Short Disordered Epitope of CRTAM Ig-Like V Domain as a Potential Target for Blocking Antibodies
Abstract
:1. Introduction
2. Results and Discussion
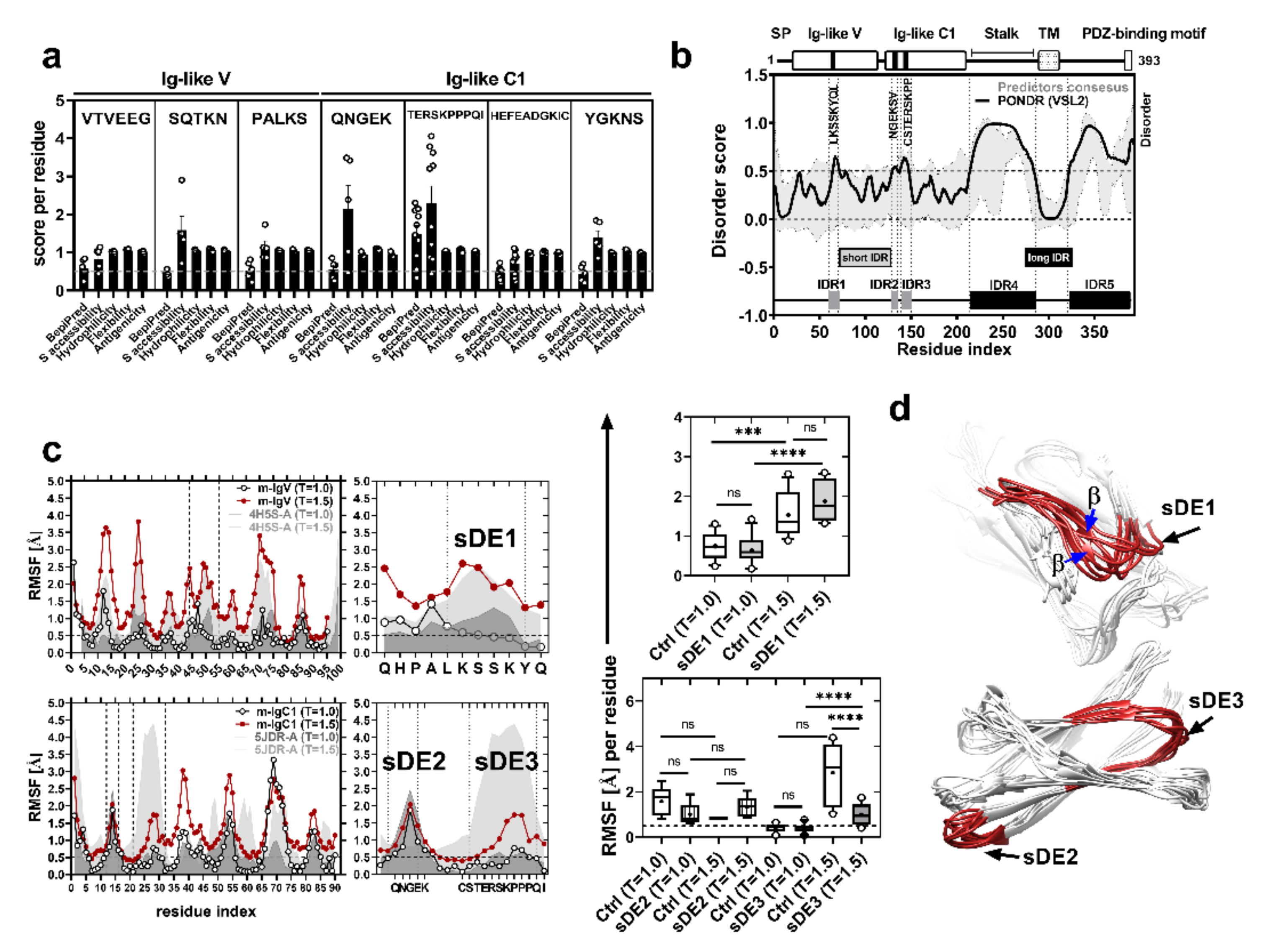
2.1. CRTAM Protein Has Short Disordered Epitopes That Are Conserved in Mammals
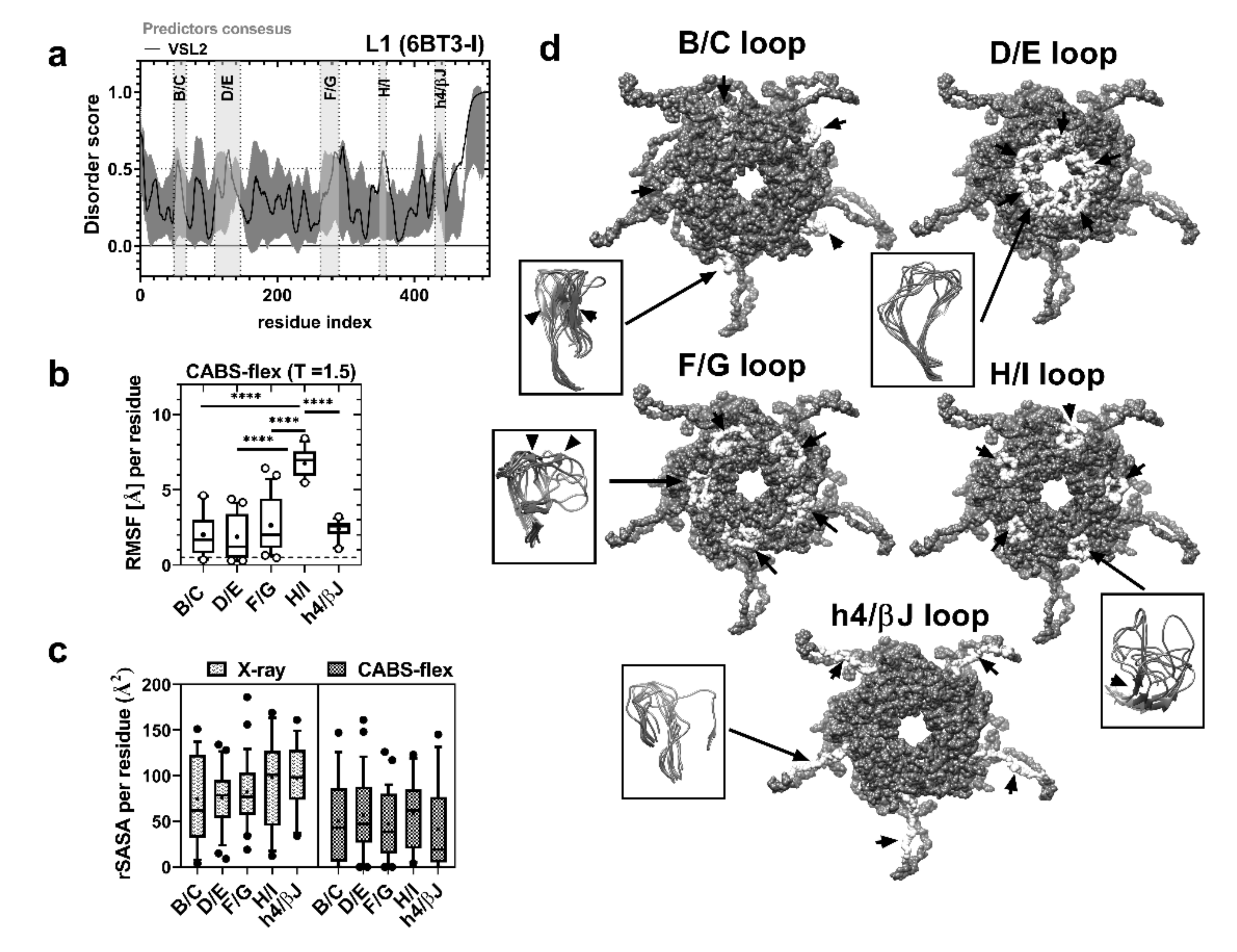
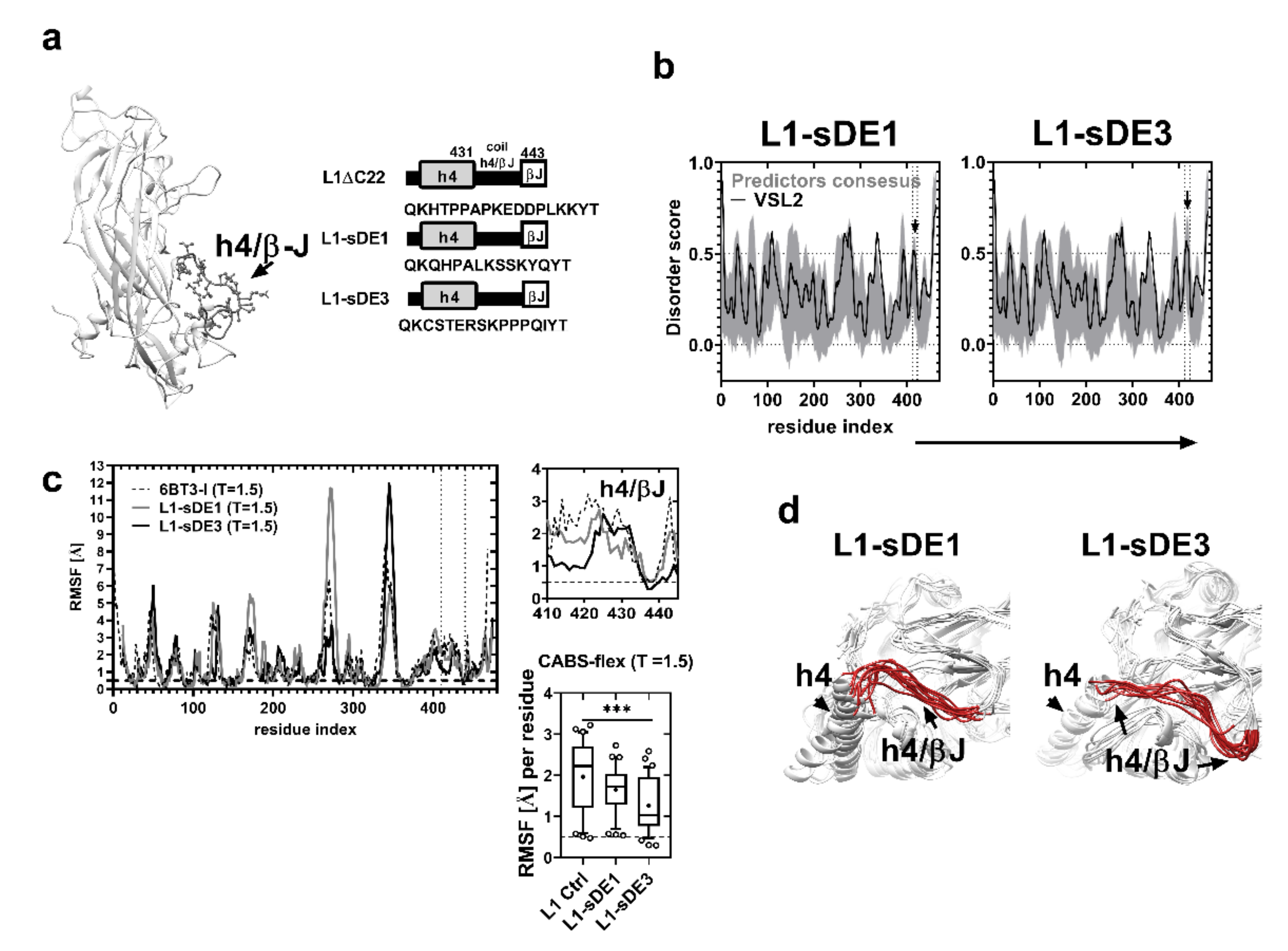
2.2. sDE1 Has a Flexible Loop Structure
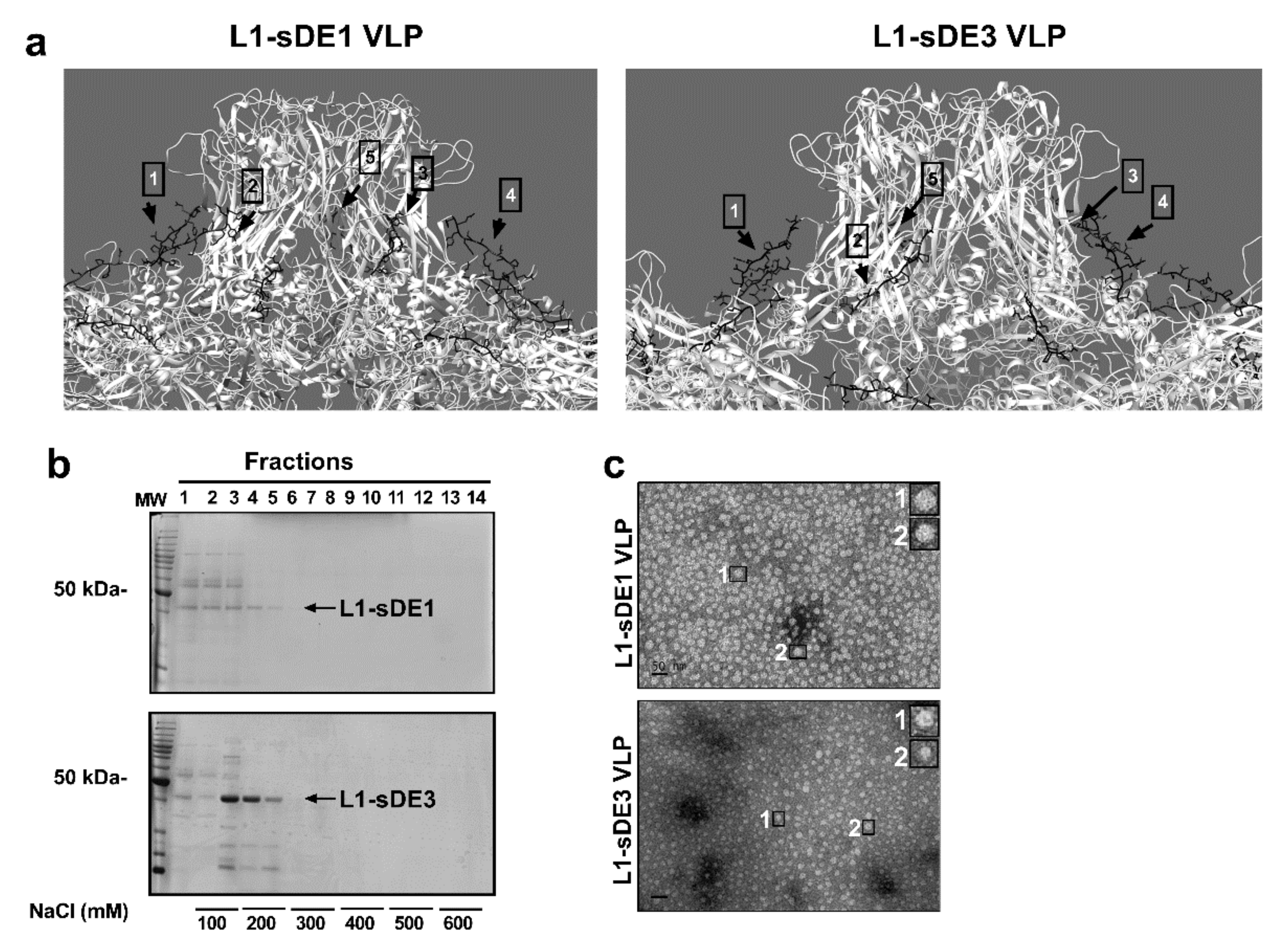
2.3. Virus-Like Particles as Carriers of Short Disordered Epitopes
2.4. Chimeric Constructs Were Assembled into Virus-Like Particles
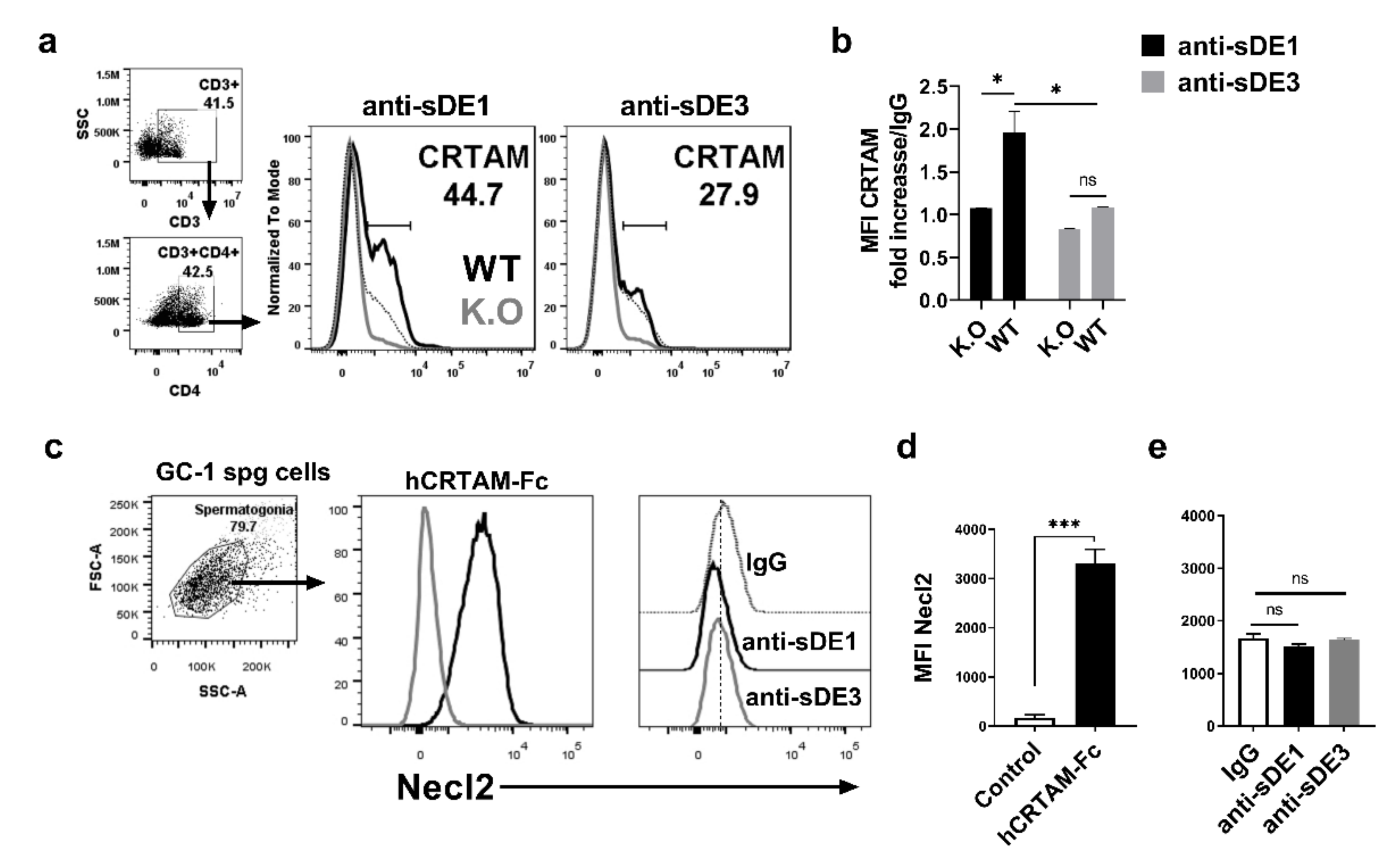
2.5. Chimeric L1 Particles Induce Specific Antibodies That Recognize CRTAM
3. Material and Methods
3.1. Animal Models
3.2. Cell Culture
3.3. B-Cell Epitope and Disorder Predictions
3.4. Multiple Alignment of Sequences
3.5. Structural Modeling Prediction and Validation
3.6. Molecular Dynamics and Relative SASA Analysis
3.7. Construction and Expression of the Chimeric L1 Gene
3.8. Extraction and Purification of Viral Protein
3.9. Characterization of Chimeric Virus-Like Particles (VLPs)
3.10. Immunization with Chimeric VLPs
3.11. Polyclonal Antibodies and Recombinant Protein Purification
3.12. Flow Cytometry
3.13. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Dunker, A.K.; Obradovic, Z. The protein trinity—linking function and disorder. Nat. Biotechnol. 2001, 19, 805–806. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins: Struct. Funct. Bioinform. 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Dyson, H.J. Making Sense of Intrinsically Disordered Proteins. Biophys. J. 2016, 110, 1013–1016. [Google Scholar] [CrossRef] [Green Version]
- Yoon, M.-K.; Mitrea, D.M.; Ou, L.; Kriwacki, R.W. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem. Soc. Trans. 2012, 40, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic Disorder in Transcription Factors. Biochemistry 2006, 45, 6873–6888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dembinski, H.; Wismer, K.; Balasubramaniam, D.; Gonzalez, H.A.; Alverdi, V.; Iakoucheva, L.M.; Komives, E.A. Predicted disorder-to-order transition mutations in IκBα disrupt function. Phys. Chem. Chem. Phys. 2014, 16, 6480–6485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Fishbain, S.; Inobe, T.; Israeli, E.; Chavali, S.; Yu, H.; Kago, G.; Babu, M.M.; Matouschek, A. Sequence composition of disordered regions fine-tunes protein half-life. Nat. Struct. Mol. Biol. 2015, 22, 214–221. [Google Scholar] [CrossRef] [Green Version]
