Effective RNA Knockdown Using CRISPR-Cas13a and Molecular Targeting of the EML4-ALK Transcript in H3122 Lung Cancer Cells
Abstract
:1. Introduction
2. Results
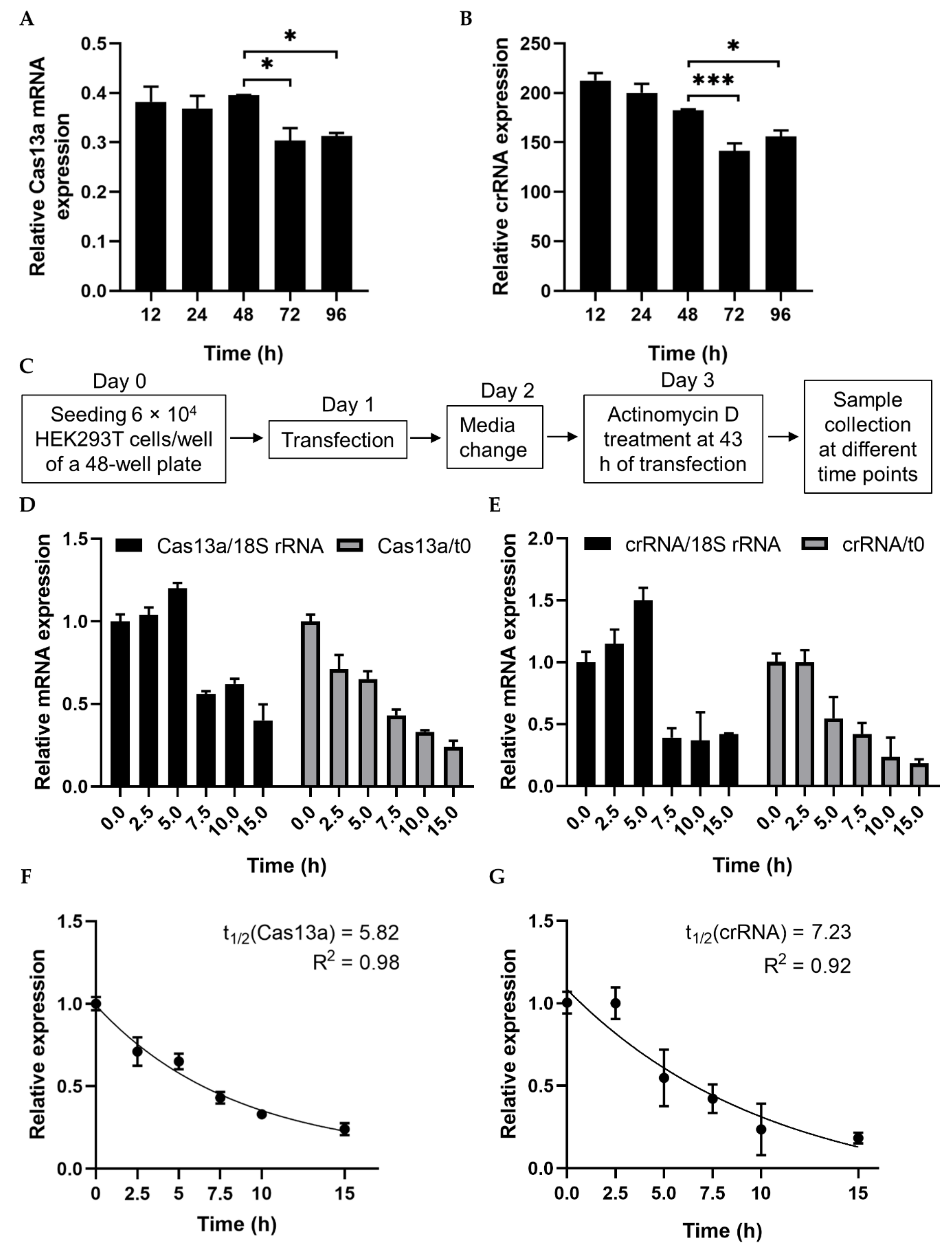
2.1. Analysis of RNA Stability
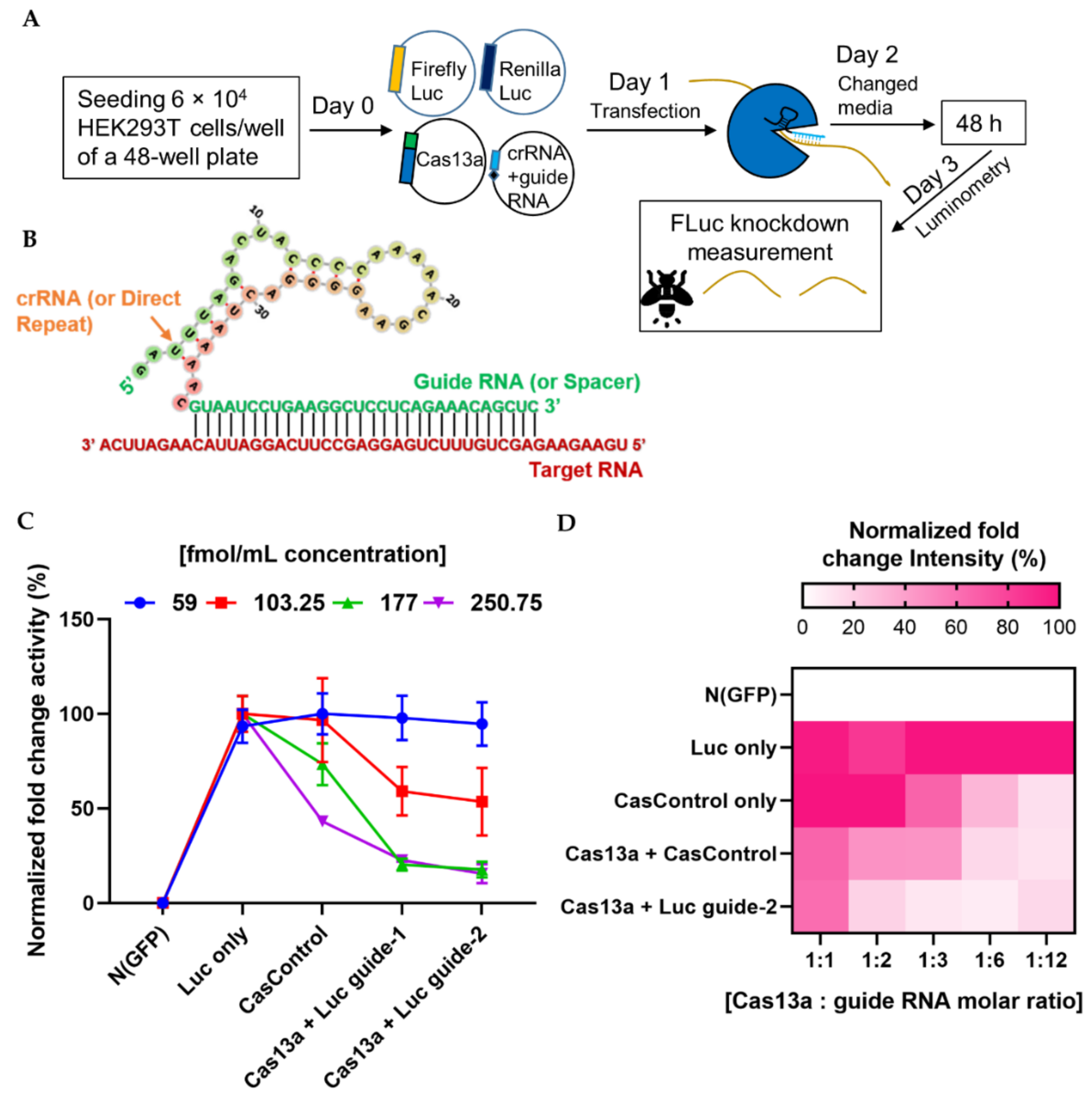
2.2. Optimizing the Quantity of Cas13a for Efficient RNA Knockdown
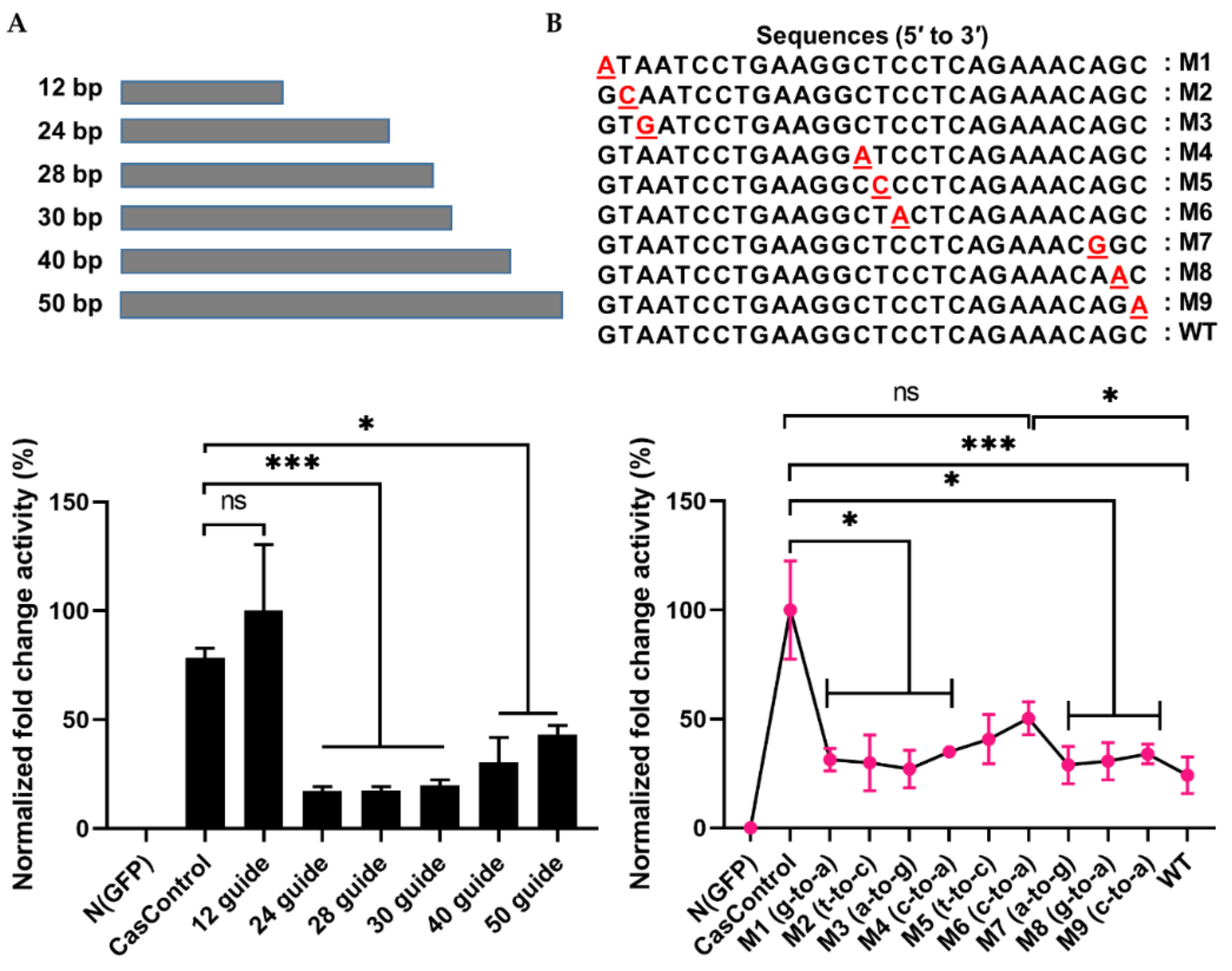
2.3. Optimal Condition for RNA Knockdown and Preference of 5′ crRNA for Cas13a Activity
2.4. Selectivity of Cas13a in Guide RNA Design
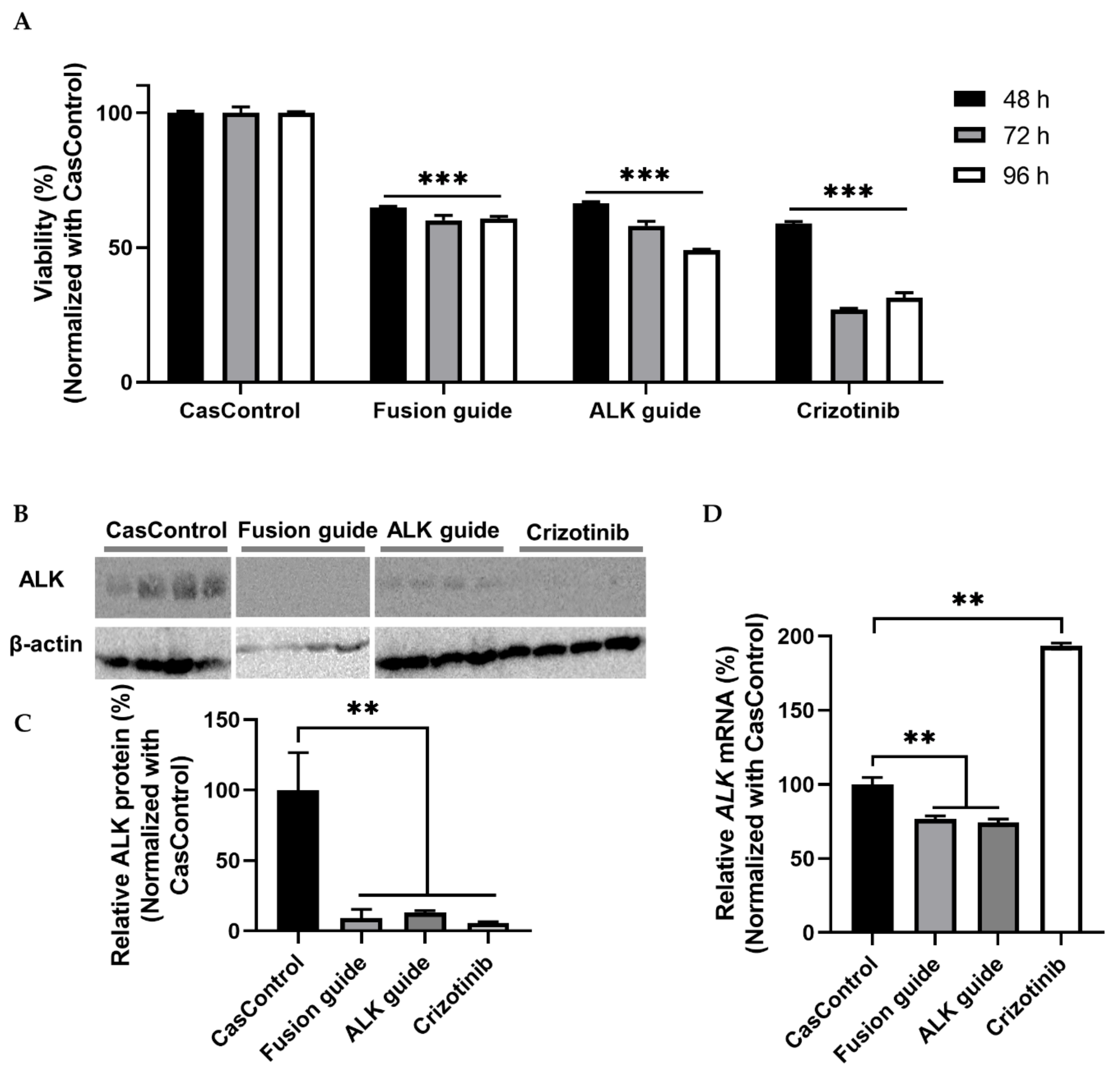
2.5. Validation of Effective Knockdown: mCherry and Molecular Targeting of EML4-ALK Oncofusion Genes
3. Discussion
4. Materials and Methods
4.1. Plasmid Vectors
4.2. Guide RNA Design
4.3. Sanger Sequencing
4.4. Cell Culture and Transfection
4.5. RNA Stability Assay
4.6. Total RNA Extraction
4.7. cDNA Synthesis
4.8. RT-PCR Quantification
4.9. SYBR Green-Based qPCR
4.10. Luciferase Luminometry Assay
4.11. MTT Assay and Western Blot Analyses
4.12. Bioinformatics and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| crRNA | CRISPR RNA |
| EML4-ALK | Echinoderm Microtubule-associated protein-like 4-Anaplastic Lymphoma Kinase |
| RNAi | Ribonucleic Acid interference |
| FLuc | Firefly Luciferase |
| ActD | Actinomycin D |
References
- Lieberman, J. Tapping the RNA world for therapeutics. Nat. Struct. Mol. Biol. 2018, 25, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Manfredsson, F.P.; Lewin, A.S.; Mandel, R.J. RNA knockdown as a potential therapeutic strategy in Parkinson’s disease. Gene Ther. 2006, 13, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Rossi, J.J. RNAi mechanisms and applications. BioTechniques 2008, 44, 613–616. [Google Scholar] [CrossRef]
