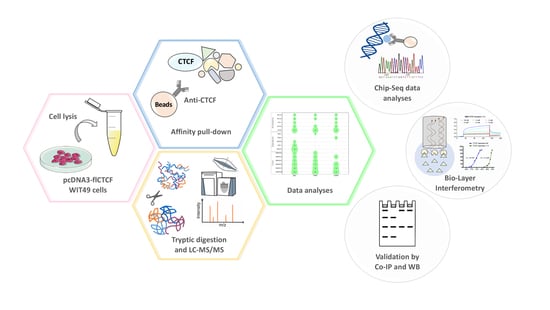
Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF
Abstract
:1. Introduction
2. Results
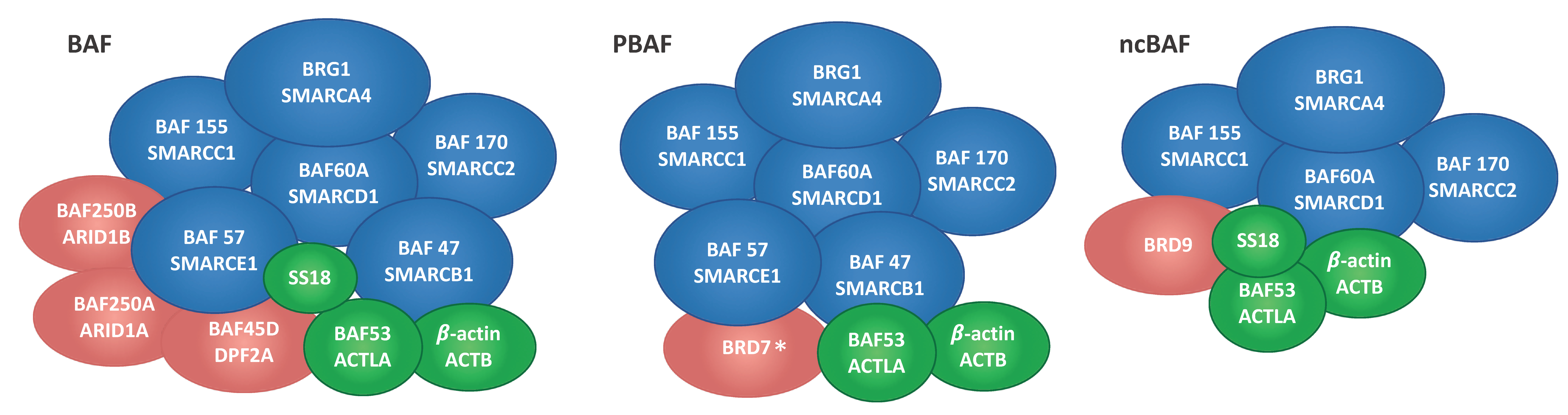
2.1. Immunoprecipitation and Identification of CCCTC-Binding Factor (CTCF)-Interacting Members of the Switch/Sucrose Nonfermentable (SWI/SNF) Complex
2.2. Validation by Western Blot
2.3. Genomic Co-Occupancy by CTCF and SWI/SNF Subunits
2.4. CTCF Directly Interacts with the BRK Domain of BRG1
3. Discussion
4. Materials and Methods
4.1. Sample Preparation for Mass Spectrometry Analysis
4.2. High Resolution Nano-LC−Tandem Mass Spectrometry
4.3. MS Data Processing
4.4. Bioinformatic Analyses
4.5. Validations by Western Blot Analyses
4.6. ChIP-seq Data Analysis
4.7. Chemical Synthesis of the BRK Domain of Human BRG1
4.8. Recombinant CTCF Zinc Finger Domains 1–11 and 4–8 Expression and Purification
4.9. Binding of BRK-BRG1 to Recombinant CTFC Multiple Zinc Finger Domains by Biolayer Interferometry (BLI)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AP-MS | Affinity-purification mass spectrometry |
| BAF | BRG1/BRM associated factor |
| CTCF | CCCTC-binding factor |
| CHD | Chromodomain helicase DNA-binding |
| IP | Immunoprecipitation |
| ncBAF | Non-canonical BAF |
| PBAF | Polybromo-associated BAF |
| PSM | Peptide-to-spectra matches |
| SWI/SNF | Switch/sucrose nonfermentable |
| TADs | Topological associated domains |
References
- Ghirlando, R.; Felsenfeld, G. CTCF: Making the right connections. Genes Dev. 2016, 30, 881–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, J.A.; Felsenfeld, G. We gather together: Insulators and genome organization. Curr. Opin. Genet. Dev. 2007, 17, 400–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlatanova, J.; Caiafa, P. CTCF and its protein partners: Divide and rule? J. Cell Sci. 2009, 122 Pt 9, 1275–1284. [Google Scholar] [CrossRef] [Green Version]
- Holwerda, S.J.; de Laat, W. CTCF: The protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusufzai, T.M.; Tagami, H.; Nakatani, Y.; Felsenfeld, G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 2004, 13, 291–298. [Google Scholar] [CrossRef]
- Chernukhin, I.V.; Shamsuddin, S.; Robinson, A.F.; Carne, A.F.; Paul, A.; El-Kady, A.I.; Lobanenkov, V.V.; Klenova, E.M. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 2000, 275, 29915–29921. [Google Scholar] [CrossRef] [Green Version]
- Donohoe, M.E.; Zhang, L.F.; Xu, N.; Shi, Y.; Lee, J.T. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell 2007, 25, 43–56. [Google Scholar] [CrossRef]
- Defossez, P.A.; Kelly, K.F.; Filion, G.J.; Perez-Torrado, R.; Magdinier, F.; Menoni, H.; Nordgaard, C.L.; Daniel, J.M.; Gilson, E. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J. Biol. Chem. 2005, 280, 43017–43023. [Google Scholar] [CrossRef] [Green Version]
- De La Rosa-Velazquez, I.A.; Rincon-Arano, H.; Benitez-Bribiesca, L.; Recillas-Targa, F. Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res. 2007, 67, 2577–2585. [Google Scholar] [CrossRef] [Green Version]
- Majumder, P.; Gomez, J.A.; Boss, J.M. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J. Biol. Chem. 2006, 281, 18435–18443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, P.; Gomez, J.A.; Chadwick, B.P.; Boss, J.M. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 2008, 205, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Chernukhin, I.; Shamsuddin, S.; Kang, S.Y.; Bergstrom, R.; Kwon, Y.W.; Yu, W.; Whitehead, J.; Mukhopadhyay, R.; Docquier, F.; Farrar, D.; et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol. Cell. Biol. 2007, 27, 1631–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishihara, K.; Oshimura, M.; Nakao, M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell 2006, 23, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Religa, T.L.; Freund, S.M.; Bycroft, M. Solution structure of the BRK domains from CHD7. J. Mol. Biol. 2007, 371, 1135–1140. [Google Scholar] [CrossRef]