- Van der Lee, R.; Lang, B.; Kruse, K.; Gsponer, J.; de Groot, N.S.; Huynen, M.A.; Matouschek, A.; Fuxreiter, M.; Babu, M.M. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014, 8, 1832–1844. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Uversky, V.N. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 2008, 4, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef]
- Anbo, H.; Sato, M.; Okoshi, A.; Fukuchi, S. Functional Segments on Intrinsically Disordered Regions in Disease-Related Proteins. Biomolecules 2019, 9, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koehler Leman, J.; Ulmschneider, M.B.; Gray, J.J. Computational modeling of membrane proteins. Proteins 2014, 83, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugge, K.; Brakti, I.; Fernandes, C.B.; Dreier, J.E.; Lundsgaard, J.E.; Olsen, J.G.; Skriver, K.; Kragelund, B.B. Interactions by Disorder—A Matter of Context. Front. Mol. Biosci. 2020, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; Oldfield, C.J.; Mohan, A.; Radivojac, P.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Characterization of Molecular Recognition Features, MoRFs, and Their Binding Partners. J. Proteome Res. 2007, 6, 2351–2366. [Google Scholar] [CrossRef] [Green Version]
- MacRaild, C.A.; Seow, J.; Das, S.C.; Norton, R.S. Disordered epitopes as peptide vaccines. Pept. Sci. Hoboken J. 2018, 110, e24067. [Google Scholar] [CrossRef] [Green Version]
- Ramamurthy, M.; Sankar, S.; Abraham, A.M.; Nandagopal, B.; Sridharan, G. B cell epitopes in the intrinsically disordered regions of neuraminidase and hemagglutinin proteins of H5N1 and H9N2 avian influenza viruses for peptide-based vaccine development. J. Cell. Biochem. 2019, 120, jcb.29017. [Google Scholar] [CrossRef]
- Kennedy, J.; Vicari, A.P.; Saylor, V.; Zurawski, S.M.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Zlotnik, A. A molecular analysis of NKT cells: identification of a class-I restricted T cell-associated molecule (CRTAM). J. Leukoc. Biol. 2000, 67, 725–734. [Google Scholar] [CrossRef]
- Yeh, J.-H.H.; Sidhu, S.S.; Chan, A.C. Regulation of a Late Phase of T Cell Polarity and Effector Functions by Crtam. Cell 2008, 132, 846–859. [Google Scholar] [CrossRef] [Green Version]
- Patiño-Lopez, G.; Hevezi, P.; Lee, J.; Willhite, D.; Verge, G.M.; Lechner, S.M.; Ortiz-Navarrete, V.; Zlotnik, A. Human class-I restricted T cell associated molecule is highly expressed in the cerebellum and is a marker for activated NKT and CD8+ T lymphocytes. J. Neuroimmunol. 2006, 171, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Medina-Contreras, O.; Soldevila, G.; Patiño-Lopez, G.; Canche-Pool, E.; Valle-Rios, R.; Ortiz-Navarrete, V. Role of CRTAM during mouse early T lymphocytes development. Dev. Comp. Immunol. 