- Mroweh, M.; Decaens, T.; Marche, P.; Jilkova, Z.M.; Clément, F. Modulating the Crosstalk between the Tumor and Its Microenvironment Using RNA Interference: A Treatment Strategy for Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 5250. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Rogge, M. TargetingRNA: A Transformative Therapeutic Strategy. Clin. Transl. Sci. 2019, 12, 98–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aagaard, L.; Rossi, J.J. RNAi therapeutics: Principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007, 59, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, Y.; Takeyama, Y.; Mizuno, T.; Satoh, F.; Asanuma, H. Investigation of Strand-Selective Interaction of SNA-Modified siRNA with AGO2-MID. Int. J. Mol. Sci. 2020, 21, 5218. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [Green Version]
- Filippov-Levy, N.; Reich, R.; Davidson, B. The Biological and Clinical Role of the Long Non-Coding RNA LOC642852 in Ovarian Carcinoma. Int. J. Mol. Sci. 2020, 21, 5237. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; East-Seletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R. Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR–Cas Systems. J. Mol. Biol. 2019, 431, 66–87. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [Green Version]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef] [Green Version]
- Burmistrz, M.; Krakowski, K.; Krawczyk-Balska, A. RNA-Targeting CRISPR–Cas Systems and Their Applications. Int. J. Mol. Sci. 2020, 21, 1122. [Google Scholar] [CrossRef] [Green Version]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.-I.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef]
