Functional Comparison of Blood-Derived Human Neural Progenitor Cells
Abstract
:1. Introduction
2. Results
2.1. Donor Selection and NPC Generation
2.2. Characterization of NPCs Generated by Different Methods
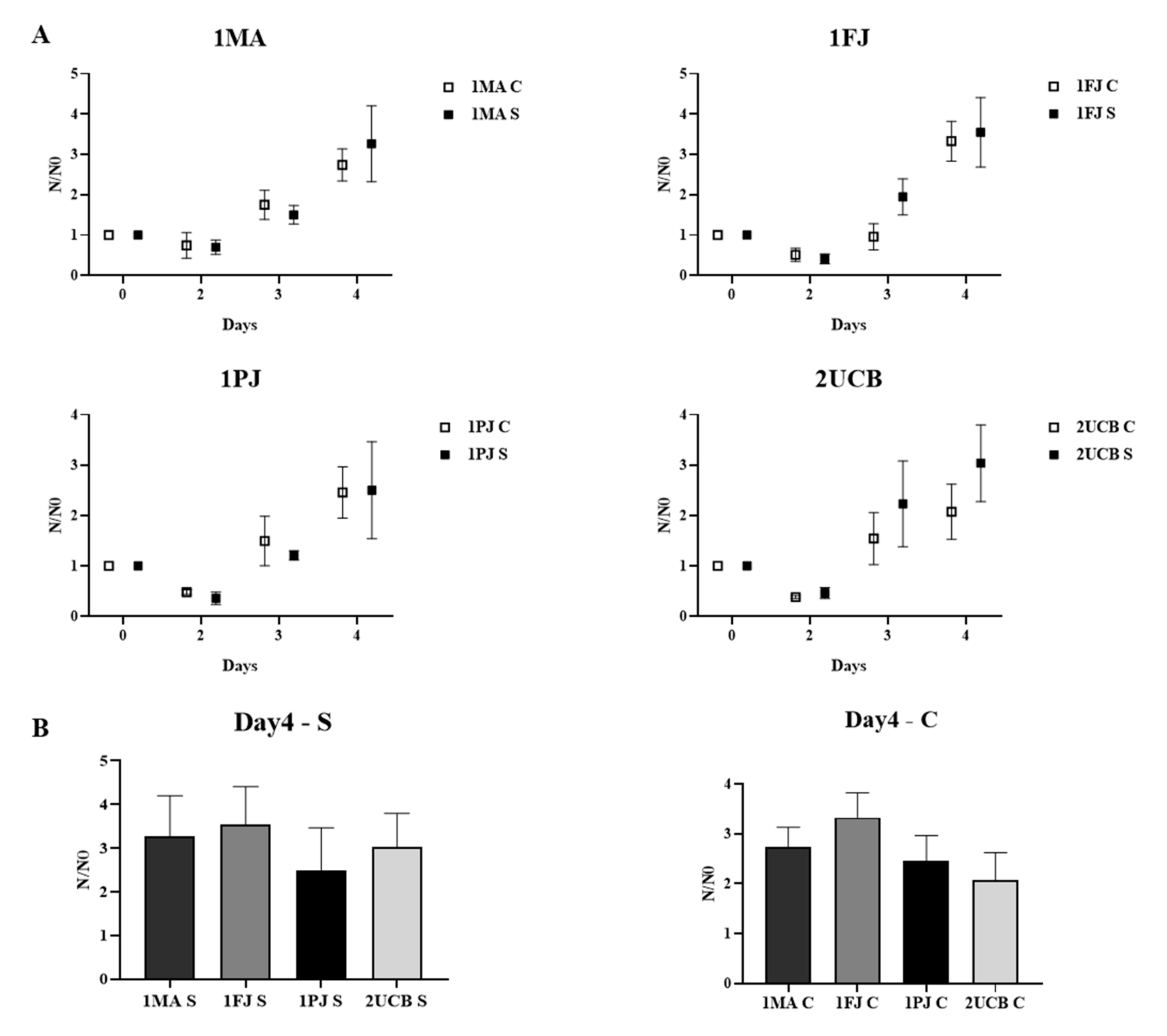
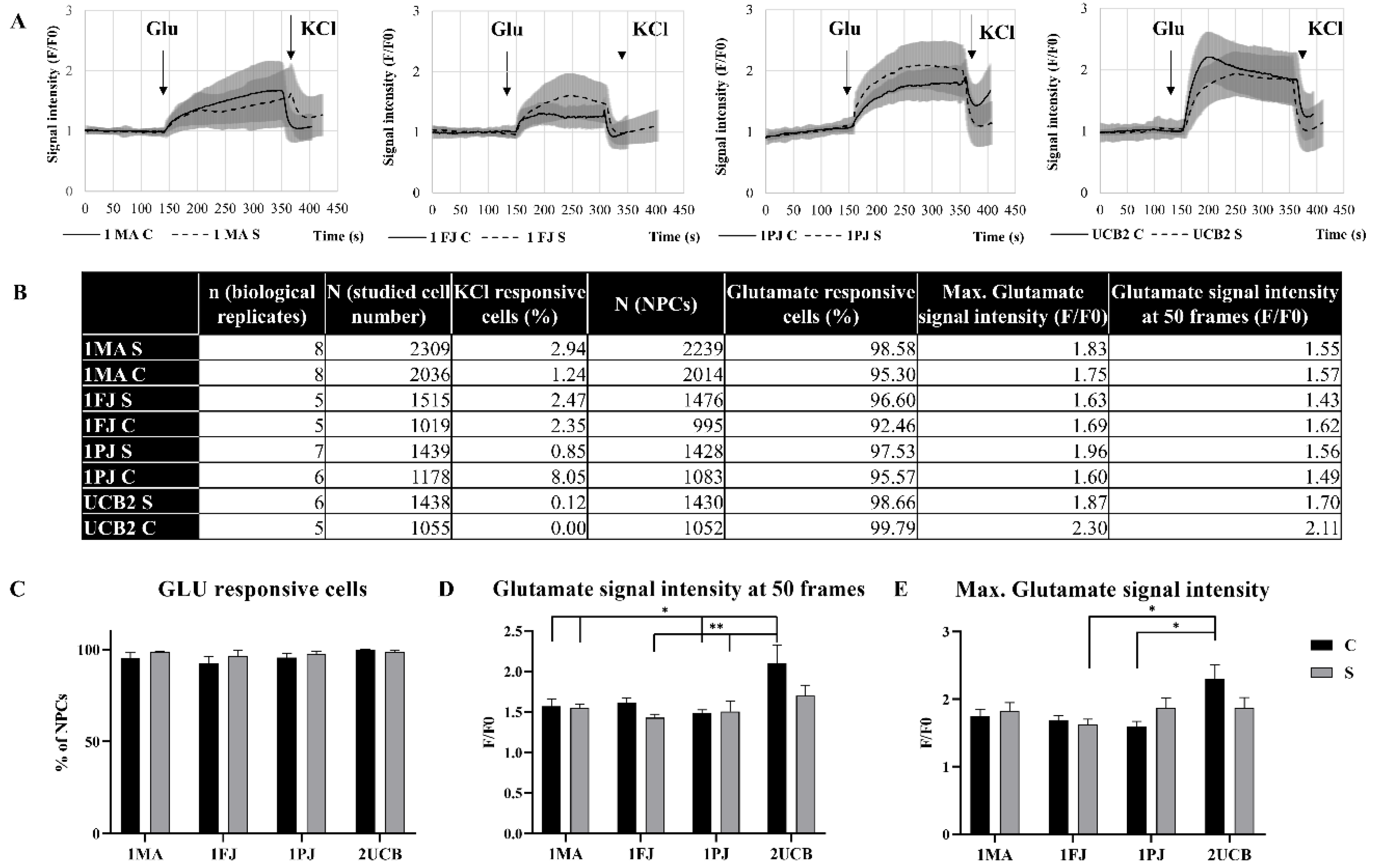
2.3. Functional Comparison of Classic and Shortcut NPCs
3. Discussion
4. Materials and Methods
4.1. Cell Culturing and Differentiation
4.2. Immunocytochemical Staining
4.3. Gene Expression Analysis
4.4. Growth Curve Measurements
4.5. Scratch Test
4.6. ROS Test
4.7. Mitotracker Assay
4.8. Calcium Signal Measurements
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Ascl1 | Achaete-scute homolog 1 |
| BDNF | Brain-Derived Neurotrophic Factor |
| Brn2 | Brain2 |
| BSA | Bovine serum albumin |
| CASD | Cell-activation and signalling-directed lineage conversion |
| c-Myc | Avian myelocytomatosis virus oncogene cellular homolog |
| cNPCs | Classical NPC |
| CNS | Central nervous system |
| DAPI | 2-(4-amidinophenyl)-1H-indole-6-carboxamidine |
| DG | Dentate gyrus |
| DKK1 | Dickkopf-related protein 1 |
| DPBS | Dulbecco’s modified PBS |
| FGF2 | Fibroblast growth factor 2 |
| FOXG1 | Forkhead box protein G1 |
| HBSS | Hanks’ balanced salt solution |
| hPSC | Human pluripotent stem cell |
| iN | Induced neurons |
| iPSC | Induced pluripotent stem cell |
| Klf4 | Kruppel-like factor 4 |
| MAP2 | Microtubule-associated protein 2 |
| MEF | Mouse embryonic fibroblasts |
| Myt1l | Myelin transcription factor 1 |
| NPCs | Neural progenitor cells |
| Oct3/4 | Octamer-binding transcription factor 3/4 |
| PAX6 | Paired box protein 6 |
| PBMC | Peripheral blood mononuclear cells |
| PFA | Paraformaldehyde |
| ROS | Reactive oxygen species |
| RPLP0 | Ribosomal Protein Lateral Stalk Subunit P0 |
| RT-PCR | Real-time quantitative PCR |
| SCZ | Schizophrenia |
| sNPC | Shortcut NPC |
| SOX2 | SRY (sex determining region Y)-box 2 |
| UCB | Umbilical cord blood |
References
