Secretory Peptides as Bullets: Effector Peptides from Pathogens against Antimicrobial Peptides from Soybean
Abstract
:1. Introduction
2. Introduction to Soybean–Microbial Pathogen Interactions
2.1. Soybean–Fungus Interactions
2.2. Soybean–Oomycete Interactions
2.3. Soybean–Bacterium Interactions
2.4. Soybean–Virus Interactions
3. Introduction to the Innate Immunity of Plants
4. Compatibility of Released Molecules from Plants and Pathogens Determine Disease Susceptibility
4.1. PAMP Sequence Polymorphism Influences Plant Susceptibility
4.2. Post Translational Modifications of PAMPs Could Influence the Virulence
4.3. Peptides Play Important Roles in the Defense Responses of Soybean
5. Peptides Secreted by Soybean Pathogenic Microbes during the Attack
5.1. Pathogenic Effector Peptides Repress the Immune Responses of Soybean
5.2. Peptides Secreted by Soybean Pathogens Cause Sudden Death of the Host
5.3. Effector Peptides Secreted by Soybean Pathogens Influence the Epigenetics of the Host
5.3.1. Effects on Histone Modification
5.3.2. Effects on mRNA Regulation
5.3.3. The Possible Roles of lncRNAs in Transcription Regulation
5.4. Effector Peptides Secreted by Soybean Pathogens Affect Phytohormone Biosynthesis in the Host
6. Plant Antimicrobial Peptides
6.1. Introduction to Plant Antimicrobial Peptides
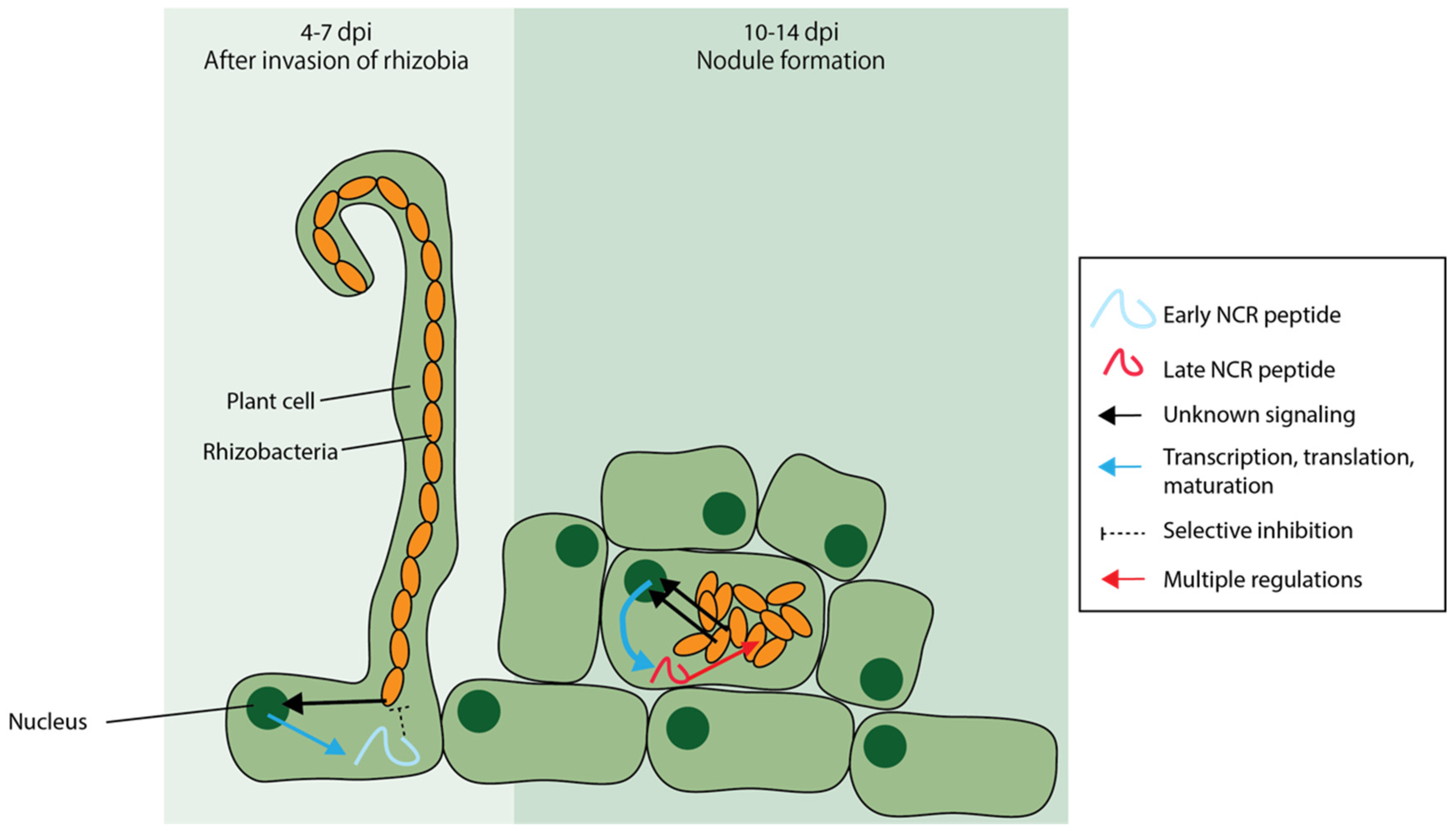
6.2. Nodule-Specific Cysteine-Rich (NCR) Peptides Are AMPs Unique to Certain Legumes
7. AMPs Employed by Soybeans to Defend against Microbial Pathogens
7.1. Soybean AMPs
7.2. AMPs Secreted by Soybean-Associated Microbes
7.2.1. Endophytes
7.2.2. Rhizospheric Microbes
8. The Potential Application of Soybean Antimicrobial Peptides and Soybean-Associated Microbes as Biopesticides
9. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Hartman, G.L.; West, E.D.; Herman, T.K. Crops that feed the world 2. Soybean—Worldwide production, use, and constraints caused by pathogens and pests. Food Sec. 2011, 3, 5–17. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Nahashon, S.N.; Kilonzo-Nthenge, A.K. Advances in soybean and soybean by-products in monogastric nutrition and health. In Soybean and Nutrition; El-Shemy, H., Ed.; InTech: Rijeka, Croatia, 2011; pp. 125–156. [Google Scholar]
- SoyStats. Available online: http://soystats.com/ (accessed on 10 September 2020).