- Sabir, S.R.; Yeoh, S.; Jackson, G.; Bayliss, R. EML4-ALK Variants: Biological and Molecular Properties, and the Implications for Patients. Cancers 2017, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Lin, E.; Li, L.; Guan, Y.; Soriano, R.; Rivers, C.S.; Mohan, S.; Pandita, A.; Tang, J.; Modrusan, Z. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol. Cancer Res. 2009, 7, 1466–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katayama, R.; Khan, T.M.; Benes, C.; Lifshits, E.; Ebi, H.; Rivera, V.M.; Shakespeare, W.C.; Iafrate, A.J.; Engelman, J.A.; Shaw, A.T. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc. Natl. Acad. Sci. USA 2011, 108, 7535–7540. [Google Scholar] [CrossRef] [Green Version]
- Zou, H.Y.; Li, Q.; Lee, J.H.; Arango, M.E.; McDonnell, S.R.; Yamazaki, S.; Koudriakova, T.B.; Alton, G.; Cui, J.J.; Kung, P.-P.; et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007, 67, 4408–4417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golding, B.; Luu, A.; Jones, R.; Viloria-Petit, A.M. The function and therapeutic targeting of anaplastic lymphoma kinase (ALK) in non-small cell lung cancer (NSCLC). Mol. Cancer 2018, 17, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuchipudi, S.V.; Tellabati, M.; Nelli, R.K.; White, G.A.; Perez, B.B.; Sebastian, S.; Slomka, M.J.; Brookes, S.M.; Brown, I.H.; Dunham, S.P.; et al. 18S rRNAis a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 2012, 9, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Liu, X.; Zhou, J.; Yang, C.; Wang, G.; Tan, Y.; Wu, Y.; Zhang, S.; Yi, K.; Kang, C. The CRISPR-Cas13a Gene-Editing System Induces Collateral Cleavage of RNA in Glioma Cells. Adv. Sci. 2019, 6, 1901299. [Google Scholar] [CrossRef] [Green Version]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630. [Google Scholar] [CrossRef] [Green Version]