- Marino, M.M.; Rega, C.; Russo, R.; Valletta, M.; Gentile, M.T.; Esposito, S.; Baglivo, I.; De Feis, I.; Angelini, C.; Xiao, T.; et al. Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF. J. Biol. Chem. 2019, 294, 861–873. [Google Scholar] [CrossRef] [Green Version]
- Trotter, K.W.; Archer, T.K. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 2008, 6, e004. [Google Scholar] [CrossRef] [Green Version]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [Green Version]
- Mashtalir, N.; D’Avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; McKenzie, Z.M.; Kubiak, R.L.; St Pierre, R.; et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175, 1272–1288.e20. [Google Scholar] [CrossRef] [Green Version]
- Allen, M.D.; Bycroft, M.; Zinzalla, G. Structure of the BRK domain of the SWI/SNF chromatin remodeling complex subunit BRG1 reveals a potential role in protein-protein interactions. Protein Sci. 2020, 29, 1047–1053. [Google Scholar] [CrossRef]
- Ong, C.T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoury, A.; Achinger-Kawecka, J.; Bert, S.A.; Smith, G.C.; French, H.J.; Luu, P.L.; Peters, T.J.; Du, Q.; Parry, A.J.; Valdes-Mora, F.; et al. Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains. Nat. Commun. 2020, 11, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichols, M.H.; Corces, V.G. A CTCF Code for 3D Genome Architecture. Cell 2015, 162, 703–705. [Google Scholar] [CrossRef] [Green Version]
- Braccioli, L.; de Wit, E. CTCF: A Swiss-army knife for genome organization and transcription regulation. Essays Biochem. 2019, 63, 157–165. [Google Scholar]
- Barutcu, A.R.; Lian, J.B.; Stein, J.L.; Stein, G.S.; Imbalzano, A.N. The connection between BRG1, CTCF and topoisomerases at TAD boundaries. Nucleus (Austin Tex.) 2017, 8, 150–155. [Google Scholar] [CrossRef] [Green Version]
- Pugacheva, E.M.; Kubo, N.; Loukinov, D.; Tajmul, M.; Kang, S.; Kovalchuk, A.L.; Strunnikov, A.V.; Zentner, G.E.; Ren, B.; Lobanenkov, V.V. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. USA 2020, 117, 2020–2031. [Google Scholar] [CrossRef] [Green Version]
- Hansen, A.S. CTCF as a boundary factor for cohesin-mediated loop extrusion: Evidence for a multi-step mechanism. Nucleus (Austin Tex.) 2020, 11, 132–148. [Google Scholar] [CrossRef]
- Hansen, A.S.; Pustova, I.; Cattoglio, C.; Tjian, R.; Darzacq, X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 2017, 6, e25776. [Google Scholar] [CrossRef]
- Euskirchen, G.M.; Auerbach, R.K.; Davidov, E.; Gianoulis, T.A.; Zhong, G.N.; Rozowsky, J.; Bhardwaj, N.; Gerstein, M.B.; Snyder, M. Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches. PLoS Genet. 2011, 7, e1002008. [Google Scholar] [CrossRef] [Green Version]
- Michel, B.C.; D’Avino, A.R.; Cassel, S.H.; Mashtalir, N.; McKenzie, Z.M.; McBride, M.J.; Valencia, A.M.; Zhou, Q.; Bocker, M.; Soares, L.M.M.; et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 2018, 20, 1410–1420. [Google Scholar] [CrossRef]