- Verma, S.; Nizam, S.; Verma, P. Biotic and abiotic stress signaling in plants. In Stress Signaling in Plants: Genomics and Proteomics Perspective; Sarwat, M., Ahmad, A., Abdin, M.Z., Eds.; Springer Science: New York, NY, USA, 2013; pp. 25–49. [Google Scholar]
- Athow, K. Soybean pest management. J. Am. Oil Chem. Soc. 1981, 58, 130–135. [Google Scholar] [CrossRef]
- Strange, R.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef]
- Bandara, A.; Weerasooriya, D.; Bradley, C.; Allen, T.; Esker, P. Dissecting the economic impact of soybean diseases in the United States over two decades. PLoS ONE 2020, 15, e0231141. [Google Scholar] [CrossRef] [Green Version]
- Whitham, S.A.; Qi, M.; Innes, R.W.; Ma, W.; Lopes-Caitar, V.; Hewezi, T. Molecular soybean-pathogen interactions. Annu. Rev. Phytopathol. 2016, 54, 443–468. [Google Scholar] [CrossRef]
- Hartman, G.; Hill, C. Diseases of soybean and their management. In The Soybean: Botany, Production and Uses; Singh, G., Ed.; CABI Publishing: Wallingford, UK, 2010; pp. 276–299. [Google Scholar]
- Chang, C.; Tian, L.; Ma, L.; Li, W.; Nasir, F.; Li, X.; Tran, P.L.-S.; Tian, C. Differential responses of molecular mechanisms and physiochemical characters in wild and cultivated soybeans against invasion by the pathogenic Fusarium oxysporum Schltdl. Physiol. Plant. 2019, 166, 1008–1025. [Google Scholar] [CrossRef]
- Jiang, C.-J.; Sugano, S.; Ochi, S.; Kaga, A.; Ishimoto, M. Evaluation of Glycine max and Glycine soja for resistance to Calonectria ilicicola. Agronomy 2020, 10, 887. [Google Scholar] [CrossRef]
- Lam, H.-M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.-L.; Li, M.-W.; He, W.; Qin, N.; Wang, B.; et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef]
- Kim, M.Y.; Van, K.; Kang, Y.J.; Kim, K.H.; Lee, S.-H. Tracing soybean domestication history: From nucleotide to genome. Breed. Sci. 2012, 61, 445–452. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Qiu, X.; Kang, J.; Wang, Y.; Chen, H.; Huang, J.; Qiu, M.; Zhao, Y.; Kong, G.; Ma, Z.; et al. A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr. Biol. 2017, 27, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Silverstein, K.A.T.; Gao, L.; Walton, J.D.; Nallu, S.; Guhlin, J.; Young, N.D. Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application). BMC Bioinform. 2013, 14, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranathunge, N.; Mongkolporn, O.; Ford, R.; Taylor, P. Colletotrichum truncatum pathosystem on Capsicum spp: Infection, colonization and defence mechanisms. Aust. J. Plant Physiol. 2012, 41, 463–473. [Google Scholar] [CrossRef]
- Jiang, R.H.Y.; Tyler, B.M. Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 2012, 50, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S. Plant pathogens: Oomycetes (water mold). In Encyclopedia of Microbiology; Schaechter, M., Ed.; Academic Press: Cambridge, MA, USA, 2009; pp. 689–695. ISBN 978-0-12-373944-5. [Google Scholar]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–280. [Google Scholar] [CrossRef] [Green Version]
- Senthilkumar, M.; Govindasamy, V.; Dureja, P.; Annapurna, K. Purification and partial characterization of antifungal peptides from soybean endophyte-Penibacillus sp. strain HKA-15. J. Plant Biochem. Biotechnol. 2007, 16, 131–134. [Google Scholar] [CrossRef]
- Senthilkumar, M.; Govindasamy, V.; Annapurna, K. Role of antibiosis in suppression of charcoal rot disease by soybean endophyte Paenibacillus sp. HKA-15. Curr. Microbiol. 2007, 55, 25–29. [Google Scholar] [CrossRef]
- De Almeida Lopes, K.B.; Carpentieri-Pipolo, V.; Fira, D.; Balatti, P.A.; López, S.M.Y.; Oro, T.H.; Pagliosa, E.S.; Degrassi, G. Screening of bacterial endophytes as potential biocontrol agents against soybean diseases. J. Appl. Microbiol. 2018, 125, 1466–1481. [Google Scholar] [CrossRef] [Green Version]
- León, M.; Yaryura, P.M.; Montechchia, M.S.; Hernández, A.I.; Correa, O.S.; Pucheu, N.L.; Kerber, N.L.; García, A.F. Antifungal activity of selected indigenous Pseudomonas and Bacillus from the soybean rhizosphere. Int. J. Microbiol. 2009, 2009, 572049. [Google Scholar] [CrossRef] [Green Version]
- Maget-Dana, R.; Thimon, L.; Peypoux, F.; Ptak, M. Surfactin/iturin A interactions may explain the synergistic effect of surfactin on the biological properties of iturin A. Biochimie 1992, 74, 1047–1051. [Google Scholar] [CrossRef]
- Wang, G.H.; Zhou, K.Q.; Jin, J.; Pan, X.W.; Liu, X.B.; Luo, Y.H. Antagonism on organism BRF-1 against soybean root rot. Soybean Sci. 2004, 23, 188–191. [Google Scholar]
- Chen, X.; Wang, G.; Xu, M.; Jin, J.; Liu, X. Antifungal peptide produced by Paenibacillus polymyxa BRF-1 isolated from soybean rhizosphere. Afr. J. Microbiol. Res. 2010, 4, 2692–2698. [Google Scholar]
- Pawlowski, M.; Hartman, G. Infection mechanisms and colonization patterns of fungi associated with soybean. In Fungal Pathogenicity; Sultan, S., Ed.; IntechOpen: London, UK, 2016. [Google Scholar]
