Responses of DNA Mismatch Repair Proteins to a Stable G-Quadruplex Embedded into a DNA Duplex Structure
Abstract
:1. Introduction
2. Results
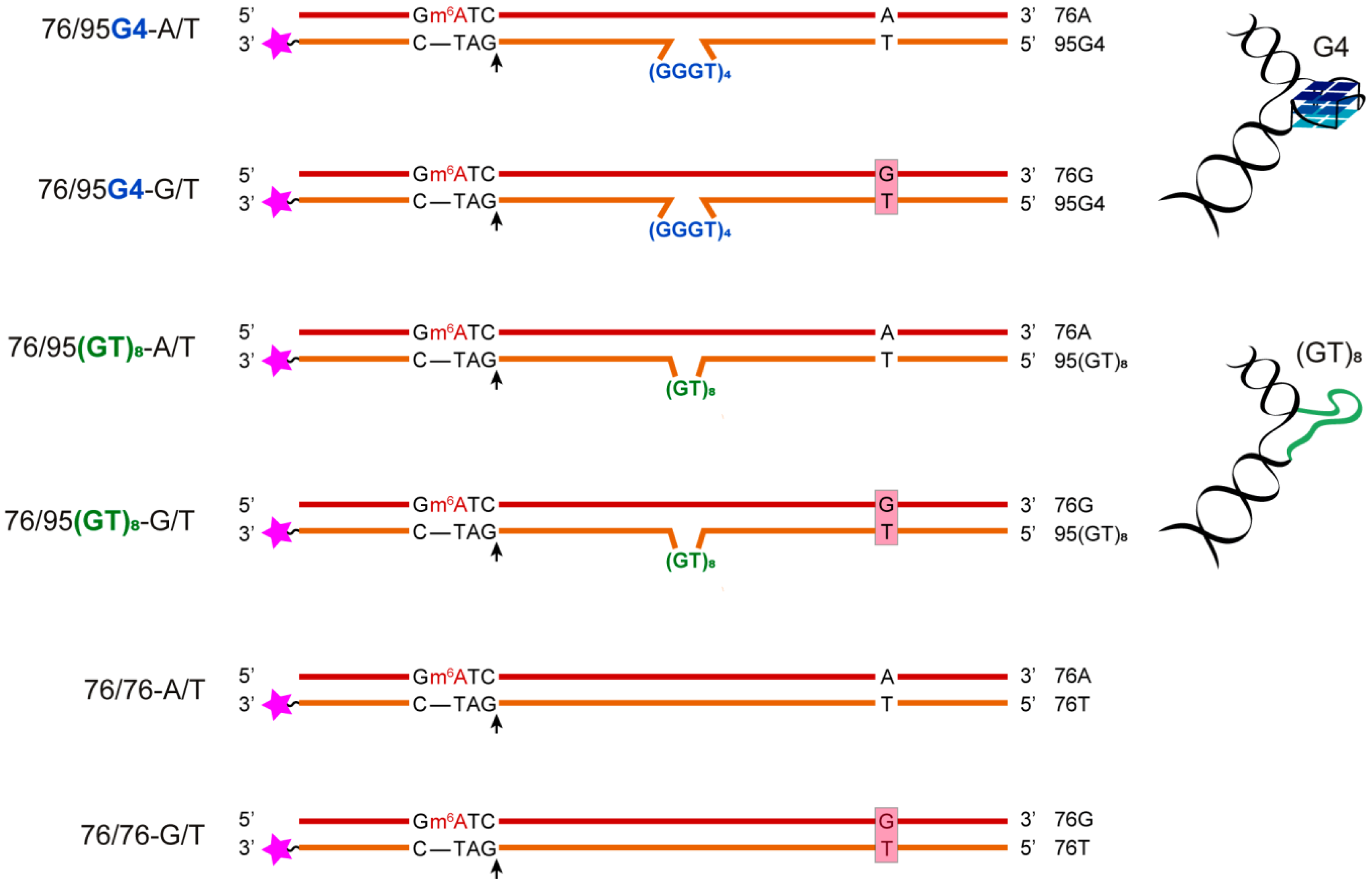
2.1. Design of the DNA Models
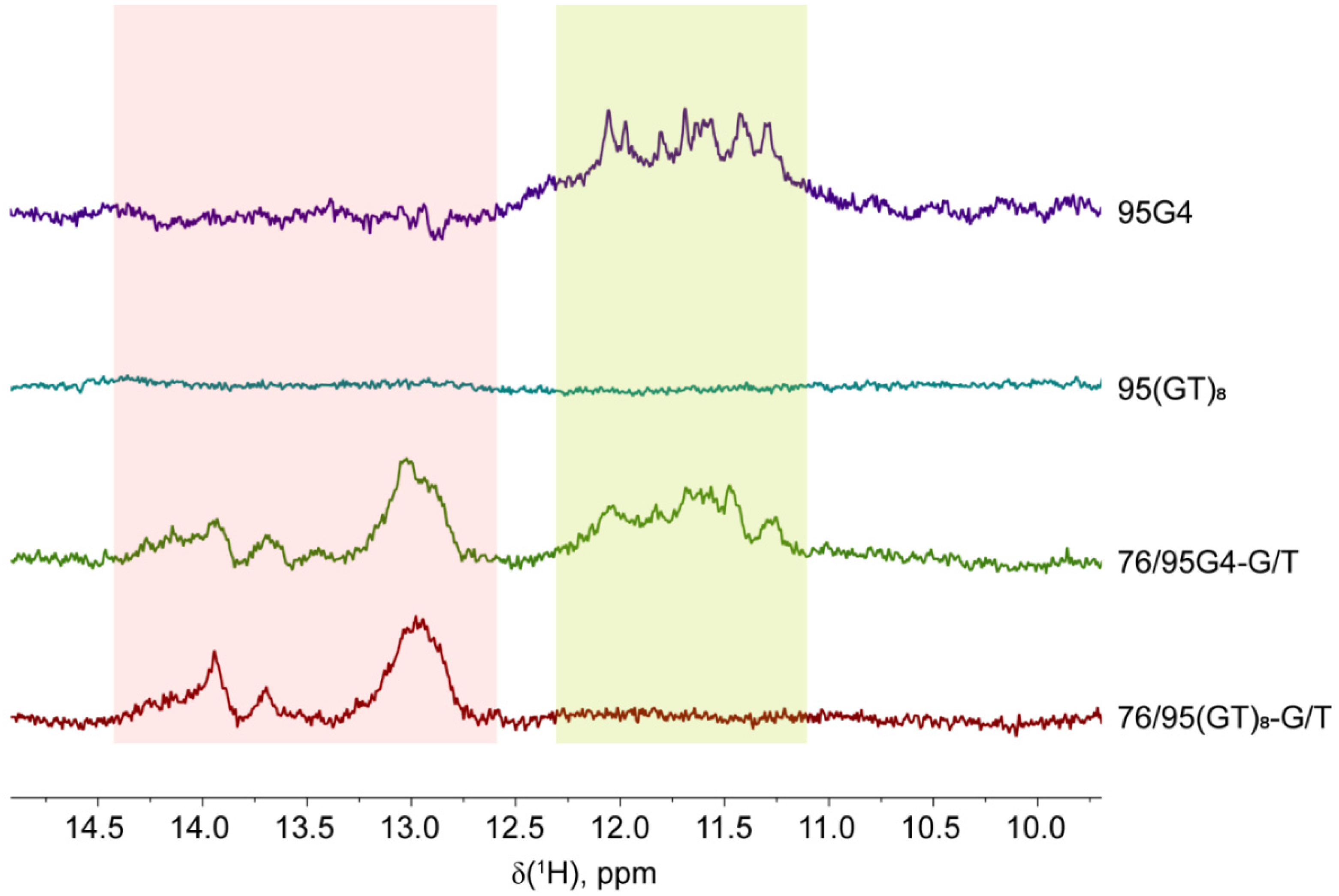
2.2. Monitoring of G4 and Duplex Structures within the Long DNA Constructs by 1H NMR Spectroscopy
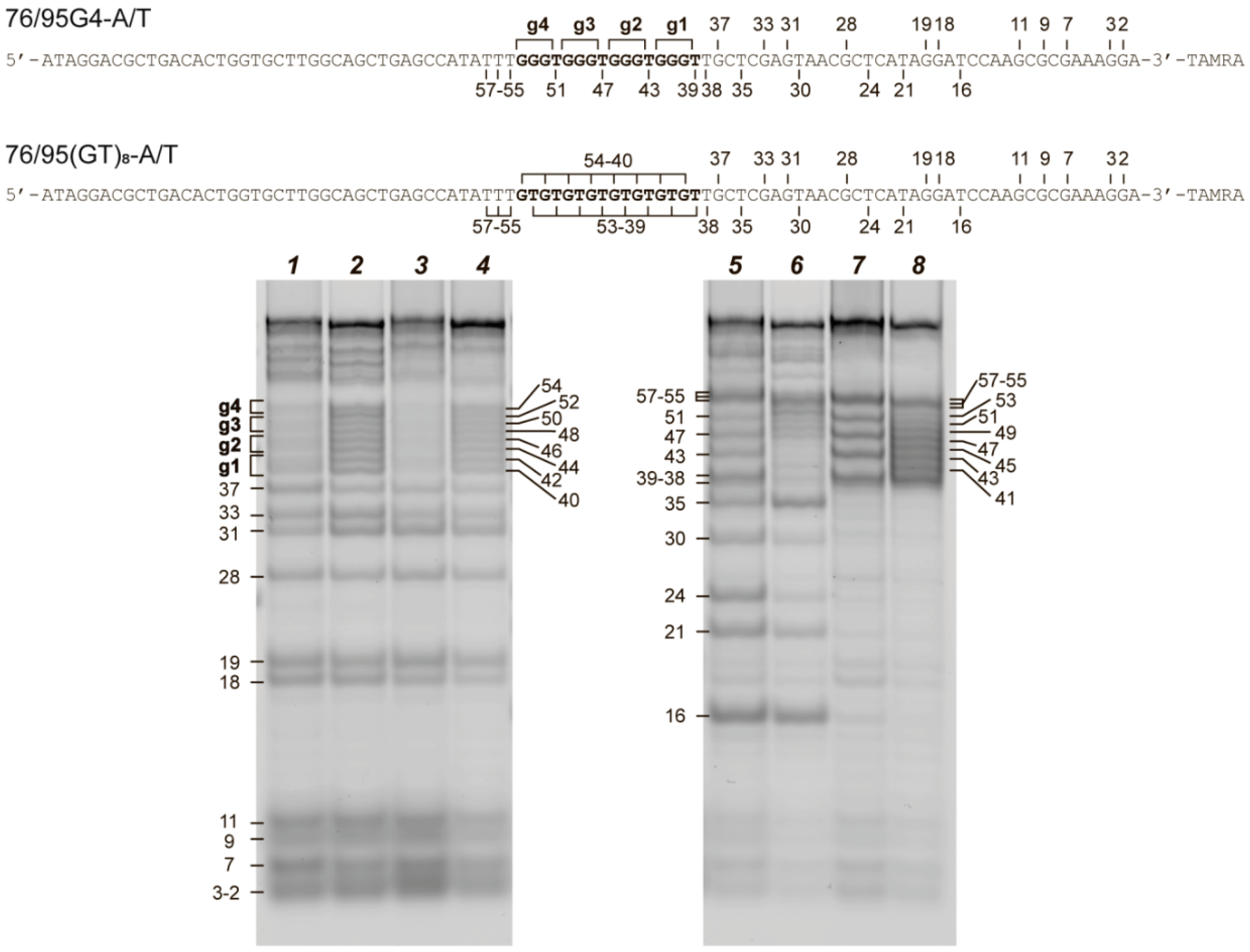
2.3. Chemical Probing Assays Clearly Indicate the Existence of G4 and Duplex Structures in 76/95 DNA Models
2.4. CD and UV Spectroscopy Confirm the Existence of a Parallel G4 Structure Folded within the DNA Double Helix
2.5. Dual-Label Fluorescent DNA Probes Are Informative for Detecting Duplex Formation in 41/22 Models rather than G4 Folding
2.6. An Intramolecular G4 Tightly Binds MutS Regardless of a Nucleotide Cofactor and Stimulates MutS ATPase Activity
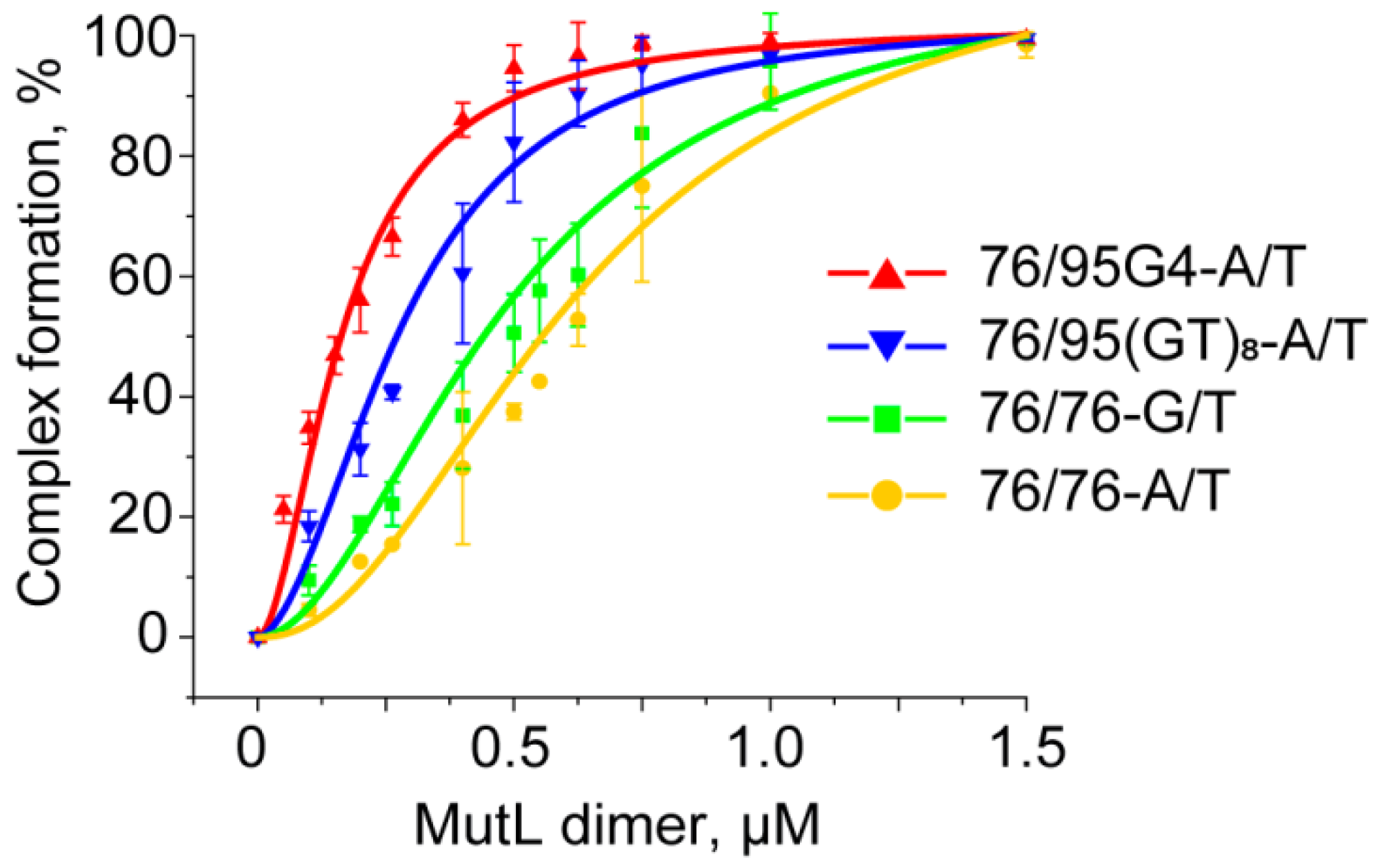
2.7. Highly Effective Binding of G4 DNA to ecMutL Is Discovered
2.8. A G4 Unlike a G/T Mismatch Does Not Activate the MMR System
2.9. A G4 Does not Prevent the Mismatch-Dependent Activation of E. coli MMR
3. Discussion
4. Materials and Methods
4.1. DNA Oligonucleotides
4.2. DNA Duplexes
4.3. Purification of Recombinant Proteins
4.4. 1H NMR Spectroscopy
4.5. A Footprinting Assay
4.6. UV Melting Experiments
4.7. CD Measurements
4.8. A Fluorescence Experiment
4.9. DNA-Binding Activity of EcMutS and RsMutS
4.10. Colorimetric Determination of EcMutS ATPase Activity
4.11. DNA-Binding Activity of EcMutL
4.12. Hydrolysis of Plasmid DNA by RsMutL
4.13. DNA Cleavage by E. coli MMR Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [Green Version]
- Eddy, J.; Vallur, A.C.; Varma, S.; Liu, H.; Reinhold, W.C.; Pommier, Y.; Maizels, N. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 2011, 39, 4975–4983. [Google Scholar] [CrossRef] [Green Version]
- Todd, A.K.; Neidle, S. Mapping the sequences of potential guanine quadruplex motifs. Nucleic Acids Res. 2011, 39, 4917–4927. [Google Scholar] [CrossRef] [Green Version]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Yadav, V.K.; Basundra, R.; Kumar, A.; Chowdhury, S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009, 37, 4194–4204. [Google Scholar] [CrossRef] [Green Version]
- Sissi, C.; Gatto, B.; Palumbo, M. The evolving world of protein–G-quadruplex recognition: A medicinal chemist’s perspective. Biochimie 2011, 93, 1219–1230. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- Dolinnaya, N.G.; Ogloblina, A.M.; Yakubovskaya, M.G. Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: Overview 50 years after their discovery. Biochemistry 2016, 81, 1602–1649. [Google Scholar] [CrossRef]