2010, 34, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Arase, N.; Takeuchi, A.; Unno, M.; Hirano, S.; Yokosuka, T.; Arase, H.; Saito, T. Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int. Immunol. 2005, 17, 1227–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, A.; Badr, M.E.S.G.; Miyauchi, K.; Ishihara, C.; Onishi, R.; Guo, Z.; Sasaki, Y.; Ike, H.; Takumi, A.; Tsuji, N.M.; et al. CRTAM determines the CD4 + cytotoxic T lymphocyte lineage. J. Exp. Med. 2016, 213, 123–138. [Google Scholar] [CrossRef]
- Beristain-Covarrubias, N.; Canche-Pool, E.B.; Ramirez-Velazquez, C.; Barragan-Galvez, J.C.; Gomez-Diaz, R.A.; Ortiz-Navarrete, V. Class I-Restricted T Cell-Associated Molecule Is a Marker for IFN-γ-Producing iNKT Cells in Healthy Subjects and Patients with Type 1 Diabetes. J. Interferon Cytokine Res. 2017, 37, 39–49. [Google Scholar] [CrossRef]
- Serrano, C.; Galán, S.; Rubio, J.F.; Candelario-Martínez, A.; Montes-Gómez, A.E.; Chánez-Paredes, S.; Cedillo-Barrón, L.; Schnoor, M.; Meraz-Ríos, M.A.; Villegas-Sepúlveda, N.; et al. Compartmentalized Response of IL-6/STAT3 Signaling in the Colonic Mucosa Mediates Colitis Development. J. Immunol. (Baltimore, Md. 1950) 2019, 202, 1239–1249. [Google Scholar] [CrossRef]
- Ramirez-Velazquez, C.; Beristain-Covarrubias, N.; Guido-Bayardo, L.; Ortiz-Navarrete, V. Peripheral blood T cells and neutrophils from asthma patients express class-I MHC-restricted T cell-associated molecule. Allergy Asthma Clin. Immunol. Off. J. Can. Soc. Allergy Clin. Immunol. 2014, 10, 46. [Google Scholar] [CrossRef] [Green Version]
- Galibert, L.; Diemer, G.S.; Liu, Z.; Johnson, R.S.; Smith, J.L.; Walzer, T.; Comeau, M.R.; Rauch, C.T.; Wolfson, M.F.; Sorensen, R.A.; et al. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for Class-I-restricted T-cell-associated molecule. J. Biol. Chem. 2005, 280, 21955–21964. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Lu, G.; Qi, J.; Li, Y.; Zhang, Z.; Zhang, B.; Fan, Z.; Yan, J.; Gao, G.F. Competition of Cell Adhesion and Immune Recognition: Insights into the Interaction between CRTAM and Nectin-like 2. Structure 2013, 21, 1430–1439. [Google Scholar] [CrossRef] [Green Version]
- Dessarthe, B.; Thedrez, A.; Latouche, J.-B.; Cabillic, F.; Drouet, a.; Daniel, P.; de La Pintiere, C.T.; Catros, V.; Toutirais, O. CRTAM Receptor Engagement by Necl-2 on Tumor Cells Triggers Cell Death of Activated V 9V 2 T Cells. J. Immunol. 2013, 190, 4868–4876. [Google Scholar] [CrossRef] [Green Version]