- Barutcu, A.R.; Lajoie, B.R.; Fritz, A.J.; McCord, R.P.; Nickerson, J.A.; van Wijnen, A.J.; Lian, J.B.; Stein, J.L.; Dekker, J.; Stein, G.S.; et al. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016, 26, 1188–1201. [Google Scholar] [CrossRef] [Green Version]
- Ross-Innes, C.S.; Brown, G.D.; Carroll, J.S. A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genom. 2011, 12, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolstorukov, M.Y.; Sansam, C.G.; Lu, P.; Koellhoffer, E.C.; Helming, K.C.; Alver, B.H.; Tillman, E.J.; Evans, J.A.; Wilson, B.G.; Park, P.J.; et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. USA 2013, 110, 10165–10170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.H.; Murphy, M.R.; Lee, J.S.; Chung, J.H. Targeting a SWI/SNF-related chromatin remodeling complex to the beta-globin promoter in erythroid cells. Proc. Natl. Acad. Sci. USA 1999, 96, 12311–12315. [Google Scholar] [CrossRef] [Green Version]
- Peterson, C.L.; Workman, J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000, 10, 187–192. [Google Scholar] [CrossRef]
- Bracken, A.P.; Brien, G.L.; Verrijzer, C.P. Dangerous liaisons: Interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev. 2019, 33, 936–959. [Google Scholar] [CrossRef]
- Cote, J.; Quinn, J.; Workman, J.L.; Peterson, C.L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 1994, 265, 53–60. [Google Scholar] [CrossRef]
- Doerks, T.; Copley, R.R.; Schultz, J.; Ponting, C.P.; Bork, P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002, 12, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Renda, M.; Baglivo, I.; Burgess-Beusse, B.; Esposito, S.; Fattorusso, R.; Felsenfeld, G.; Pedone, P.V. Critical DNA binding interactions of the insulator protein CTCF: A small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 2007, 282, 33336–33345. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, H.; Wang, D.; Horton, J.R.; Zhang, X.; Corces, V.G.; Cheng, X. Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol. Cell 2017, 66, 711–720.e3. [Google Scholar] [CrossRef] [Green Version]
- Yin, M.; Wang, J.; Wang, M.; Li, X.; Zhang, M.; Wu, Q.; Wang, Y. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res. 2017, 27, 1365–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, J.; Crossley, M. Mammalian Kruppel-like transcription factors: More than just a pretty finger. Trends Biochem. Sci. 1999, 24, 236–240. [Google Scholar] [CrossRef]
- Lutz, M.; Burke, L.J.; Barreto, G.; Goeman, F.; Greb, H.; Arnold, R.; Schultheiss, H.; Brehm, A.; Kouzarides, T.; Lobanenkov, V.; et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000, 28, 1707–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alipour, E.; Marko, J.F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012, 40, 11202–11212. [Google Scholar] [CrossRef] [Green Version]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [Green Version]
- Haarhuis, J.H.I.; van der Weide, R.H.; Blomen, V.A.; Yanez-Cuna, J.O.; Amendola, M.; van Ruiten, M.S.; Krijger, P.H.L.; Teunissen, H.; Medema, R.H.; van Steensel, B.; et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 2017, 169, 693–707.e14. [Google Scholar] [CrossRef] [Green Version]
- Sanborn, A.L.; Rao, S.S.; Huang, S.C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef] [Green Version]