- Bryan, T.M. Mechanisms of DNA replication and repair: Insights from the study of G-quadruplexes. Molecules 2019, 24, 3439. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, O.; Bourdoncle, A.; Boulé, J.B.; Brosh, R.M.; Mergny, J.-L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef] [Green Version]
- Burra, S.; Marasco, D.; Malfatti, M.C.; Antoniali, G.; Esposito, V.; Demple, B.; Galeone, A.; Tell, G. Human AP-endonuclease (Ape1) activity on telomeric G4 structures is modulated by acetylable lysine residues in the N-terminal sequence. DNA Repair 2019, 73, 129–143. [Google Scholar] [CrossRef]
- Halabi, A.; Ditch, S.; Wang, J.; Grabczyk, E. DNA mismatch repair complex MutSβ promotes GAA·TTC repeat expansion in human cells. J. Biol. Chem. 2012, 287, 29958–29967. [Google Scholar] [CrossRef] [Green Version]
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef]
- Perevoztchikova, S.A.; Romanova, E.A.; Oretskaya, T.S.; Friedhoff, P.; Kubareva, E.A. Modern aspects of the structural and functional organization of the DNA mismatch repair system. Acta Nat. 2013, 5, 17–34. [Google Scholar] [CrossRef]
- Corrette-Bennett, S.E.; Mohlman, N.L.; Rosado, Z.; Miret, J.J.; Hess, P.M.; Parker, B.O.; Lahue, R.S. Efficient repair of large DNA loops in Saccharomyces cerevisiae. Nucleic Acids Res. 2001, 29, 4134–4143. [Google Scholar] [CrossRef] [Green Version]
- McCulloch, S.D.; Gu, L.; Li, G.-M. Bi-directional processing of DNA loops by mismatch repair-dependent and -independent pathways in human cells. J. Biol. Chem. 2003, 278, 3891–3896. [Google Scholar] [CrossRef] [Green Version]
- Ranjha, L.; Anand, R.; Cejka, P. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to holliday junctions. J. Biol. Chem. 2014, 289, 5674–5686. [Google Scholar] [CrossRef] [Green Version]
- Larson, E.D.; Duquette, M.L.; Cummings, W.J.; Streiff, R.J.; Maizels, N. MutSα binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 2005, 15, 470–474. [Google Scholar] [CrossRef] [Green Version]
- Ehrat, E.A.; Johnson, B.R.; Williams, J.D.; Borchert, G.M.; Larson, E.D. G-quadruplex recognition activities of E. coli MutS. BMC Mol. Biol. 2012, 13, 23. [Google Scholar] [CrossRef] [Green Version]
- Rawal, P.; Bhadra, V.; Kummarasetti, R.; Ravindran, J.; Kumar, N.; Halder, K.; Sharma, R.; Mukerji, M.; Das, S.K. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Res. 2006, 16, 644–655. [Google Scholar] [CrossRef] [Green Version]
- Augusto-Pinto, L.; Da Silva, C.G.R.; De Oliveira Lopes, D.; Machado-Silva, A.; Machado, C.R. Escherichia coli as a model system to study DNA repair genes of eukaryotic organisms. Genet. Mol. Res. 2003, 2, 77–91. [Google Scholar]
- Duquette, M.L.; Handa, P.; Vincent, J.A.; Taylor, A.F.; Maizels, N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004, 18, 1618–1629. [Google Scholar] [CrossRef] [Green Version]