- Langenbach, C.; Campe, R.; Beyer, S.F.; Mueller, A.N.; Conrath, U. Fighting Asian soybean rust. Front. Plant Sci. 2016, 7, 797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goellner, K.; Loehrer, M.; Langenbach, C.; Conrath, U.; Koch, E.; Schaffrath, U. Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol. Plant Pathol. 2010, 11, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kelly, H.Y.; Dufault, N.S.; Walker, D.R.; Isard, S.A.; Schneider, R.W.; Giesler, L.J.; Wright, D.L.; Marois, J.J.; Hartman, G.L. From select agent to an established pathogen: The response to Phakopsora pachyrhizi (soybean rust) in North America. Phytopathology 2015, 105, 905–916. [Google Scholar] [CrossRef] [Green Version]
- Hoppe, H.H.; Koch, E. Defense reactions in host and nonhost plants against the soybean rust fungus (Phakopsora pachyrhizi Syd.). J. Phytopathol. 1989, 125, 77–88. [Google Scholar] [CrossRef]
- Takamatsu, S.; Shin, H.-D.; Paksiri, U.; Limkaisang, S.; Taguchi, Y.; Binh, N.T.; Sato, Y. Two Erysiphe species associated with recent outbreak of soybean powdery mildew: Results of molecular phylogenetic analysis based on nuclear rDNA sequences. Mycoscience 2002, 43, 333–341. [Google Scholar] [CrossRef]
- Prins, T.; Tudzynski, P.; von Tiedemann, A.; Tudzynski, B.; ten Have, A.; Hansen, M.E.; Tenberge, K.; van Kan, J. AL Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In Fungal Pathology; Kronstad, J., Ed.; Springer: Dordrecht, The Netherlands, 2000. [Google Scholar]
- van Kan, J.A.L. Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006, 11, 247–253. [Google Scholar] [CrossRef]
- Laluk, K.; Mengiste, T. Necrotroph attacks on plants: Wanton destruction or covert extortion. Arab. Book 2010, 8, e0136. [Google Scholar] [CrossRef] [Green Version]
- Malvick, D.; Impulliti, A. Detection and quantification of Phialophora gregata in soybean and soil samples with a quantitative, real-time PCR assay. Plant Dis. 2007, 91, 724–736. [Google Scholar] [CrossRef]
- Chehri, K.; Salleh, B.; Zakaria, L. Fusarium virguliforme, a soybean sudden death syndrome fungus in Malaysian soil. Autralasian Plant Dis. Notes 2014, 9, 128. [Google Scholar] [CrossRef] [Green Version]
- Fenille, R.C.; de Souza, N.L.; Kuramae, E.E. Characterization of Rhizoctonia solani associated with soybean in Brazil. Eur. J. Plant Pathol. 2002, 108, 783–792. [Google Scholar] [CrossRef]
- Coser, S.M.; Chowda Reddy, R.; Zhang, J.; Mueller, D.S.; Mengistu, A.; Wise, K.A.; Allen, T.W.; Singh, A.; Singh, A. Genetic architecture of charcoal rot (Macrophomina phaseolina) resistance in soybean revealed using a diverse panel. Front. Plant Sci. 2017, 8, 1626. [Google Scholar] [CrossRef] [PubMed]
- Hartman, G.; Rupe, J.; Sikora, E.; Domier, L.; Davis, J.; Steffey, K. Part I. Infectious Diseases. In Compendium of Soybean Diseases and Pests, 5th ed.; Hartman, G., Rupe, J., Sikora, E., Domier, L., Davis, J., Steffey, K., Eds.; APS Publications: St. Paul, MN, USA, 2016; ISBN 978-0-89054-475-4. [Google Scholar]
- Manandhar, J.; Kunwar, I.; Singh, T.; Hartman, G.; Sinclair, J. Penetration and infection of soybean leaf tissues by Colletotrichum truncatum and Glomerella glycines. Phytopathology 1985, 75, 704–708. [Google Scholar] [CrossRef]
- Chen, L.; Chu, C.; Liu, C.; Chen, R.; Tsay, J. PCR-based detection and differentiation of anthracnose pathogens, Colletotrichum gloeosporioides and C. truncatum, from vegetable soybean in Taiwan. J. Phytopathol. 2006, 154, 654–662. [Google Scholar] [CrossRef]
- Tyler, B.M. Phytophthora sojae: Root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007, 8, 1–8. [Google Scholar] [CrossRef]
- Rojas, A.J.; Jacobs, J.L.; Napieralski, S.; Karaj, B.; Bradley, C.A.; Chase, T.; Esker, P.D.; Giesler, L.J.; Jardine, D.J.; Malvick, D.K.; et al. Oomycete species associated with soybean seedlings in North America—Part I: Identification and pathogenicity characterization. Phytopathology 2016, 107, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Tyler, B.M.; Wu, M.; Wang, J.; Cheung, W.; Morris, P.F. Chemotactic preferences and strain variation in the response of Phytophthora sojae zoospores to host isoflavones. Appl. Environ. Microbiol. 1996, 62, 2811–2817. [Google Scholar] [CrossRef] [Green Version]
- Enkerli, K.; Mims, C.W.; Hahn, M.G. Ultrastructure of compatible and incompatible interactions of soybean roots infected with the plant pathogenic oomycete Phytophthora sojae. Can. J. Bot. 1997, 75, 1493–1508. [Google Scholar] [CrossRef]