- Bayart, E. Technological Overview of iPS Induction from Human Adult Somatic Cells. Curr. Gene Ther. 2013, 13, 73–92. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Ang, C.E.; Chanda, S.; Olmos, V.H.; Haag, D.; Levinson, D.F.; Sudhof, T.C.; Wernig, M. Transdifferentiation of human adult peripheral blood T cells into neurons. Proc. Natl. Acad. Sci. USA 2018, 115, 6470–64753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nat. Cell Biol. 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Drouin-Ouellet, J.; Pircs, K.; Barker, R.A.; Jakobsson, J.; Parmar, M. Direct Neuronal Reprogramming for Disease Modeling Studies Using Patient-Derived Neurons: What Have We Learned? Front. Neurosci. 2017, 11, 530. [Google Scholar] [CrossRef] [Green Version]
- Hussein, S.M.I.; Puri, M.C.; Tonge, P.D.; Benevento, M.; Corso, A.J.; Clancy, J.L.; Mosbergen, R.; Li, M.; Lee, D.-S.; Cloonan, N.; et al. Genome-wide characterization of the routes to pluripotency. Nat. Cell Biol. 2014, 516, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Flitsch, L.J.; Brüstle, O. Evolving principles underlying neural lineage conversion and their relevance for biomedical translation. F1000Research 2019, 8, 1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Wang, S.; Bai, Y.; Wu, J.; Fu, L.; Li, M.; Xu, Q.; Xu, Z.-Q.D.; Zhang, Y.; Chen, Z. Conversion of adult human peripheral blood mononuclear cells into induced neural stem cell by using episomal vectors. Stem Cell Res. 2016, 16, 236–242. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Choi, E.; Monaco, M.C.G.; Major, E.O.; Medynets, M.; Nath, A. Direct Induction of Human Neural Stem Cells from Peripheral Blood Hematopoietic Progenitor Cells. J. Vis. Exp. 2015, 95, e52298. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.; Huang, N.; Yu, J.; Jares, A.; Yang, J.; Zieve, G.; Avila, C.; Jiang, X.; Zhang, X.-B.; Ma, Y. Direct Conversion of Cord Blood CD34+ Cells Into Neural Stem Cells by OCT4. Stem Cells Transl. Med. 2015, 4, 755–763. [Google Scholar] [CrossRef]