- Pavlović, M.D.; Jandrlić, D.R.; Mitić, N.S. Epitope distribution in ordered and disordered protein regions. Part B - Ordered regions and disordered binding sites are targets of T- and B-cell immunity. J. Immunol. Methods 2014, 407, 90–107. [Google Scholar] [CrossRef]
- MacRaild, C.A.; Zachrdla, M.; Andrew, D.; Krishnarjuna, B.; Nováček, J.; Žídek, L.; Sklenář, V.; Richards, J.S.; Beeson, J.G.; Anders, R.F.; et al. Conformational dynamics and antigenicity in the disordered malaria antigen merozoite surface protein 2. PLoS ONE 2015, 10, e0119899. [Google Scholar] [CrossRef] [PubMed]
- Adda, C.G.; MacRaild, C.A.; Reiling, L.; Wycherley, K.; Boyle, M.J.; Kienzle, V.; Masendycz, P.; Foley, M.; Beeson, J.G.; Norton, R.S.; et al. Antigenic Characterization of an Intrinsically Unstructured Protein, Plasmodium falciparum Merozoite Surface Protein 2. Infect. Immun. 2012, 80, 4177–4185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi, M.; Bang, G.; Tougan, T.; Palacpac, N.M.Q.; Arisue, N.; Aoshi, T.; Matsumoto, Y.; Ishii, K.J.; Egwang, T.G.; Druilhe, P.; et al. Protective Epitopes of the Plasmodium falciparum SERA5 Malaria Vaccine Reside in Intrinsically Unstructured N-Terminal Repetitive Sequences. PLoS ONE 2014, 9, e98460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwong, P.D.; Doyle, M.L.; Casper, D.J.; Cicala, C.; Leavitt, S.A.; Majeed, S.; Steenbeke, T.D.; Venturi, M.; Chaiken, I.; Fung, M.; et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 2002, 420, 678–682. [Google Scholar] [CrossRef]
- MacRaild, C.A.; Richards, J.S.; Anders, R.F.; Norton, R.S. Antibody Recognition of Disordered Antigens. Structure (Lond. Engl. 1993) 2016, 24, 148–157. [Google Scholar] [CrossRef] [Green Version]
- Garay, E.; Patiño-López, G.; Islas, S.; Alarcón, L.; Canche-Pool, E.; Valle-Rios, R.; Medina-Contreras, O.; Granados, G.; Chávez-Munguía, B.; Juaristi, E.; et al. CRTAM: A molecule involved in epithelial cell adhesion. J. Cell. Biochem. 2010, 111, 111–122. [Google Scholar] [CrossRef]
- Olugbile, S.; Kulangara, C.; Bang, G.; Bertholet, S.; Suzarte, E.; Villard, V.; Frank, G.; Audran, R.; Razaname, A.; Nebie, I.; et al. Vaccine potentials of an intrinsically unstructured fragment derived from the blood stage-associated Plasmodium falciparum protein PFF0165c. Infect. Immun. 2009, 77, 5701–5709. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.-J.; Leng, C.-H.; Lien, S.-P.; Chi, H.-Y.; Huang, C.-Y.; Lin, C.-L.; Lian, W.-C.; Chen, C.-J.; Hsieh, S.-L.; Chong, P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 2006, 24, 3100–3108. [Google Scholar] [CrossRef]