- Tolin, S.A.; Lacy, G.H. Viral, bacterial, and phytoplasmal diseases of soybean. In Soybeans: Improvement, Production, and Uses; Shibles, R.M., Harper, J.E., Wilson, R.F., Shoemaker, R.C., Eds.; The American Society of Agronomy, Inc.; Crop Science Society of America, Inc.; Soil Science Society of America, Inc.: Madison, WI, USA, 2004. [Google Scholar]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Baker, C.; Chitrakar, R.; Obulareddy, N.; Panchal, S.; Williams, P.; Melotto, M. Molecular battles between plant and pathogenic bacteria in the phyllosphere. Braz. J. Med. Biol. Res. 2010, 43, 698–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, X.-F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar]
- Alfano, J.R.; Collmer, A. TYPE III SECRETION SYSTEM EFFECTOR PROTEINS: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.; Lim, S.M.; Shaw, P.D. Cloning and characterization of pathogenicity genes from Xanthomonas campestris pv. glycines. J. Bacteriol. 1992, 174, 1923–1931. [Google Scholar] [CrossRef] [Green Version]
- Chatnaparat, T.; Prathuangwong, S.; Ionescu, M.; Lindow, S.E. XagR, a LuxR homolog, contributes to the virulence of Xanthomonas axonopodis pv. glycines to soybean. MPMI 2012, 25, 1104–1117. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.H.; Whitham, S.A. Control of virus diseases in soybeans. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar]
- Liu, J.-Z.; Fang, Y.; Pang, H. The current status of the soybean-Soybean Mosaic Virus (SMV) pathosystem. Front. Microbiol. 2016, 7, 1906. [Google Scholar] [CrossRef]
- Rupe, J.; Luttrell, R.G. Effect of pests and diseases on soybean quality. In Soybeans. Chemistry, Production, Processing and Utilization; Johnson, L.A., White, P.J., Galloway, R., Eds.; Academic Press: Cambridge, MA, USA; AOCS Press: Urbana, IL, USA, 2008; pp. 93–116. [Google Scholar]
- Song, Y.; Li, C.; Zhao, L.; Karthikeyan, A.; Li, N.; Li, K.; Zhi, H. Disease spread of a popular soybean mosaic virus strain (SC7) in southern China and effects on two susceptible soybean cultivars. Philipp. Agric. Sci. 2016, 99, 355–364. [Google Scholar]
- Goodman, R.M.; Bowers, G.R., Jr.; Paschal, E., II. Identification of soybean germplasm lines and cultivars with low incidence of soybean mosaic virus transmission through seed. Crop Sci. 1979, 19, 264–267. [Google Scholar] [CrossRef]
- Hajimorad, M.; Domier, L.; Tolin, S.; Whitham, S.; Saghai Maroof, M. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jossey, S.; Hobbs, H.; Domier, L. Role of soybean mosaic virus-encoded proteins in seed and aphid transmission in soybean. Phytopathology 2013, 103, 801–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C.; Oldroyd, G.E.D. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Ku, Y.-S.; Contador, C.A.; Lam, H.-M. The impacts of domestication and agricultural practices on legume nutrient acquisition through symbiosis with Rhizobia and rrbuscular mycorrhizal fungi. Front. Genet. 2020, 11, 583954. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Inheritance of pathogenecity in a cross between physiologic races 22 and 24 of Melampsora lini. Phytopathology 1942, 32, 653–699. [Google Scholar]
- Flor, H.H. Host-parasite interaction in flax rust- its genetics and other implications. Phytopathology 1955, 45, 680–685. [Google Scholar]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Rouxel, T.; Balesdent, M.-H. Avirulence Genes. In Encyclopedia of Life Sciences (ELS); John Wiley & Sons, Ltd.: Chichester, UK, January 2010. [Google Scholar]
- Medzhitov, R.; Janeway, C.A. Innate immunity: The virtues of a nonclonal system of recognition. Cell 1997, 91, 295–298. [Google Scholar] [CrossRef] [Green Version]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef]
- Schwessinger, B.; Ronald, P.C. Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant Biol. 2012, 63, 451–482. [Google Scholar] [CrossRef] [Green Version]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.-M.; Chai, J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 341, 1387–1391. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, Y.; Zhou, Z.; Zhou, J.M. Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 2016, 59, 878–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhou, J.M. Plant immunity triggered by microbial molecular signatures. Mol. Plant 2010, 3, 783–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fürst-Jansen, J.M.R.; de Vries, S.; de Vries, J. Evo-physio: On stress responses and the earliest land plants. J. Exp. Bot. 2020, 71, 3254–3269. [Google Scholar] [CrossRef] [Green Version]
- de Vries, S.; de Vries, J.; von Dahlen, J.K.; Gould, S.B.; Archibald, J.M.; Rose, L.E.; Slamovits, C.H. On plant defense signaling networks and early land plant evolution. Commun. Integr. Biol. 2018, 11, 1–14. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plantĝ€” pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.; Verma, S.; Reddy, V.P.; Sharma, D. Hypersensitive responses in plants. Agric. Rev. 2019, 40, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, M.E.; Pennell, R.I.; Meijer, P.-J.; Ishikawa, A.; Dixon, R.A.; Lamb, C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 1998, 92, 773–784. [Google Scholar] [CrossRef] [Green Version]