- Haridhasapavalan, K.K.; Borgohain, M.P.; Dey, C.; Saha, B.; Narayan, G.; Kumar, S.; Thummer, R.P. An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene 2019, 686, 146–159. [Google Scholar] [CrossRef]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; Devine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, R.; Weinberger, D.R. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 2017, 18, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, D.R. Implications of Normal Brain Development for the Pathogenesis of Schizophrenia. Arch. Gen. Psychiatry 1987, 44, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Moslem, M.; Olive, J.; Falk, A. Stem cell models of schizophrenia, what have we learned and what is the potential? Schizophr. Res. 2019, 210, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Ardhanareeswaran, K.; Mariani, J.; Coppola, G.; Abyzov, A.; Vaccarino, F.M. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat. Rev. Neurol. 2017, 13, 265–278. [Google Scholar] [CrossRef]
- Hoffmann, A.; Ziller, M.; Spengler, D. Progress in iPSC-Based Modeling of Psychiatric Disorders. Int. J. Mol. Sci. 2019, 20, 4896. [Google Scholar] [CrossRef] [Green Version]
- Murai, K.; Sun, G.; Ye, P.; Tian, E.; Yang, S.; Cui, Q.; Sun, G.; Trinh, D.; Sun, O.; Hong, T.; et al. The TLX-miR-219 cascade regulates neural stem cell proliferation in neurodevelopment and schizophrenia iPSC model. Nat. Commun. 2016, 7, 10965. [Google Scholar] [CrossRef] [Green Version]
- Srikanth, P.; Han, K.; Callahan, D.G.; Makovkina, E.; Muratore, C.R.; Lalli, M.A.; Zhou, H.; Boyd, J.D.; Kosik, K.S.; Selkoe, D.J.; et al. Genomic DISC1 Disruption in hiPSCs Alters Wnt Signaling and Neural Cell Fate. Cell Rep. 2015, 12, 1414–1429. [Google Scholar] [CrossRef] [Green Version]
- Topol, A.; Zhu, S.; Tran, N.; Simone, A.; Fang, G.; Brennand, K.J. Altered WNT Signaling in Human Induced Pluripotent Stem Cell Neural Progenitor Cells Derived from Four Schizophrenia Patients. Biol. Psychiatry 2015, 78, e29–e34. [Google Scholar] [CrossRef] [Green Version]
- Yoon, K.-J.; Nguyen, H.N.; Ursini, G.; Zhang, F.; Kim, N.-S.; Wen, Z.; Makri, G.; Nauen, D.; Shin, J.H.; Park, Y.; et al. Modeling a Genetic Risk for Schizophrenia in iPSCs and Mice Reveals Neural Stem Cell Deficits Associated with Adherens Junctions and Polarity. Cell Stem Cell 2014, 15, 79–91. [Google Scholar] [CrossRef] [Green Version]