- Stavropoulos, I.; Khaldi, N.; Davey, N.E.; O’Brien, K.; Martin, F.; Shields, D.C. Protein Disorder and Short Conserved Motifs in Disordered Regions Are Enriched near the Cytoplasmic Side of Single-Pass Transmembrane Proteins. PLoS ONE 2012, 7, e44389. [Google Scholar] [CrossRef] [Green Version]
- Minezaki, Y.; Homma, K.; Nishikawa, K. Intrinsically Disordered Regions of Human Plasma Membrane Proteins Preferentially Occur in the Cytoplasmic Segment. J. Mol. Biol. 2007, 368, 902–913. [Google Scholar] [CrossRef] [PubMed]
- López, C.A.; Sethi, A.; Goldstein, B.; Wilson, B.S.; Gnanakaran, S. Membrane-mediated regulation of the intrinsically disordered CD3ϵ cytoplasmic tail of the TCR. Biophys. J. 2015, 108, 2481–2491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mészáros, B.; Dosztányi, Z.; Simon, I. Disordered Binding Regions and Linear Motifs-Bridging the Gap between Two Models of Molecular Recognition. PLoS ONE 2012, 7, 16–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasahara, K.; Shiina, M.; Higo, J.; Ogata, K.; Nakamura, H. Phosphorylation of an intrinsically disordered region of Ets1 shifts a multi-modal interaction ensemble to an auto-inhibitory state. Nucleic Acids Res. 2018, 46, 2243–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trudeau, T.; Nassar, R.; Cumberworth, A.; Wong, E.T.C.; Woollard, G.; Gsponer, J. Structure and Intrinsic Disorder in Protein Autoinhibition. Structure 2013, 21, 332–341. [Google Scholar] [CrossRef] [Green Version]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Lin, D.Y.-W.; Tanaka, Y.; Iwasaki, M.; Gittis, A.G.; Su, H.-P.; Mikami, B.; Okazaki, T.; Honjo, T.; Minato, N.; Garboczi, D.N. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. USA 2008, 105, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Wei, H.; Wang, X.; Bai, Y.; Wang, P.; Wu, J.; Jiang, X.; Wang, Y.; Cai, H.; Xu, T.; et al. Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade. Cell Discov. 2017, 3, 17004. [Google Scholar] [CrossRef] [Green Version]
- DeForte, S.; Uversky, V.N. Resolving the ambiguity: Making sense of intrinsic disorder when PDB structures disagree. Protein Sci. 2016, 25, 676–688. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, T.; Romero, P.R.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in the Protein Data Bank. J. Biomol. Struct. Dyn. 2007, 24, 325–342. [Google Scholar] [CrossRef]
- Monroy-García, A.; Gómez-Lim, M.A.; Weiss-Steider, B.; Hernández-Montes, J.; Huerta-Yepez, S.; Rangel-Santiago, J.F.; Santiago-Osorio, E.; Mora García, M.d.L. Immunization with an HPV-16 L1-based chimeric virus-like particle containing HPV-16 E6 and E7 epitopes elicits long-lasting prophylactic and therapeutic efficacy in an HPV-16 tumor mice model. Arch. Virol. 2014, 159, 291–305. [Google Scholar] [CrossRef]