- Monakhova, M.V.; Penkina, A.I.; Pavlova, A.V.; Lyaschuk, A.M.; Kucherenko, V.V.; Alexeevski, A.V.; Lunin, V.G.; Friedhoff, P.; Klug, G.; Oretskaya, T.S.; et al. Endonuclease activity of MutL protein of the Rhodobacter sphaeroides mismatch repair system. Biochemistry (Moscow) 2018, 83, 281–293. [Google Scholar] [CrossRef]
- Risitano, A.; Fox, K.R. Stability of intramolecular DNA quadruplexes: Comparison with DNA duplexes. Biochemistry 2003, 42, 6507–6513. [Google Scholar] [CrossRef]
- Mendoza, O.; Elezgaray, J.; Mergny, J.-L. Kinetics of quadruplex to duplex conversion. Biochimie 2015, 118, 225–233. [Google Scholar] [CrossRef]
- Tatarinova, O.; Tsvetkov, V.; Basmanov, D.; Barinov, N.; Smirnov, I.; Timofeev, E.; Kaluzhny, D.; Chuvilin, A.; Klinov, D.; Varizhuk, A.; et al. Comparison of the ‘chemical’ and ‘structural’ approaches to the optimization of the thrombin-binding aptamer. PLoS ONE 2014, 9, e89383. [Google Scholar] [CrossRef] [Green Version]
- Ogloblina, A.M.; Bannikova, V.A.; Khristich, A.N.; Oretskaya, T.S.; Yakubovskaya, M.G.; Dolinnaya, N.G. Parallel G-quadruplexes formed by guanine-rich microsatellite repeats inhibit human topoisomerase I. Biochemistry 2015, 80, 1026–1038. [Google Scholar] [CrossRef]
- Adrian, M.; Heddi, B.; Phan, A.T. NMR spectroscopy of G-quadruplexes. Methods 2012, 57, 11–24. [Google Scholar] [CrossRef]
- Patel, D.J. NMR studies of the structure and stability of 1:2 actinomycin D d-pG-C complex in aqueous solution. BBA Nucleic Acids Protein Synth. 1976, 442, 98–108. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 324, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bui, C.T.; Rees, K.; Cotton, R.G.H. Permanganate oxidation reactions of DNA: Perspective in biological studies. Nucleosides Nucleotides Nucleic Acids 2003, 22, 1835–1855. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Hurley, L.H. Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods Mol. Biol. 2010, 608, 65–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowiect, J.A.; Zhang, L.; Sasse-Dwight, S.; Gralla, J.D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J. Mol. Biol. 1987, 196, 101–111. [Google Scholar] [CrossRef]
- Bailly, C.; Marchand, C.; Nguyen, C.H.; Bisagni, E.; Garestier, T.; Hélène, C.; Waring, M.J. Localized chemical reactivity in double-stranded DNA associated with the intercalative binding of benzo[e]pyridoindole and benzo[g]pyridoindole triple-helix-stabilizing ligands. Eur. J. Biochem. 1995, 232, 66–76. [Google Scholar] [CrossRef]
- Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef]