- Gao, Q.-M.; Zhu, S.; Kachroo, P.; Kachroo, A. Signal regulators of systemic acquired resistance. Front. Plant Sci. 2015, 6, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, Y.; Huffaker, A. Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 2011, 14, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Narváez-Vásquez, J.; Ryan, C.A. The cellular localization of prosystemin: A functional role for phloem parenchyma in systemic wound signaling. Planta 2004, 218, 360–369. [Google Scholar] [PubMed]
- Farmer, E.E.; Johnson, R.R.; Ryan, C.A. Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 1992, 98, 995–1002. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.W.; Klessig, D.F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lygin, A.V.; Hill, C.B.; Zernova, O.V.; Crull, L.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. Response of soybean pathogens to glyceollin. Phytopathology 2010, 100, 897–903. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.N.; Liu, D.D.; Zhong, C.L.; Xu, H.Y.; Yang, S.; Fang, Y.; Ran, J.; Liu, J.Z. Silencing GmFLS2 enhances the susceptibility of soybean to bacterial pathogen through attenuating the activation of GmMAPK signaling pathway. Plant Sci. 2020, 292, 110386. [Google Scholar] [CrossRef]
- Huet, G. Breeding for resistances to Ralstonia solanacearum. Front. Plant Sci. 2014, 5, 715. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Balaceanu, A.; Rufian, J.S.; Segonzac, C.; Zhao, A.; Morcillo, R.J.L.; Macho, A.P. An immune receptor complex evolved in soybean to perceive a polymorphic bacterial flagellin. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Takeuchi, K.; Taguchi, F.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 2003, 185, 6658–6665. [Google Scholar]
- Taguchi, F.; Takeuchi, K.; Katoh, E.; Murata, K.; Suzuki, T.; Marutani, M.; Kawasaki, T.; Eguchi, M.; Katoh, S.; Kaku, H.; et al. Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell. Microbiol. 2006, 8, 923–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearce, G.; Yamaguchi, Y.; Barona, G.; Ryan, C.A. A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 2010, 107, 14921–14925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, R.; Segorbe, D.; Prusky, D.; Di Pietro, A. How alkalinization drives fungal pathogenicity. PLoS ONE 2017, 13, e1006621. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Barona, G.; Ryan, C.A.; Pearce, G. GmPep914, an eight-amino acid peptide isolated from soybean leaves, activates defense-related genes. Plant Physiol. 2011, 156, 932–942. [Google Scholar] [CrossRef]
- Lee, M.W.; Huffaker, A.; Crippen, D.; Robbins, R.T.; Goggin, F.L. Plant elicitor peptides promote plant defences against nematodes in soybean. Mol. Plant Pathol. 2018, 19, 858–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evangelisti, E.; Gogleva, A.; Hainaux, T.; Doumane, M.; Tulin, F.; Quan, C.; Yunusov, T.; Floch, K.; Schornack, S. Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol. 2017, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Li, X.; Cui, H.; He, P.; Thilmony, R.; Chintamanani, S.; Zwiesler-Vollick, J.; Gopalan, S.; Tang, X.; Zhou, J.M. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 2006, 103, 19200–19205. [Google Scholar] [CrossRef] [Green Version]
- Selote, D.; Kachroo, A. RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol. 2010, 153, 1199–1211. [Google Scholar] [CrossRef] [Green Version]
- Selote, D.; Kachroo, A. RIN4-like proteins mediate resistance protein-derived soybean defense against Pseudomonas syringae. Plant Signal. Behav. 2010, 5, 1453–1456. [Google Scholar] [CrossRef] [Green Version]
- Kessens, R.; Ashfield, T.; Kim, S.H.; Innes, R.W. Determining the GmRIN4 requirements of the soybean disease resistance proteins Rpg1b and Rpg1r using a Nicotiana glutinosa-based agroinfiltration system. PLoS ONE 2014, 9, e108159. [Google Scholar] [CrossRef] [Green Version]
- Chung, E.H.; El-Kasmi, F.; He, Y.; Loehr, A.; Dangl, J.L. A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 2014, 16, 484–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashfield, T.; Redditt, T.; Russell, A.; Kessens, R.; Rodibaugh, N.; Galloway, L.; Kang, Q.; Podicheti, R.; Innes, R.W. Evolutionary relationship of disease resistance genes in soybean and Arabidopsis specific for the Pseudomonas syringae effectors AvrB and AvrRpm1. Plant Physiol. 2014, 166, 235–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, A.R.; Ashfield, T.; Innes, R.W. Pseudomonas syringae effector AvrPphB suppresses AvrB-induced activation of RPM1 but not AvrRpm1-induced activation. Mol. Plant-Microbe Interact. 2015, 28, 727–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, F.; Golstein, C.; Ade, J.; Stoutemyer, M.; Dixon, J.E.; Innes, R.W. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 2003, 301, 1230–1233. [Google Scholar] [CrossRef]