- Paz De la Rosa, G.; Monroy-García, A.; Mora-García, M.D.L.; Peña, C.G.R.; Hernández-Montes, J.; Weiss-Steider, B.; Gómez-Lim, M.A. An HPV 16 L1-based chimeric human papilloma virus-like particles containing a string of epitopes produced in plants is able to elicit humoral and cytotoxic T-cell activity in mice. Virol. J. 2009, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dale, C.J.J.; Liu, X.S.; De Rose, R.; Purcell, D.F.J.J.; Anderson, J.; Xu, Y.; Leggatt, G.R.; Frazer, I.H.; Kent, S.J. Chimeric Human Papilloma Virus–Simian/Human Immunodeficiency Virus Virus-like-Particle Vaccines: Immunogenicity and Protective Efficacy in Macaques. Virology 2002, 301, 176–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.; Li, N.; Zhou, J.; Chen, Y.; Jiang, M.; Qi, Y.; Liu, H.; Liu, Y.; Liu, D.; Zhao, J.; et al. Mapping the B cell epitopes within the major capsid protein L1 of human papillomavirus type 16. Int. J. Biol. Macromol. 2018, 118, 1354–1361. [Google Scholar] [CrossRef]
- Fukui, A.; Matsueda, S.; Kawano, K.; Tsuda, N.; Komatsu, N.; Shichijo, S.; Sasada, T.; Hattori, S.; Ushijima, K.; Itoh, K.; et al. Identification of B cell epitopes reactive to human papillomavirus type-16L1- derived peptides. Virol. J. 2012, 9, 199. [Google Scholar] [CrossRef] [Green Version]
- Tamarozzi, E.R.; Giuliatti, S. Understanding the role of intrinsic disorder of viral proteins in the oncogenicity of different types of HPV. Int. J. Mol. Sci. 2018, 19, 198. [Google Scholar] [CrossRef] [Green Version]
- Kumar, D.; Singh, A.; Kumar, P.; Uversky, V.N.; Rao, C.D.; Giri, R. Understanding the penetrance of intrinsic protein disorder in rotavirus proteome. Int. J. Biol. Macromol. 2020, 144, 892–908. [Google Scholar] [CrossRef]
- Giri, R.; Kumar, D.; Sharma, N.; Uversky, V.N. Intrinsically Disordered Side of the Zika Virus Proteome. Front. Cell. Infect. Microbiol. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Lyngdoh, D.L.; Shukla, H.; Sonkar, A.; Anupam, R.; Tripathi, T. Portrait of the Intrinsically Disordered Side of the HTLV-1 Proteome. ACS Omega 2019, 4, 10003–10018. [Google Scholar] [CrossRef]
- Bissett, S.L.; Godi, A.; Beddows, S. The DE and FG loops of the HPV major capsid protein contribute to the epitopes of vaccine-induced cross-neutralising antibodies. Sci. Rep. 2016, 6, 39730. [Google Scholar] [CrossRef] [Green Version]