- Dolinnaya, N.G.; Yuminova, A.V.; Spiridonova, V.A.; Arutyunyan, A.M.; Kopylov, A.M. Coexistence of G-quadruplex and duplex domains within the secondary structure of 31-mer DNA thrombin-binding aptamer. J. Biomol. Struct. Dyn. 2012, 30, 524–531. [Google Scholar] [CrossRef]
- Liu, J.; Hanne, J.; Britton, B.M.; Bennett, J.; Kim, D.; Lee, J.; Fishel, R. Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature 2016, 539, 583–587. [Google Scholar] [CrossRef] [Green Version]
- Natrajan, G.; Lamers, M.H.; Enzlin, J.H.; Winterwerp, H.H.K.; Perrakis, A.; Sixma, T.K. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: A common recognition mode for diverse substrates. Nucleic Acids Res. 2003, 31, 4814–4821. [Google Scholar] [CrossRef] [Green Version]
- Acharya, S.; Foster, P.L.; Brooks, P.; Fishel, R.; Hall, J.; Street, E.T.; Cremieux, G. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell 2003, 12, 233–246. [Google Scholar] [CrossRef]
- Hingorani, M.M. Mismatch binding, ADP-ATP exchange and intramolecular signaling during mismatch repair. DNA Repair 2016, 38, 24–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessmer, I.; Yang, Y.; Zhai, J.; Du, C.; Hsieh, P.; Hingorani, M.M.; Erie, D.A. Mechanism of MutS searching for DNA mismatches and signaling repair. J. Biol. Chem. 2008, 283, 36646–36654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junop, M.S.; Obmolova, G.; Rausch, K.; Hsieh, P.; Yang, W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 2001, 7, 1–12. [Google Scholar] [CrossRef]
- Sancar, A.; Hearst, J.E. Molecular matchmakers. Science 1993, 259, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Kadyrov, F.A.; Dzantiev, L.; Constantin, N.; Modrich, P. Endonucleolytic function of MutLα in human mismatch repair. Cell 2006, 126, 297–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadyrov, F.A.; Holmes, S.F.; Arana, M.E.; Lukianova, O.A.; O’Donnell, M.; Kunkel, T.A.; Modrich, P. Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J. Biol. Chem. 2007, 282, 37181–37190. [Google Scholar] [CrossRef] [Green Version]
- Bende, S.M.; Grafström, R.H. The DNA binding properties of the MutL protein isolated from Escherichia coli. Nucleic Acids Res. 1991, 19, 1549–1555. [Google Scholar] [CrossRef] [Green Version]
- Fukui, K.; Iino, H.; Baba, S.; Kumasaka, T.; Kuramitsu, S.; Yano, T. Crystal structure and DNA-binding property of the ATPase domain of bacterial mismatch repair endonuclease MutL from Aquifex aeolicus. BBA Proteins Proteom. 2017, 1865, 1178–1187. [Google Scholar] [CrossRef]
- Hall, M.C.; Wang, H.; Erie, D.A.; Kunkel, T.A. High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J. Mol. Biol. 2001, 312, 637–647. [Google Scholar] [CrossRef] [Green Version]