- Selote, D.; Robin, G.P.; Kachroo, A. GmRIN4 protein family members function nonredundantly in soybean race-specific resistance against Pseudomonas syringae. New Phytol. 2013, 197, 1225–1235. [Google Scholar] [CrossRef]
- Zhou, H.; Lin, J.; Johnson, A.; Morgan, R.L.; Zhong, W.; Ma, W. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe 2011, 9, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Isoflavonoid accumulation in soybean hairy roots upon treatment with Fusarium solani. Plant Physiol. Biochem. 2004, 42, 671–679. [Google Scholar] [CrossRef]
- Hartman, G.L.; Chang, H.X.; Leandro, L.F. Research advances and management of soybean sudden death syndrome. Crop Prot. 2015, 73, 60–66. [Google Scholar] [CrossRef]
- Brar, H.K.; Swaminathan, S.; Bhattacharyya, M.K. The Fusarium virguliforme toxin FvTox1 causes foliar sudden death syndrome-like symptoms in soybean. Mol. Plant-Microbe Interact. 2011, 24, 1179–1188. [Google Scholar] [CrossRef] [Green Version]
- Brar, H.K.; Bhattacharyya, M.K. Expression of a single-chain variable-fragment antibody against a Fusarium virguliforme toxin peptide enhances tolerance to sudden death syndrome in transgenic soybean plants. Mol. Plant-Microbe Interact. 2012, 25, 817–824. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.X.; Domier, L.L.; Radwan, O.; Yendrek, C.R.; Hudson, M.E.; Hartman, G.L. Identification of multiple phytotoxins produced by Fusarium virguliforme including a phytotoxic effector (fvnis1) associated with sudden death syndrome foliar symptoms. Mol. Plant-Microbe Interact. 2016, 29, 96–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.X.; Tan, R.; Hartman, G.L.; Wen, Z.; Sang, H.; Domier, L.L.; Whitham, S.A.; Wang, D.; Chilvers, M.I. Characterization of soybean STAY-GREEN genes in susceptibility to foliar chlorosis of sudden death syndrome. Plant Physiol. 2019, 180, 711–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wang, H.; Jing, M.; Zhu, J.; Guo, B.; Wang, Y.; Lin, Y.; Chen, H.; Kong, L.; Ma, Z.; et al. A Phytophthora effector recruits a host cytoplasmic transacetylase into nuclear speckles to enhance plant susceptibility. eLife 2018, 7, e40039. [Google Scholar] [CrossRef]
- Qiao, Y.; Shi, J.; Zhai, Y.; Hou, Y.; Ma, W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. USA 2015, 112, 18. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Gu, L.; Zhang, Y.; Yan, T.; Kong, G.; Kong, L.; Guo, B.; Qiu, M.; Wang, Y.; Jing, M.; et al. An oomycete plant pathogen reprograms host pre-mRNA splicing to subvert immunity. Nat. Commun. 2017, 8, 2051. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ye, W.; Wang, Y. Genome-wide identification of long non-coding RNAs suggests a potential association with effector gene transcription in Phytophthora sojae. Mol. Plant Pathol. 2018, 19, 2177–2186. [Google Scholar] [CrossRef] [Green Version]
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Wang, Y.; Guo, B.; Jing, M.; Zhou, H.; Li, Y.; Wang, H.; Huang, J.; Wang, Y.; Ye, W.; et al. The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 2019, 222, 425–437. [Google Scholar] [CrossRef]
- Jing, M.; Guo, B.; Li, H.; Yang, B.; Wang, H.; Kong, G.; Zhao, Y.; Xu, H.; Wang, Y.; Ye, W.; et al. A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant Binding immunoglobulin Proteins. Nat. Commun. 2016, 7, 11685. [Google Scholar] [CrossRef]
- Wang, H.; Ren, Y.; Zhou, J.; Du, J.; Hou, J.; Jiang, R.; Wang, H.; Tian, Z.; Xie, C. The cell death triggered by the nuclear localized RxLR effector PITG_22798 from Phytophthora infestans is suppressed by the effector AVR3b. Int. J. Mol. Sci. 2017, 18, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, F.; He, Q.; Armstrong, M.; Giuliani, L.M.; Boevink, P.C.; Zhang, W.; Tian, Z.; Birch, P.R.J.; Gilroy, E.M. The potato MAP3K StVIK is required for the Phytophthora infestans RXLR effector Pi17316 to promote disease. Plant Physiol. 2018, 177, 398–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Zhang, Y.; Li, H.; Zhang, Z.; Sheng, G.; Li, Y.; Xing, Y.; Huang, S.; Tao, H.; Kuan, T.; et al. The RXLR effector PcAvh1 Is required for full virulence of Phytophthora capsici. MPMI 2019, 32, 986–1000. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; Gu, B.; Zhao, H.; Jia, J.; Fan, G.; Meng, Y.; DU, Y.; Shan, W. An RXLR effector secreted by Phytophthora parasitica is a virulence factor and triggers cell death in various plants. Mol. Plant Pathol. 2019, 20, 356–371. [Google Scholar] [CrossRef] [Green Version]
- Dalio, R.J.D.; Maximo, H.J.; Oliveira, T.S.; Dias, R.O.; Breton, M.C.; Felizatti, H.; Machado, M. Phytophthora parasitica effector PpRxLR2 suppresses Nicotiana benthamiana immunity. MPMI 2018, 31, 481–493. [Google Scholar] [CrossRef] [Green Version]