- Brennand, K.J.; Savas, J.N.; Kim, Y.; Tran, N.; Di Simone, A.; Hashimototorii, K.; Beaumont, K.G.; Kim, H.J.; Topol, A.; Ladran, I.; et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry 2015, 20, 361–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.X.; Di Giorgio, F.P.; Yao, J.; Marchetto, M.C.; Brennand, K.; Wright, R.; Mei, A.; McHenry, L.; Lisuk, D.; Grasmick, J.M.; et al. Modeling Hippocampal Neurogenesis Using Human Pluripotent Stem Cells. Stem Cell Rep. 2014, 2, 295–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vőfély, G.; Berecz, T.; Szabó, E.; Szebényi, K.; Hathy, E.; Orbán, T.I.; Sarkadi, B.; Homolya, L.; Marchetto, M.C.; Réthelyi, J.; et al. Characterization of calcium signals in human induced pluripotent stem cell-derived dentate gyrus neuronal progenitors and mature neurons, stably expressing an advanced calcium indicator protein. Mol. Cell. Neurosci. 2018, 88, 222–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hathy, E.; Szabó, E.; Vincze, K.; Haltrich, I.; Kiss, E.; Varga, N.; et al. Generation of 1103 multiple iPSC clones from a male schizophrenia patient carrying de novo 1104 mutations in genes KHSRP, LRRC7, and KIR2DL1, and his parents. Stem Cell Res. 2020. submitted, under revision. [Google Scholar]
- Mulligan, K.A.; Cheyette, B.N. Neurodevelopmental Perspectives on Wnt Signaling in Psychiatry. Mol. Neuropsychiatry 2017, 2, 219–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cory, G. Scratch-Wound Assay. Methods Mol. Biol. 2011, 769, 25–30. [Google Scholar] [CrossRef]
- Goff, D.C.; Coyle, J.T. The Emerging Role of Glutamate in the Pathophysiology and Treatment of Schizophrenia. Am. J. Psychiatry 2001, 158, 1367–1377. [Google Scholar] [CrossRef]
- Dauvermann, M.R.; Lee, G.; Dawson, N. Glutamatergic regulation of cognition and functional brain connectivity: Insights from pharmacological, genetic and translational schizophrenia research. Br. J. Pharmacol. 2017, 174, 3136–3160. [Google Scholar] [CrossRef]
- Suzuki, I.K.; Vanderhaeghen, P. Is this a brain which I see before me? Modeling human neural development with pluripotent stem cells. Development 2015, 142, 3138–3150. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-N.; Lu, S.-Y.; Yao, J. Application of induced pluripotent stem cells to understand neurobiological basis of bipolar disorder and schizophrenia. Psychiatry Clin. Neurosci. 2017, 71, 579–599. [Google Scholar] [CrossRef] [Green Version]