- Groothuizen, F.S.; Winkler, I.; Cristóvão, M.; Fish, A.; Winterwerp, H.H.; Reumer, A.; Marx, A.D.; Hermans, N.; Nicholls, R.A.; Murshudov, G.N.; et al. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. Elife 2015, 4, e06744. [Google Scholar] [CrossRef] [PubMed]
- Ban, C.; Junop, M.; Yang, W. Transformation of MutL by ATP binding and hydrolysis: A switch in DNA mismatch repair. Cell 1999, 97, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Au, K.G.; Welsh, K.; Modrich, P. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 1992, 267, 12142–12148. [Google Scholar]
- Fukui, K. DNA mismatch repair in eukaryotes and bacteria. J. Nucleic Acids 2010, 2010, 260512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipps, H.J.; Rhodes, D. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Q.M.; Wang, Y.R.; Xi, X.G.; Hou, X.M. DNA-unwinding activity of Saccharomyces cerevisiae Pif1 is modulated by thermal stability, folding conformation, and loop lengths of G-quadruplex DNA. J. Biol. Chem. 2018, 293, 18504–18513. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.M.; Monsell, K.R.; Noulin, F.; Toyin Famodimu, M.; Smargiasso, N.; Damblon, C.; Horrocks, P.; Merrick, C.J. G-quadruplex DNA motifs in the malaria parasite Plasmodium falciparum and their potential as novel antimalarial drug targets. Antimicrob. Agents Chemother. 2018, 62, e01828-17. [Google Scholar] [CrossRef] [Green Version]
- Perrone, R.; Lavezzo, E.; Riello, E.; Manganelli, R.; Palù, G.; Toppo, S.; Provvedi, R.; Richter, S.N. Mapping and characterization of G-quadruplexes in Mycobacterium tuberculosis gene promoter regions. Sci. Rep. 2017, 7, 5743. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.K.; Jain, N.; Shankar, U.; Tawani, A.; Sharma, T.K.; Kumar, A. Characterization of highly conserved G-quadruplex motifs as potential drug targets in Streptococcus pneumoniae. Sci. Rep. 2019, 9, 1791. [Google Scholar] [CrossRef]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef] [Green Version]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More than just a kink in microbial genomes. Trends Microbiol. 2018, 27, 148–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartas, M.; Čutová, M.; Brázda, V.; Kaura, P.; Šťastný, J.; Kolomazník, J.; Coufal, J.; Goswami, P.; Červeň, J.; Pečinka, P. The presence and localization of G-quadruplex forming sequences in the domain of bacteria. Molecules 2019, 24, 1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahoon, L.A.; Seifert, S.H. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 2009, 325, 764–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; Mccauley, P.; Boutell, J.M.; Di Antonio, M.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holder, I.T.; Hartig, J.S. A matter of location: Influence of G-quadruplexes on Escherichia coli gene expression. Chem. Biol. 2014, 21, 1511–1521. [Google Scholar] [CrossRef] [Green Version]