- Matić, S.; Rinaldi, R.; Masenga, V.; Noris, E. Efficient production of chimeric Human papillomavirus 16 L1 protein bearing the M2e influenza epitope in Nicotiana benthamiana plants. BMC Biotechnol. 2011, 11, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thrane, S.; Janitzek, C.M.; Agerbæk, M.; Ditlev, S.B.; Resende, M.; Nielsen, M.A.; Theander, T.G.; Salanti, A.; Sander, A.F. A novel virus-like particle based vaccine platform displaying the placental malaria antigen VAR2CSA. PLoS ONE 2015, 10, e0143071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Castro, R.; Acero Galindo, G.; García Salcedo, Y.; Uribe Campero, L.; Vazquez Perez, V.; Carrillo-Tripp, M.; Gevorkian, G.; Gomez Lim, M.A. Plant-based chimeric HPV-virus-like particles bearing amyloid-β epitopes elicit antibodies able to recognize amyloid plaques in APP-tg mouse and Alzheimer’s disease brains. Inflammopharmacology 2018, 26, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, T.; Liu, H.; Hao, Y.; Liao, G.; Xu, X. Displaying 31RG-1 peptide on the surface of HPV16 L1 by use of a human papillomavirus chimeric virus-like particle induces cross-neutralizing antibody responses in mice. Hum. Vaccines Immunother. 2018, 14, 2025–2033. [Google Scholar] [CrossRef]
- Chen, X.S.; Garcea, R.L.; Goldberg, I.; Casini, G.; Harrison, S.C. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 2000, 5, 557–567. [Google Scholar] [CrossRef]
- Maclean, J.; Koekemoer, M.; Olivier, A.J.; Stewart, D.; Hitzeroth, I.I.; Rademacher, T.; Fischer, R.; Williamson, A.-L.L.; Rybicki, E.P. Optimization of human papillomavirus type 16 (HPV-16) L1 expression in plants: Comparison of the suitability of different HPV-16 L1 gene variants and different cell-compartment localization. J. Gen. Virol. 2007, 88, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Chu, K.B.; Kang, H.J.; Quan, F.S. Virus-like particles containing multiple antigenic proteins of Toxoplasma gondii induce memory T cell and B cell responses. PLoS ONE 2019, 14, e0220865. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, A.; Sun, X.; Sha, W.; Ai, K.; Pan, G.; Zhou, C.; Zhou, H.; Cong, H.; He, S. Immunogenicity of a virus-like-particle vaccine containing multiple antigenic epitopes of toxoplasma gondii against acute and chronic toxoplasmosis in mice. Front. Immunol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Lui, W.-Y. Transforming Growth Factor-β1 (TGF-β1) Regulates Cell Junction Restructuring via Smad-Mediated Repression and Clathrin-Mediated Endocytosis of Nectin-like Molecule 2 (Necl-2). PLoS ONE 2013, 8, e64316. [Google Scholar] [CrossRef] [Green Version]