- Hewes, A.M.; Sansbury, B.M.; Barth, S.; Tarcic, G.; Kmiec, E.B. gRNA Sequence Heterology Tolerance Catalyzed by CRISPR/Cas in an In Vitro Homology-Directed Repair Reaction. Mol. Ther. Nucleic Acids 2020, 20, 568–579. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Takezawa, K.; Okamoto, I.; Nishio, K.; Jänne, P.A.; Nakagawa, K. Role of ERK-BIM and STAT3-Survivin Signaling Pathways in ALK Inhibitor-Induced Apoptosis in EML4-ALK-Positive Lung Cancer. Clin. Cancer Res. 2011, 17, 2140–2148. [Google Scholar] [CrossRef] [Green Version]
- Katayama, R.; Shaw, A.T.; Khan, T.M.; Mino-Kenudson, M.; Solomon, B.J.; Halmos, B.; Jessop, N.A.; Wain, J.C.; Yeo, A.T.; Benes, C.; et al. Mechanisms of Acquired Crizotinib Resistance in ALK-Rearranged Lung Cancers. Sci. Transl. Med. 2012, 4, 120ra17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, R.; Wang, Y.; Voeller, D.; Gower, A.; Kim, I.-K.; Zhang, Y.-W.; Giaccone, G. CRKL mediates EML4-ALK signaling and is a potential therapeutic target for ALK-rearranged lung adenocarcinoma. Oncotarget 2016, 7, 29199–29210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrustanovic, G.; Olivas, V.; Pazarentzos, E.; Tulpule, A.; Asthana, S.; Blakely, C.M.; Okimoto, R.A.; Lin, L.; Neel, D.S.; Sabnis, A.J.; et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK–positive lung cancer. Nat. Med. 2015, 21, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Saifullah; Tsukahara, T. Genotyping of single nucleotide polymorphisms using the SNP-RFLP method. Biosci. Trends 2018, 12, 240–246. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, K.; Takeuchi, S.; Arai, S.; Katayama, R.; Nanjo, S.; Tanimoto, A.; Nishiyama, A.; Nakagawa, T.; Taniguchi, H.; Suzuki, T.; et al. Epithelial-to-Mesenchymal Transition Is a Mechanism of ALK Inhibitor Resistance in Lung Cancer Independent of ALK Mutation Status. Cancer Res. 2019, 79, 1658–1670. [Google Scholar] [CrossRef]
- Koivunen, J.P.; Mermel, C.; Zejnullahu, K.; Murphy, C.; Lifshits, E.; Holmes, A.J.; Choi, H.G.; Kim, J.; Chiang, D.; Thomas, R.; et al. EML4-ALK Fusion Gene and Efficacy of an ALK Kinase Inhibitor in Lung Cancer. Clin. Cancer Res. 2008, 14, 4275–4283. [Google Scholar] [CrossRef] [Green Version]
- Lugowski, A.; Nicholson, B.; Rissland, O.S. Determining mRNA half-lives on a transcriptome-wide scale. Methods 2018, 137, 90–98. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schagat, T.; Paguio, A.; Kopish, K. Normalizing genetic reporter assays: Approaches and considerations for increasing consistency and statistical significance. Cell Notes 2007, 17, 9–12. [Google Scholar]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Fusion | Copy Number | Cancer Type | Patients (n) |
|---|---|---|---|
| EML4-ALK | diploid | Lung adenocarcinoma | 103 |
| EML4-ALK | gain | Lung adenocarcinoma | 49 |
| EML4-ALK | diploid | Papillary renal cell carcinoma | 18 |
| EML4-ALK | shallowdel | Papillary thyroid cancer | 18 |
| STRN-ALK | shallowdel | Papillary renal cell carcinoma | 44 |
| STRN-ALK | deepdel | Papillary thyroid cancer | 7 |
| TPM1-ALK | diploid | Bladder urothelial carcinoma | 89 |