- Kota, S.; Dhamodharan, V.; Pradeepkumar, P.I.; Misra, H.S. G-quadruplex forming structural motifs in the genome of Deinococcus radiodurans and their regulatory roles in promoter functions. Appl. Microbiol. Biotechnol. 2015, 99, 9761–9769. [Google Scholar] [CrossRef]
- Mishra, S.K.; Shankar, U.; Jain, N.; Sikri, K.; Tyagi, J.S.; Sharma, T.K.; Mergny, J.; Kumar, A. Characterization of G-quadruplex motifs in espB, espK, and cyp51 genes of Mycobacterium tuberculosis as potential drug targets. Mol. Ther. Nucleic Acid. 2019, 16, 698–706. [Google Scholar] [CrossRef] [Green Version]
- Koole, W.; Van Schendel, R.; Karambelas, A.E.; Van Heteren, J.T.; Okihara, K.L.; Tijsterman, M. A polymerase theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 2014, 5, 3216. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Dos Santos, N.; Silvester, J.; Wei, V.; Garcia, J.; et al. Cx-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 14432. [Google Scholar] [CrossRef]
- Gray, L.T.; Vallur, A.C.; Eddy, J.; Maizels, N. G-quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014, 10, 313–318. [Google Scholar] [CrossRef]
- Rachwal, P.A.; Findlow, I.S.; Werner, J.M.; Brown, T.; Fox, K.R. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007, 35, 4214–4222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreig, A.; Calvert, J.; Sanoica, J.; Cullum, E.; Tipanna, R.; Myong, S. G-quadruplex formation in double strand DNA probed by NMM and CV fluorescence. Nucleic Acids Res. 2015, 43, 7961–7970. [Google Scholar] [CrossRef] [PubMed]
- Sekibo, D.A.T.; Fox, K.R. The effects of DNA supercoiling on G-quadruplex formation. Nucleic Acids Res. 2017, 45, 12069–12079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogloblina, A.M.; Khristich, A.N.; Karpechenko, N.Y.; Semina, S.E.; Belitsky, G.A.; Dolinnaya, N.G.; Yakubovskaya, M.G. Multi-targeted effects of G4-aptamers and their antiproliferative activity against cancer cells. Biochimie 2018, 145, 163–173. [Google Scholar] [CrossRef]
- Magbanua, E.; Zivkovic, T.; Hansen, B.; Beschorner, N.; Meyer, C.; Lorenzen, I.; Grötzinger, J.; Hauber, J.; Torda, A.E.; Mayer, G.; et al. d(GGGT)4 and r(GGGU)4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biol. 2013, 10, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Guédin, A.; De Cian, A.; Gros, J.; Lacroix, L.; Mergny, J.L. Sequence effects in single-base loops for quadruplexes. Biochimie 2008, 90, 686–696. [Google Scholar] [CrossRef]
- OligoAnalyzerTM Tool. Available online: https://www.idtdna.com/calc/analyzer (accessed on 30 August 2020).