- Ramada, M.H.S.; Brand, G.D.; Abrão, F.Y.; Oliveira, M.; Cardozo Filho, J.L.; Galbieri, R.; Gramacho, K.P.; Prates, M.V.; Bloch, C., Jr. Encrypted antimicrobial peptides from plant proteins. Sci. Rep. 2017, 7, 13263. [Google Scholar] [CrossRef]
- Basrai, M.A.; Hieter, P.; Boeke, J.D. Small open reading frames: Beautiful needles in the haystack. Genome Res. 1997, 7, 768–771. [Google Scholar] [CrossRef]
- Yuan, S.L.; Li, R.; Chen, H.F.; Zhang, C.J.; Chen, L.M.; Hao, Q.N.; Chen, S.L.; Shan, Z.H.; Yang, Z.L.; Zhang, X.J.; et al. RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2. Sci. Rep. 2017, 7, 42248. [Google Scholar] [CrossRef]
- Lin, X.; Lin, W.; Ku, Y.-S.; Wong, F.; Li, M.-W.; Lam, H.-M.; Ngai, S.-M.; Chan, T.-F. Analysis of soybean long non-coding RNAs reveals a subset of small peptide-coding transcripts. Plant Physiol. 2020, 182, 1359–1374. [Google Scholar] [CrossRef] [Green Version]
- Farrokhi, N.; Whitelegge, J.P.; Brusslan, J.A. Plant peptides and peptidomics. Plant Biotechnol. J. 2008, 6, 105–134. [Google Scholar] [CrossRef]
- Benko-Iseppon, A.M.; Galdino, S.L.; Calsa, T., Jr.; Kido, E.A.; Tossi, A.; Belarmino, L.C.; Crovella, S. Overview on plant antimicrobial peptides. Curr. Protein Pept. Sci. 2010, 11, 181–188. [Google Scholar]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial peptides from plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef] [PubMed]
- Tavormina, P.; De Barbara, C.; Nikonorova, N.; De Smet, I.; Cammue, B.P.A. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverstein, K.A.T.; Moskal, W.A., Jr.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef] [PubMed]
- Kereszt, A.; Mergaert, P.; Kondorosi, E. Bacteroid development in legume nodules: Evolution of mutual benefit or of sacrificial victims? Mol. Plant-Microbe Interact. 2011, 24, 1300–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Velde, W.; Zehirov, G.; Szatmari, A.; Debreczeny, M.; Ishihara, H.; Kevei, Z.; Farkas, A.; Mikulass, K.; Nagy, A.; Tiricz, H.; et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 2010, 327, 1122–1127. [Google Scholar] [CrossRef]
- Czernic, P.; Gully, D.; Cartieaux, F.; Moulin, L.; Guefrachi, I.; Patrel, D.; Pierre, O.; Fardoux, J.; Chaintreuil, C.; Nguyen, P.; et al. Convergent evolution of endosymbiont differentiation in Dalbergioid and Inverted Repeat-Lacking clade legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 2015, 169, 1254–1265. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, S.; Liu, J.; Terecskei, K.; Ábrahám, E.; Gombár, A.; Domonkos, Á.; Szücs, A.; Körmöczi, P.; Wang, T.; et al. Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6854–6859. [Google Scholar]
- Mergaert, P.; Nikovics, K.; Kelemen, Z.; Maunoury, N.; Vaubert, D.; Kondorosi, A.; Kondorosi, E. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine Motifs. Plant Physiol. 2003, 132, 161–173. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Wang, Q.; Fedorova, E.; Liu, J.; Qin, Q.; Zheng, Q.; Price, P.A.; Pan, H.; Wang, D.; Griffitts, J.S.; et al. Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6848–6853. [Google Scholar] [CrossRef] [Green Version]
- Tiricz, H.; Szücs, A.; Farkas, A.; Pap, B.; Lima, R.M.; Maróti, G.; Kondorosi, É.; Kereszt, A. Antimicrobial nodule-specific cysteine-rich peptides induces membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl. Environ. Microbiol. 2013, 79, 6737–6746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkas, A.; Maróti, G.; Kereszt, A.; Kondorosi, É. Comparative analysis of the bacterial membrane disruption effect of two natural plant antimicrobial peptides. Front. Microbiol. 2017, 8, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ördögh, L.; Vörös, A.; Nagy, I.; Kondorosi, É.; Kereszt, A. Symbiotic plant peptides eliminate Candida albicans both in vitro and in an epithelial infection model and inhibit the proliferation of immortalized human cells. J. Biomed. Biotechnol. 2014, 2014, 320796. [Google Scholar]
- Nallu, S.; Silverstein, K.A.T.; Samac, D.A.; Bucciarelli, B.; Vance, C.P.; Vandenbosch, K.A. Regulatory patterns of a large family of defensin-like genes expressed in nodules of Medicago truncatula. PLoS ONE 2013, 8, e60355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933–D937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand, G.D.; Magalhães, M.T.Q.; Tinoco, M.L.P.; Aragão, F.J.L.; Nicoli, J.; Kelly, S.M.; Cooper, A.; Bloch, C., Jr. Probing protein sequences as sources for encrypted antimicrobial peptides. PLoS ONE 2012, 7, e45848. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.L.; Ouellette, A.J.; Satchell, D.P.; Ayabe, T.; López-Boado, Y.S.; Stratman, J.L.; Hultgren, S.J.; Matrisian, L.M.; Parks, W.C. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 1999, 286, 113–118. [Google Scholar] [CrossRef]
- Pak, J.H.; Liu, C.Y.; Huangpu, J.; Graham, J.S. Construction and characterization of the soybean leaf metalloproteinase cDNA. FEBS Lett. 1997, 404, 283–288. [Google Scholar] [CrossRef] [Green Version]