| PPP4R3B-ALK | shallowdel | Rectal adenocarcinoma | 136 |
| ACTG2-ALK | amp | Leiomyosarcoma | 51 |
| ALK-STK39 | shallowdel | Cutaneous melanoma | 171 |
| KCNQ5-ALK | Cutaneous melanoma | 506 | |
| MALAT1-ALK | diploid | Papillary thyroid cancer | 31 |
| GTF2IRD1-ALK | diploid | Papillary thyroid cancer | 12 |
| ALK-GALNT14 | gain | Uterine serous carcinoma/uterine papillary serous carcinoma | 124 |
| ALK-SCEL | gain | Uterine endometrioid carcinoma | 156 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saifullah; Sakari, M.; Suzuki, T.; Yano, S.; Tsukahara, T. Effective RNA Knockdown Using CRISPR-Cas13a and Molecular Targeting of the EML4-ALK Transcript in H3122 Lung Cancer Cells. Int. J. Mol. Sci. 2020, 21, 8904. https://doi.org/10.3390/ijms21238904
Saifullah, Sakari M, Suzuki T, Yano S, Tsukahara T. Effective RNA Knockdown Using CRISPR-Cas13a and Molecular Targeting of the EML4-ALK Transcript in H3122 Lung Cancer Cells. International Journal of Molecular Sciences. 2020; 21(23):8904. https://doi.org/10.3390/ijms21238904
Chicago/Turabian StyleSaifullah, Matomo Sakari, Takeshi Suzuki, Seiji Yano, and Toshifumi Tsukahara. 2020. "Effective RNA Knockdown Using CRISPR-Cas13a and Molecular Targeting of the EML4-ALK Transcript in H3122 Lung Cancer Cells" International Journal of Molecular Sciences 21, no. 23: 8904. https://doi.org/10.3390/ijms21238904
APA StyleSaifullah, Sakari, M., Suzuki, T., Yano, S., & Tsukahara, T. (2020). Effective RNA Knockdown Using CRISPR-Cas13a and Molecular Targeting of the EML4-ALK Transcript in H3122 Lung Cancer Cells. International Journal of Molecular Sciences, 21(23), 8904. https://doi.org/10.3390/ijms21238904