- Feng, G.; Winkler, M.E. Single-step purifications of His6-MutH, His6-MutL and His6-MutS repair proteins of Escherichia coli K-12. Biotechniques 1995, 19, 956–965. [Google Scholar]
- ProtParam Tool. Available online: https://web.expasy.org/protparam (accessed on 30 August 2020).
- Rowlands, M.G.; Newbatt, Y.M.; Prodromou, C.; Pearl, L.H.; Workman, P.; Aherne, W. High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal. Biochem. 2004, 327, 176–183. [Google Scholar] [CrossRef]
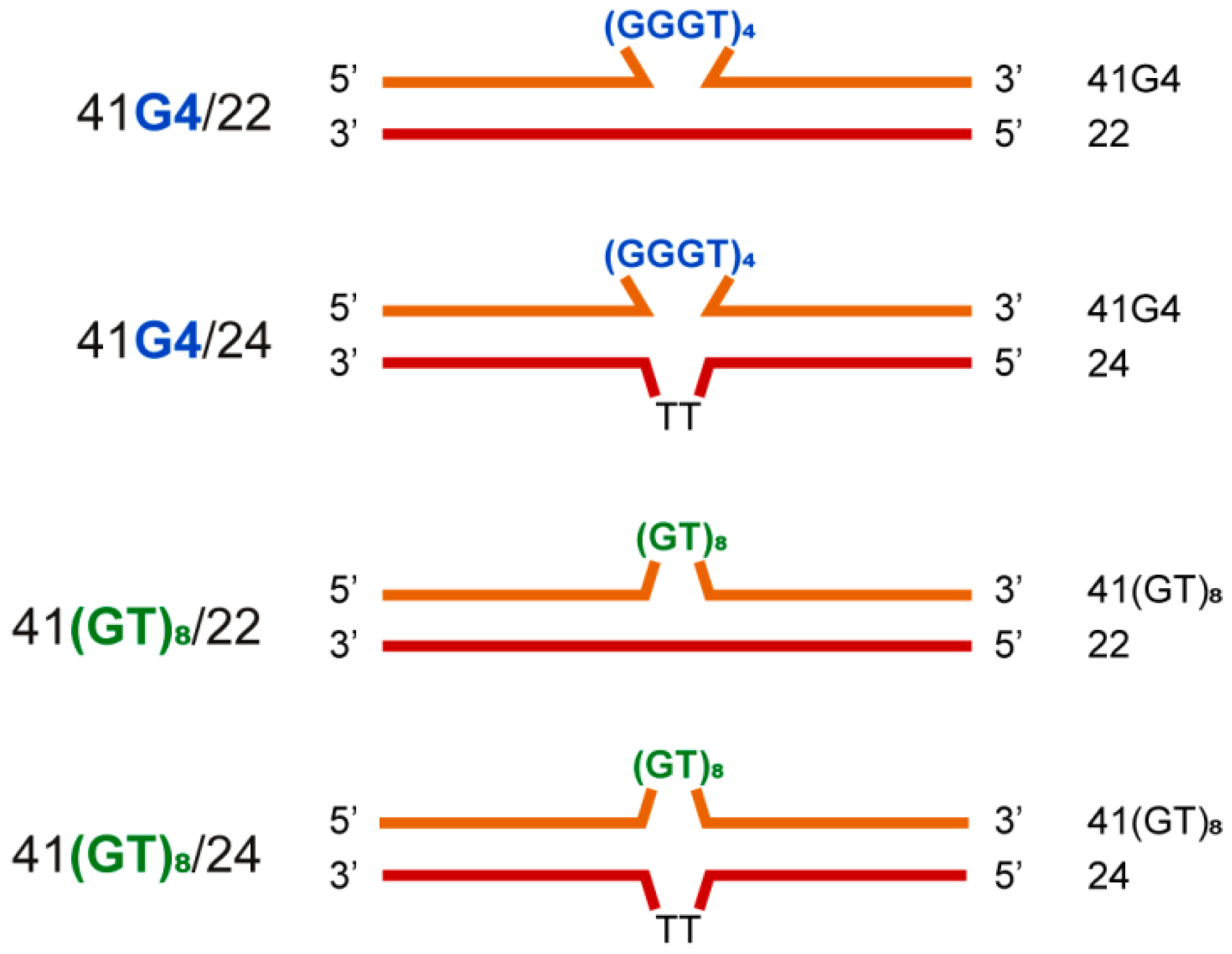
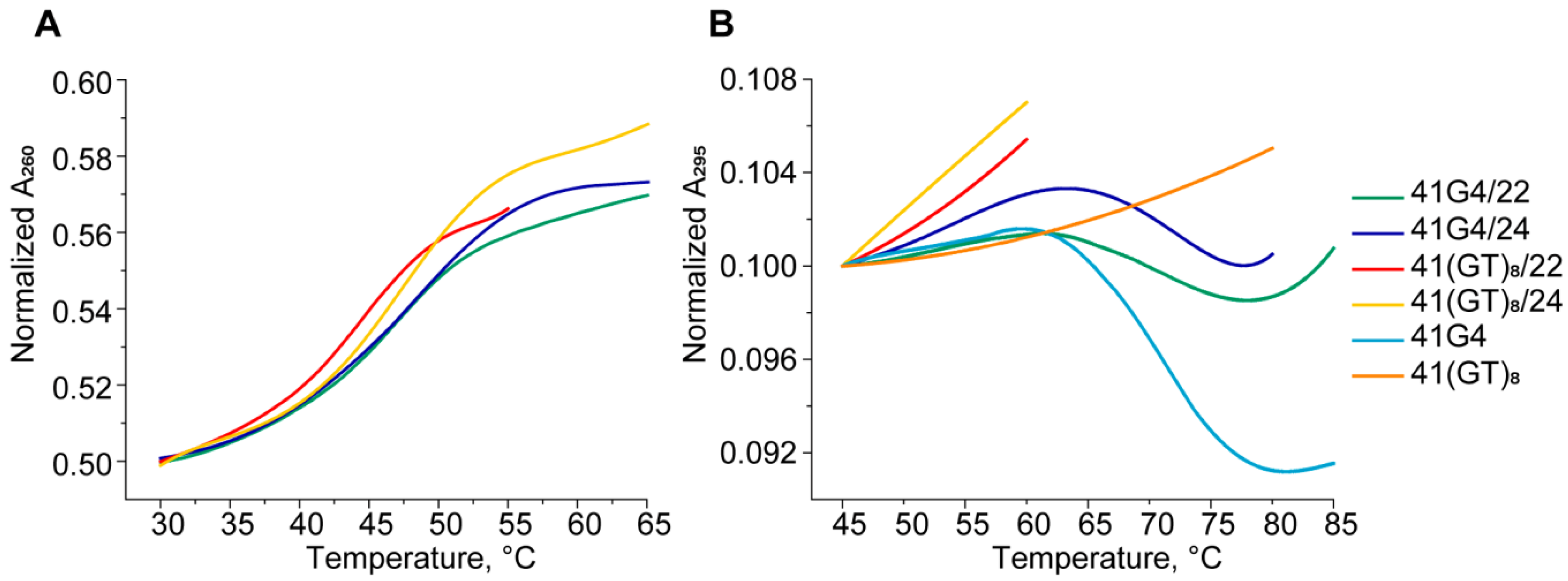




| DNA Samples | Tm of DNA Duplex, °C ± 1 | Tm of G4, °C ± 1 |
|---|---|---|
| 41G4 | -a | 71 |
| 41(GT)8 | -a | -b |
| 41G4/22 | 47 | 70 |
| 41(GT)8/22 | 44 | -b |
| 41G4/24 | 48 | 72 |
| 41(GT)8/24 | 48 | -b |
| DNA | ecMutS | rsMutS | ||||
|---|---|---|---|---|---|---|
| ADP | ATP | ATPγS | ADP | ATP | ATPγS | |
| 76/95G4-A/T | 23 ± 5 | 32 ± 4 | 31 ± 4 | 23 ± 3 | 23 ± 3 | 46 ± 5 |
| 76/76-G/T | 35 ± 3 | 38 ± 4 | 70 ± 10 | 27 ± 4 | 60 ± 10 | 360 ± 60 |
| 76/95(GT)8-A/T | 32 ± 3 | 44 ± 6 | 80 ± 10 | 31 ± 3 | 43 ± 5 | 190 ± 20 |
| 76/76-A/T | 38 ± 4 | 50 ± 10 | 150 ± 20 | 51 ± 4 | 80 ± 30 | 470 ± 70 |
| DNA | KM, µM | kcat, min−1 |
|---|---|---|
| 76/95G4-A/T | 47 ± 13 | 17 ± 1 |
| 76/76-G/T | 39 ± 8 | 22 ± 1 |
