155. [Google Scholar] [CrossRef] [Green Version]
- Beaver, S.K.; Mesa-Tores, N.; Pey, A.L.; Timson, D.J. NQO1: A target for the treatment of cancer and neurological diseases, and a model to understand loss of function disease mechanism. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 663–676. [Google Scholar] [CrossRef]
- Lesanavičius, M.; Aliverti, A.; Šarlauskas, J.; Čėnas, N. Reactions of Plasmodium falciparum ferredoxin: NADP+ oxidoreductase with redox cycling xenobiotics: A mechanistic study. Int. J. Mol. Sci. 2020, 21, 3234. [Google Scholar] [CrossRef]
- Marcus, R.A.; Sutin, N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta 1985, 811, 265–322. [Google Scholar] [CrossRef]
- Čėnas, N.; Nemeikaitė-Čėnienė, A.; Sergedienė, E.; Nivinskas, H.; Anusevičius, Ž.; Šarlauskas, J. Quantitative structure-activity relationships in enzymatic single-electron reduction of nitroaromatic explosives: Implications for their cytotoxicity. Biochim. Biophys. Acta 2001, 1528, 31–38. [Google Scholar] [CrossRef]
- Šarlauskas, J.; Nivinskas, H.; Anusevičius, Ž.; Misevičienė, L.; Marozienė, A.; Čėnas, N. Estimation of single-electron reduction potentials (E17) of nitroaromatic compounds according to the kinetics of their single-electron reduction by flavoenzymes. Chemija 2006, 17, 31–37. [Google Scholar]
- Hay, M.P.; Gamage, S.A.; Kovacs, M.S.; Pruijn, F.B.; Anderson, R.F.; Patterson, A.V.; Wilson, W.R.; Brown, J.M.; Denny, W.A. Structure-activity relationships of 1,2,4-benzotriazine 1,4-dioxides as hypoxia-selective analogues of tirapazamine. J. Med. Chem. 2003, 46, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.F.; Shinde, S.S.; Hay, M.P.; Denny, W.A. Potentiation of the cytotoxicity of the anticancer agent tirapazamine by benzotriazine-N-oxides. The role of redox equilibria. J. Am. Chem. Soc. 2006, 128, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Cahill, A.; Jenkins, T.C.; White, I.N.H. Metabolism of 3-amino-1,2,4-benzotriazine-1,4-dioxide (SR 4233) by purified DT-diaphorase under aerobic and anaerobic conditions. Biochem. Pharmacol. 1993, 45, 321–329. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Abraham, M.H.; Zissimos, A.M. Fast calculation of van der Waals volume as a sum of atomic and bond contributions and its application to drug compounds. J. Org. Chem. 2003, 68, 7368–7373. [Google Scholar] [CrossRef]
- Öllinger, K.; Brunmark, A. Effects of hydroxyl substituent position on 1,4-naphthoquinone toxicity to rat hepatocytes. J. Biol. Chem. 1991, 266, 21496–21503. [Google Scholar]