- Dekker, J.; Mirny, L. The 3D Genome as Moderator of Chromosomal Communication. Cell 2016, 164, 1110–1121. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Haarhuis, J.H.I.; Sedeno Cacciatore, A.; Oldenkamp, R.; van Ruiten, M.S.; Willems, L.; Teunissen, H.; Muir, K.W.; de Wit, E.; Rowland, B.D.; et al. The structural basis for cohesin-CTCF-anchored loops. Nature 2020, 578, 472–476. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 2010, 604, 55–71. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Cuddapah, S.; Jothi, R.; Schones, D.E.; Roh, T.Y.; Cui, K.; Zhao, K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009, 19, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Caporale, A.; Doti, N.; Monti, A.; Sandomenico, A.; Ruvo, M. Automatic procedures for the synthesis of difficult peptides using oxyma as activating reagent: A comparative study on the use of bases and on different deprotection and agitation conditions. Peptides 2018, 102, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Caporale, A.; Doti, N.; Sandomenico, A.; Ruvo, M. Evaluation of combined use of Oxyma and HATU in aggregating peptide sequences. J. Pept. Sci. 2017, 23, 272–281. [Google Scholar] [CrossRef] [PubMed]





| Benzonase | Untreated | ||||||
|---|---|---|---|---|---|---|---|
| Accession | Description | Name | # Peptides | # PSMs | # Peptides | # PSMs | MW (kDa) |
| P51532 | Transcription activator BRG1 | BRG1 | 30 | 47 | 29 | 48 | 185 |
| Q92922 | SWI/SNF complex subunit SMARCC1 | BAF155 | 19 | 36 | 21 | 29 | 123 |
| Q8TAQ2 | SWI/SNF complex subunit SMARCC2 | BAF170 | 16 | 25 | 15 | 19 | 133 |
| Q969G3 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 | BAF57 | 10 | 17 | 10 | 17 | 47 |
| Q96GM5 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 | BAF60A | 9 | 13 | 10 | 15 | 58 |
| Q12824 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | BAF47 | 6 | 9 | 9 | 11 | 44 |
| O14497 | AT-rich interactive domain-containing protein 1A | BAF250A | 33 | 57 | 29 | 50 | 242 |
| Q92785 | Zinc finger protein ubi-d4 | BAF45D | 10 | 15 | 10 | 15 | 44 |
| Q8NFD5 | AT-rich interactive domain-containing protein 1B | BAF250B | 2 | 2 | 3 | 3 | 236 |
| Q9H8M2 | Bromodomain-containing protein 9 | BRD9 | 1 | 1 | 3 | 3 | 67 |
| Q9NPI1 | Bromodomain-containing protein 7 * | BRD7 | 1 | 4 | 1 | 3 | 74 |
| P60709 | Actin, cytoplasmic 1 | ACTB | 19 | 78 | 12 | 33 | 42 |
| O96019 | Actin-like protein 6A | BAF53A | 4 | 7 | 5 | 7 | 47 |
| Q15532 | Protein SSXT | SS18 | 2 | 4 | 2 | 3 | 46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valletta, M.; Russo, R.; Baglivo, I.; Russo, V.; Ragucci, S.; Sandomenico, A.; Iaccarino, E.; Ruvo, M.; De Feis, I.; Angelini, C.; et al. Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF. Int. J. Mol. Sci. 2020, 21, 8950. https://doi.org/10.3390/ijms21238950
Valletta M, Russo R, Baglivo I, Russo V, Ragucci S, Sandomenico A, Iaccarino E, Ruvo M, De Feis I, Angelini C, et al. Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF. International Journal of Molecular Sciences. 2020; 21(23):8950. https://doi.org/10.3390/ijms21238950
Chicago/Turabian StyleValletta, Mariangela, Rosita Russo, Ilaria Baglivo, Veronica Russo, Sara Ragucci, Annamaria Sandomenico, Emanuela Iaccarino, Menotti Ruvo, Italia De Feis, Claudia Angelini, and et al. 2020. "Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF" International Journal of Molecular Sciences 21, no. 23: 8950. https://doi.org/10.3390/ijms21238950
APA StyleValletta, M., Russo, R., Baglivo, I., Russo, V., Ragucci, S., Sandomenico, A., Iaccarino, E., Ruvo, M., De Feis, I., Angelini, C., Iachettini, S., Biroccio, A., Pedone, P. V., & Chambery, A. (2020). Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF. International Journal of Molecular Sciences, 21(23), 8950. https://doi.org/10.3390/ijms21238950