| 76/95(GT)8-A/T | 30 ± 9 | 12 ± 1 |
| 76/76-A/T | 27 ± 8 | 15 ± 1 |
| No DNA | 35 ± 15 | 7 ± 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlova, A.V.; Monakhova, M.V.; Ogloblina, A.M.; Andreeva, N.A.; Laptev, G.Y.; Polshakov, V.I.; Gromova, E.S.; Zvereva, M.I.; Yakubovskaya, M.G.; Oretskaya, T.S.; et al. Responses of DNA Mismatch Repair Proteins to a Stable G-Quadruplex Embedded into a DNA Duplex Structure. Int. J. Mol. Sci. 2020, 21, 8773. https://doi.org/10.3390/ijms21228773
Pavlova AV, Monakhova MV, Ogloblina AM, Andreeva NA, Laptev GY, Polshakov VI, Gromova ES, Zvereva MI, Yakubovskaya MG, Oretskaya TS, et al. Responses of DNA Mismatch Repair Proteins to a Stable G-Quadruplex Embedded into a DNA Duplex Structure. International Journal of Molecular Sciences. 2020; 21(22):8773. https://doi.org/10.3390/ijms21228773
Chicago/Turabian StylePavlova, Anzhela V., Mayya V. Monakhova, Anna M. Ogloblina, Natalia A. Andreeva, Gennady Yu. Laptev, Vladimir I. Polshakov, Elizaveta S. Gromova, Maria I. Zvereva, Marianna G. Yakubovskaya, Tatiana S. Oretskaya, and et al. 2020. "Responses of DNA Mismatch Repair Proteins to a Stable G-Quadruplex Embedded into a DNA Duplex Structure" International Journal of Molecular Sciences 21, no. 22: 8773. https://doi.org/10.3390/ijms21228773