- Nemeikaitė, A.; Čėnas, N. The changes of prooxidant and antioxidant enzyme activities in bovine leukemia virus-transformed cells. Their influence on quinone cytotoxicity. FEBS Lett. 1993, 326, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-H.; Chiu, T.-Y.; Hu, M.-L. Fucoxanthin enhances HO-1 and NQO1 expression in murine hepatic BNL CL.2 cells through activation of the Nrf2/ARE system partially by its prooxidant activity. J. Agric. Food Chem. 2011, 59, 11344–11351. [Google Scholar] [CrossRef]
- Miliukienė, V.; Nivinskas, H.; Čėnas, N. Cytotoxicity of anticancer aziridinyl-substituted benzoquinones in primary mice splenocytes. Acta Biochim. Pol. 2014, 61, 833–836. [Google Scholar] [CrossRef] [Green Version]
- Grampp, G.; Jaenicke, W. ESR-spectroscopic investigation of the parallel electron and proton exchange between quinones and their radicals: Part I. Measurements at 298 K. J. Electroanal. Chem. 1987, 229, 297–303. [Google Scholar] [CrossRef]
- Tedeschi, G.; Chen, S.; Massey, V. DT-diaphorase. Redox potential, steady-state, and rapid reaction studies. J. Biol. Chem. 1995, 270, 1198–1204. [Google Scholar] [CrossRef] [Green Version]
- Faig, M.; Bianchet, M.A.; Talalay, P.; Chen, S.; Winski, S.; Ross, D.; Amzel, L.M. Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: Species comparison and structural changes with substrate binding and release. Proc. Natl. Acad. Sci. USA 2000, 97, 3177–3182. [Google Scholar] [CrossRef] [PubMed]
- Anusevičius, Ž.; Šarlauskas, J.; Nivinskas, H.; Segura-Aguilar, J.; Čėnas, N. DT-diaphorase catalyzes N-denitration and redox cycling of tetryl. FEBS Lett. 1998, 436, 144–148. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, M.F.; Hollabaugh, N.M.; Hettiarachchi, S.U.; McCarley, R.L. Human NAD(P)H: Quinone oxidoreductase type I (hNQO1) activation of quinone propionic acid trigger groups. Biochemistry 2012, 51, 8014–8026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubig, S.M.; Rathore, R.; Kochi, J.K. Steric control of electron transfer. Changeover from outer-sphere to inner-sphere mechanism in arene/quinone redox pairs. J. Am. Chem. Soc. 1999, 121, 617–626. [Google Scholar] [CrossRef]
- Yin, J.; Glaser, R.; Gates, K.S. Electron and spin-density analysis of tirapazamine reduction chemistry. Chem. Res. Toxicol. 2012, 25, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Holzman, J.L.; Crankshaw, D.L.; Peterson, F.J.; Polnaszek, C.F. The kinetics of the aerobic reduction of nitrofurantoin by NADPH-cytochrome P450 (c) reductase. Mol. Pharmacol. 1981, 20, 669–673. [Google Scholar]