- Readhead, B.; Hartley, B.J.; Eastwood, B.J.; Collier, D.A.; Evans, D.; Farias, R.; He, C.; Hoffman, G.E.; Sklar, P.; Dudley, J.T.; et al. Expression-based drug screening of neural progenitor cells from individuals with schizophrenia. Nat. Commun. 2018, 9, 4412. [Google Scholar] [CrossRef] [PubMed]
- Little, D.; Ketteler, R.; Gissen, P.; Devine, M.J. Using stem cell-derived neurons in drug screening for neurological diseases. Neurobiol. Aging 2019, 78, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.J.; Maddalena, L.A.; Robb, E.L.; Moradi, F.; Stuart, J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017, 119, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Péntek, A.; Pászty, K.; Apáti, A. Analysis of Intracellular Calcium Signaling in Human Embryonic Stem Cells. Methods Mol. Biol. 2014, 1307, 141–147. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szabó, E.; Juhász, F.; Hathy, E.; Reé, D.; Homolya, L.; Erdei, Z.; Réthelyi, J.M.; Apáti, Á. Functional Comparison of Blood-Derived Human Neural Progenitor Cells. Int. J. Mol. Sci. 2020, 21, 9118. https://doi.org/10.3390/ijms21239118
Szabó E, Juhász F, Hathy E, Reé D, Homolya L, Erdei Z, Réthelyi JM, Apáti Á. Functional Comparison of Blood-Derived Human Neural Progenitor Cells. International Journal of Molecular Sciences. 2020; 21(23):9118. https://doi.org/10.3390/ijms21239118
Chicago/Turabian StyleSzabó, Eszter, Flóra Juhász, Edit Hathy, Dóra Reé, László Homolya, Zsuzsa Erdei, János M. Réthelyi, and Ágota Apáti. 2020. "Functional Comparison of Blood-Derived Human Neural Progenitor Cells" International Journal of Molecular Sciences 21, no. 23: 9118. https://doi.org/10.3390/ijms21239118
APA StyleSzabó, E., Juhász, F., Hathy, E., Reé, D., Homolya, L., Erdei, Z., Réthelyi, J. M., & Apáti, Á. (2020). Functional Comparison of Blood-Derived Human Neural Progenitor Cells. International Journal of Molecular Sciences, 21(23), 9118. https://doi.org/10.3390/ijms21239118