- Larsen, J.E.P.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emini, E.A.; Hughes, J.V.; Perlow, D.S.; Boger, J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karplus, P.A.; Schulz, G.E. Prediction of chain flexibility in proteins. Naturwissenschaften 1985, 72, 212–213. [Google Scholar] [CrossRef]
- Parker, J.M.R.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and x-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Dunker, A.K. Natively disordered proteins: functions and predictions. Appl. Bioinform. 2004, 3, 105–113. [Google Scholar] [CrossRef]
- Keith Dunker, A.; Brown, C.J.; Obradovic, Z.B.T.-A. Identification and functions of usefully disordered proteins. In Unfolded Proteins; Academic Press: Cambridge, MA, USA, 2002; Volume 62, pp. 25–49. ISBN 0065-3233. [Google Scholar]
- Mészáros, B.; Erdős, G.; Dosztányi, Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
- Jones, D.T.; Cozzetto, D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinform. (Oxf. Engl.) 2015, 31, 857–863. [Google Scholar] [CrossRef]
- Linding, R.; Jensen, L.J.; Diella, F.; Bork, P.; Gibson, T.J.; Russell, R.B. Protein disorder prediction: implications for structural proteomics. Structure Lond. Engl. 1993 2003, 11, 1453–1459. [Google Scholar] [CrossRef] [Green Version]
- Källberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Martínez, J.A.; Gómez-Lim, M.A.; Morales-Ríos, E.; Gonzalez-y-Merchand, J.A.; Ortiz-Navarrete, V. Short Disordered Epitope of CRTAM Ig-Like V Domain as a Potential Target for Blocking Antibodies. Int. J. Mol. Sci. 2020, 21, 8798. https://doi.org/10.3390/ijms21228798
Vázquez-Martínez JA, Gómez-Lim MA, Morales-Ríos E, Gonzalez-y-Merchand JA, Ortiz-Navarrete V. Short Disordered Epitope of CRTAM Ig-Like V Domain as a Potential Target for Blocking Antibodies. International Journal of Molecular Sciences. 2020; 21(22):8798. https://doi.org/10.3390/ijms21228798
Chicago/Turabian StyleVázquez-Martínez, Julio Angel, Miguel Angel Gómez-Lim, Edgar Morales-Ríos, Jorge Alberto Gonzalez-y-Merchand, and Vianney Ortiz-Navarrete. 2020. "Short Disordered Epitope of CRTAM Ig-Like V Domain as a Potential Target for Blocking Antibodies" International Journal of Molecular Sciences 21, no. 22: 8798. https://doi.org/10.3390/ijms21228798
APA StyleVázquez-Martínez, J. A., Gómez-Lim, M. A., Morales-Ríos, E., Gonzalez-y-Merchand, J. A., & Ortiz-Navarrete, V. (2020). Short Disordered Epitope of CRTAM Ig-Like V Domain as a Potential Target for Blocking Antibodies. International Journal of Molecular Sciences, 21(22), 8798. https://doi.org/10.3390/ijms21228798