- Pechurskaja, T.A.; Harnastai, I.N.; Grabovec, I.P.; Gilep, A.A.; Usanov, S.A. Adrenodoxin supports reactions catalyzed by microsomal steroidgenetic cytochrome P450s. Biochem. Biophys. Res. Commun. 2007, 353, 598–604. [Google Scholar] [CrossRef]
- Balconi, E.; Pennati, A.; Crobu, D.; Pandini, V.; Cerutti, R.; Zanetti, G.; Aliverti, A. The ferredoxin-NADP+ reductase/ferredoxin electron transfer system of Plasmodium falciparum. FEBS J. 2009, 276, 2249–4260. [Google Scholar] [CrossRef]
- Prochaska, H.J. Purification and crystallization of rat liver NAD(P)H:quinone-acceptor oxidoreductase by cibacron blue affinity chromatography: Identification of a new and potent inhibitor. Arch. Biochem. Biophys. 1988, 267, 529–538. [Google Scholar] [CrossRef]
- Polmickaitė-Smirnova, E.; Šarlauskas, J.; Krikštopaitis, K.; Lukšienė, Ž.; Staniulytė, Z.; Anusevičius, Ž. Preliminary investigation on the antibacterial activity of antitumor drug 3-amino-1,2,4-benzotriazine-1,4-dioxide (tirapazamine) and its derivatives. Appl. Sci. 2020, 10, 4062. [Google Scholar] [CrossRef]
- Abushanab, E. Long-range hydrogen-fluorine spin-spin coupling. Further support for the “through-space” (direct) mechanism. J. Am. Chem. Soc. 1971, 93, 6532–6536. [Google Scholar] [CrossRef]
- Monge, A.; Palop, J.A.; Del Castillo, J.C.; Caldero, J.M.; Roca, J.; Romero, G.; Del Rio, J.; Lasheras, B. Novel antagonists of 5-HT3 receptors: Synthesis and biological evaluation of piperazinylquinoxaline derivatives. J. Med. Chem. 1993, 36, 2745–2750. [Google Scholar] [CrossRef] [PubMed]
- Robbins, R.F.; Schofield, K. Polyazabicyclic compounds. Part II. Further derivatives of benzo-1:2:4-triazine. J. Chem. Soc. 1957, 3186–3194. [Google Scholar] [CrossRef]
- Bodzioch, A.; Pomiklo, D.; Celeda, M.; Pietrzak, A.; Kaszynski, P. 3-Substituted benzo[e][1,2,4]triazines: Synthesis and electronic effects of the C(3) substituents. J. Org. Chem. 2019, 84, 6377–6394. [Google Scholar] [CrossRef]
- Ito, M. Microassay for studying anticellular effects of human interferons. J. Interferon. Res. 1984, 4, 603–608. [Google Scholar] [CrossRef] [PubMed]

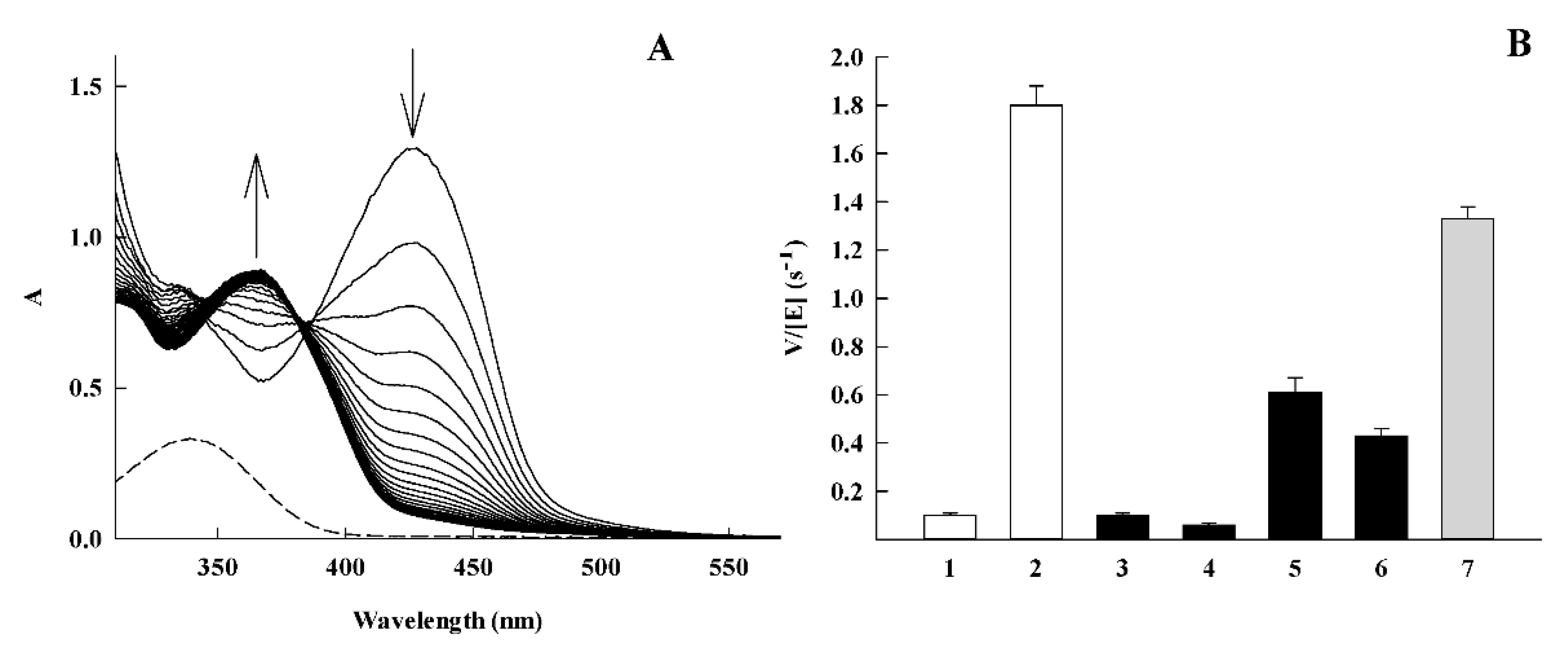
| No. | Compound | kcat (s−1) | kcat/Km (M−1.s−1) | VdWvol (Å3) a |
|---|---|---|---|---|
| 1 | 7-CF3O-tirapazamine | 0.11 ± 0.02 | 1.6 ± 0.1 × 103 | 175.8 |
| 2 | 3-CH3CONH-1,2,4-benzotriazine-1,4-dioxide | 3.40 ± 0.20 | 1.3 ± 0.1 × 104 | 172.3 |
| 3.00 ± 0.20 b | 1.0 ± 0.1 × 104 b | |||
| 3 | 3-CH3OCONH-1,2,4-benzotriazine-1,4-dioxide | 2.40 ± 0.22 | 3.0 ± 0.2 × 103 | 180.7 |
| 4 | 3-CF3SO2NH-1,2,4-benzotriazine-1,4-dioxide | 0.34 ± 0.06 | 1.0 ± 0.2 × 103 | 203.5 |
| 5 | 3-CH3SO2NH-1,2,4-benzotriazine-1,4-dioxide | 0.40 ± 0.05 | 1.2 ± 0.2 × 103 | 184.9 |
| 6 | 2-CF3-quinoxaline-1,4-dioxide | 13.5 ± 1.72 | 2.4 ± 0.2 × 104 | 168.2 |
| 7 | 2-NH2-3-CN-quinoxaline-1,4-dioxide | 0.11 ± 0.02 | 1.4 ± 0.1 × 103 | 146.2 |
| 8 | 1,2,4-Benzotriazine-1-oxide | 1.10 ± 0.12 | 2.1 ± 0.2 × 103 | 111.7 |
| 9 | 3-CH3CONH-1,2,4-benzotriazine-1-oxide | 0.06 ± 0.02 | 5.0 ± 1.0 × 102 | 163.5 |
| 10 | 1,2,4-Benzotriazine-1,4-dioxide c | 1.00 ± 0.10 | 5.1 ± 0.7 × 103 | 120.5 |
| 11 | 7-CF3-tirapazamine c | 1.30 ± 0.10 | 3.7 ± 0.4 × 103 | 167.0 |
| 12 | 7-Cl-tirapazamine c | 1.00 ± 0.10 | 4.3 ± 0.4 × 103 | 152.6 |
| 13 | 7-F-tirapazamine c | 0.70 ± 0.10 | 4.9 ± 0.6 × 103 | 143.5 |
| 14 | Tirapazamine c | 0.20 ± 0.10 | 2.4 ± 0.2 × 103 | 131.5 |
| 15 | 7-CH3-tirapazamine c | 0.30 ± 0.05 | 1.7 ± 0.1 × 103 | 148.8 |
| 16 | 7-C2H5O-tirapazamine c | 0.30 ± 0.04 | 1.0 ± 0.1 × 103 | 174.9 |
| 17 | 3-Amino-1,2,4-benzotriazine-1-oxide c | 0.10 ± 0.02 | 1.6 ± 0.2 × 103 | 122,7 |
| 18 | Quinoxaline-1,4-dioxide c | 0.60 ± 0.10 | 7.9 ± 0.9 × 103 | 126.8 |
| No. | Compound | log kcat/Km (avge) | log D | cL50 (µM) MH22a | GI50 (µM) HCT-116 |
|---|---|---|---|---|---|
| 1 | 7-CF3O-tirapazamine | 4.62 | 1.32 | 3.6 ± 0.7 | 17 ± 3.0 |
| 2 | 3-CH3CONH-1,2,4-benzotriazine-1,4-dioxide | 4.82 | −0.82 | 1.5 ± 0.3 | 2.5 ± 0.5 |
| 3 | 3-CH3OCONH-1,2,4-benzotriazine-1,4-dioxide | 4.78 | −0.08 | 9.4 ± 1.4 | 6.3 ± 1.0 |
| 4 | 3-CF3SO2NH-1,2,4-benzotriazine-1,4-dioxide | 4.41 | −0.40 | 44 ± 6.5 | 125 ± 19 |
| 5 | 3-CH3SO2NH-1,2,4-benzotriazine-1,4-dioxide | 3.67 | −2.42 | 184 ± 25 | 185 ± 23 |
| 6 | 2-CF3-quinoxaline-1,4-dioxide | 4.19 | 0.42 | 10 ± 2.0 | 12.5 ± 2.0 |
| 7 | 2-NH2-3-CN-quinoxaline-1,4-dioxide | 3.96 | −0.18 | 358 ± 52 | 125 ± 17 |
| 8 | 1,2,4-Benzotriazine-1-oxide | 3.94 | 0.45 | 168 ± 21 | 225 ± 27 |
| 9 | 3-CH3CONH-1,2,4-benzotriazine-1-oxide | 3.58 | 0.37 | ≥600 | ≥1000 |
| 10 | 1,2,4-Benzotriazine-1,4-dioxide | 5.00 | −0.70 | 11 ± 1.5 a | 18 ± 2.0 |
| 11 | 7-CF3-tirapazamine | 4.83 | 0.76 | 3.4 ± 0.4 a | 6.0 ± 1.0 a |
| 12 | 7-Cl-tirapazamine | 4.71 | 0.49 | 3.1 ± 0.5 a | 13 ± 1.5 a |
| 13 | 7-F-tirapazamine | 4.48 | 0.03 | 7.2 ± 1.0 a | 25 ± 4.0 |
| 14 | Tirapazamine | 3.84 | 0.11 | 31 ± 5.5 a 28 ± 4.0 | 75 ± 7.0 a 64 ± 7.0 |
| 15 | 7-CH3-tirapazamine | 3.82 | 0.40 | 83 ± 10 a | 50 ± 6.0 a |
| 16 | 7-C2H5O-tirapazamine | 3.65 | 0.08 | 64 ± 10 a | 60 ± 7.0 a |
| 17 | 3-Amino-1,2,4-benzotriazine-1-oxide | 3.48 | 0.30 | ≥600 a | ≥600 a |
| 18 | Quinoxaline-1,4-dioxide | 3.22 | −0.90 | 325 ± 40 a | ≥800 |
| No. | Compound | Cell Viability (%) | ||||
|---|---|---|---|---|---|---|
| No Additions | Additions: | |||||
| DESF (1.0 mM) | DPPD (2.5 µM) | BCNU (20 µM) | DIC (20 µM) | |||
| 1 | 3-CH3CONH-1,2,4-benzotriazine-1,4-dioxide, 1.5 µM | 47.2 ± 4.0 | 71.8 ± 4.0 *** | 64.9 ± 3.8 *** | 34.0 ± 3.0 ** | 83.7 ± 6.1 *** |
| 2 | 2-NH2-3-CN-quinoxaline-1,4-dioxide, 350 µM | 47.3 ± 4.2 | 83.5 ± 7.0 ** | 72.1 ± 5.7 ** | 35.3 ± 3.6 * | 71.6 ± 5.1 ** |
| 3 | 1,2,4-Benzotriazine-1,4-dioxide, 10 µM | 55.6 ± 4.5 | n.d. | n.d. | n.d. | 81.2 ± 7.4 ** |
| 4 | 3-CF3SO2NH-1,2,4-benzotriazine-1,4-dioxide, 40 µM | 55.8 ± 4.9 | n.d. | n.d. | n.d. | 79.0 ± 3.1 ** |
| 5 | 2-CF3-quinoxaline-1,4-dioxide, 10 µM | 57.6 ± 4.5 | n.d. | n.d. | n.d. | 83.1 ± 5.6 ** |
| Cell Line | a | b | c | d | r2 |
|---|---|---|---|---|---|
| 7.22 ± 0.80 | −1.36 ± 0.19 | - | - | 0.7638 | |
| MH22a | 7.07 ± 0.84 | −1.33 ± 0.20 | −0.09 ± 0.13 | - | 0.7704 |
| 8.44 ± 0.88 | −1.20 ± 0.18 | - | −0.55 ± 0.24 | 0.8266 | |
| 8.30 ± 0.88 | −1.14 ± 0.19 | −0.13 ± 0.118 | −0.59 ± 0.24 | 0.8405 | |
| 6.68 ± 0.73 | −1.19 ± 0.17 | - | - | 0.7440 | |
| HCT-116 | 6.63 ± 0.79 | −1.18 ± 0.19 | −0.03 ± 0.13 | - | 0.7448 |
| 7.74 ± 0.82 | −1.05 ± 0.17 | - | −0.48 ± 0.22 | 0.8052 | |
| 7.67 ± 0.85 | −1.02 ± 0.18 | −0.06 ± 0.11 | −0.50 ± 0.23 | 0.8093 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemeikaitė-Čėnienė, A.; Šarlauskas, J.; Misevičienė, L.; Marozienė, A.; Jonušienė, V.; Lesanavičius, M.; Čėnas, N. Aerobic Cytotoxicity of Aromatic N-Oxides: The Role of NAD(P)H:Quinone Oxidoreductase (NQO1). Int. J. Mol. Sci. 2020, 21, 8754. https://doi.org/10.3390/ijms21228754
Nemeikaitė-Čėnienė A, Šarlauskas J, Misevičienė L, Marozienė A, Jonušienė V, Lesanavičius M, Čėnas N. Aerobic Cytotoxicity of Aromatic N-Oxides: The Role of NAD(P)H:Quinone Oxidoreductase (NQO1). International Journal of Molecular Sciences. 2020; 21(22):8754. https://doi.org/10.3390/ijms21228754
Chicago/Turabian StyleNemeikaitė-Čėnienė, Aušra, Jonas Šarlauskas, Lina Misevičienė, Audronė Marozienė, Violeta Jonušienė, Mindaugas Lesanavičius, and Narimantas Čėnas. 2020. "Aerobic Cytotoxicity of Aromatic N-Oxides: The Role of NAD(P)H:Quinone Oxidoreductase (NQO1)" International Journal of Molecular Sciences 21, no. 22: 8754. https://doi.org/10.3390/ijms21228754