- Maidment, J.M.; Moore, D.; Murphy, G.P.; Murphy, G.; Clark, I.M. Matrix metalloproteinase homologues from Arabidopsis thaliana EXPRESSION AND ACTIVITY. J. Biol. Chem. 1999, 274, 34706–34710. [Google Scholar] [CrossRef] [Green Version]
- Delorme, V.G.R.; McCabe, P.F.; Kim, D.-J.; Leaver, C.J. A matrix metalloproteinase gene is expressed at the boundary of senescence and programmed cell death in cucumber. Plant Physiol. 2000, 123, 917–927. [Google Scholar] [CrossRef] [Green Version]
- Belozersky, M.A.; Dunaevsky, Y.E.; Voskoboynikova, N.E. Isolation and properties of a metalloproteinase from buckwheat (Fagopyrum esculentum) seeds. Biochem. J. 1990, 272, 677–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, J.S.; Xiong, J.; Gillikin, J.W. Purification and developmental analysis of a metalloendoproteinase from the leaves of Glycine max. Plant Physiol. 1991, 97, 786–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Dammann, C.; Bhattacharyya, M.K. The matrix metalloproteinase gene GmMMP2 is activated in response to pathogenic infections in soybean. Plant Physiol. 2001, 127, 1788–1797. [Google Scholar] [CrossRef] [PubMed]
- Onishi, M.; Tachi, H.; Kojima, T.; Shiraiwa, M.; Takahara, H. Molecular cloning and characterization of a novel salt-inducible gene encoding an acidic isoform of PR-5 protein in soybean (Glycine max [L.] Merr.). Plant Physiol. Biochem. 2006, 44, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cheng, F.; Sun, Y.; Ma, H.; Yang, X. Structure−function relationship of a novel PR-5 protein with antimicrobial activity from soy hulls. J. Agric. Food Chem. 2016, 64, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Tachi, H.; Fukuda-Yamada, K.; Kojima, T.; Shiraiwa, M.; Takahara, H. Molecular characterization of a novel soybean gene encoding a neutral PR-5 protein induced by high-salt stress. Plant Physiol. Biochem. 2009, 47, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wu, J.; Xue, A.; Li, W.; Chen, W.; Wei, L.; Lv, H.; Lin, S.; Fan, S.; Li, N.; et al. Differentially expressed genes of soybean during infection by Phytophthora sojae. J. Integr. Agric. 2012, 11, 368–377. [Google Scholar] [CrossRef]
- Fan, S.; Jiang, L.; Wu, J.; Dong, L.; Cheng, Q.; Xu, P.; Zhang, S. A Novel pathogenesis-related class 10 protein Gly m 4l, increases resistance upon Phytophthora sojae infection in soybean (Glycine max [L.] Merr.). PLoS ONE 2015, 10, e0140364. [Google Scholar] [CrossRef]
- JGI Phytozome 12. Available online: https://phytozome.jgi.doe.gov/pz/portal.html# (accessed on 31 August 2020).
- Haq, I.U.; Sarwar, M.K.; Faraz, A.; Latif, M.Z. Synthetic chemicals: Major component of plant disease management. In Plant Dsease Management Strategies for Sustainable Agriculture through Traditional and Modern Approaches; Haq, I.U., Ijaz, S., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2020; pp. 53–81. ISBN 9783030359553. [Google Scholar]
- Bernardes, M.F.F.; Pazin, M.; Pereira, L.C.; Dorta, D.J. Impact of pesticides on environmental and human health. In Toxicology Studies-Cells, Drugs and Environment; Andreazza, A.C., Scola, G., Eds.; IntechOpen: London, UK, 2015. [Google Scholar]
- Chandler, D.; Bailey, A.S.; Tatchell, G.M.; Davidson, G.; Greaves, J.; Grant, W.P. The development, regulation and use of biopesticides for integrated pest management. Phil. Trans. R. Soc. B 2011, 366, 1987–1998. [Google Scholar] [CrossRef]
- Ruiu, L. Microbial biopesticides in agroecosystems. Agronomy 2018, 8, 235. [Google Scholar] [CrossRef] [Green Version]

| Disease | Pathogen | Microbe Type |
|---|---|---|
| Anthracnose | Colletotrichum spp. | Fungus |
| Brown stem rot | Cadophora gregata (Allington and D.W. Chamb.) T.C. Harr. and McNew (syn. Phialophora gregata) | |
| Charcoal rot | Macrophomina phaseolina (Tassi) Goid | |
| Downy mildew | Pernospora manshurica (Naumov) Syd. ex Gäum | |
| Foliar blight | Rhizoctania solani J.G. Kühn | |
| Frogeye leaf spot | Cercospora sojina Hara | |
| Northern stem canker | Diaporthe phaseolorum var. caulivora Athow and Caldwell | |
| Phomopsis seed decay | Phomopsis spp./Diaporthe spp. | |
| Pod and stem blight | Diaporthe phaseolorum var. sojae (Lehman) Wehm. | |
| Purple seed stain and Cercospora leaf blight | Cercospora kikuchii (Tak. Matsumoto and Tomoy.) M.W. Gardner | |
| Rust | Phakopsora pachyrhizi Syd. and P. Syd. | |
| Sclerotinia stem rot | Sclerotinia sclerotiorum (Lib.) de Bary | |
| Septoria brown spot | Septoria glycines Hemmi | |
| Sudden death syndrome | Fusarium virguliforme O’Donnell and T. Aoki, 2003 | |
| Target leaf spot | Corynespora cassiicola (Berk. and M.A. Curtis) C.T. Wei | |
| Phytophthora root and stem rot and damping-off of seedlings | Phytophthora sojae Kaufm. and Gerd. | Oomycete |
| Damping-off of seedlings | Pythium spp. Pringsh. | |
| Downy mildew | Peronospora manshurica Syd. (Naumov) | |
| Damping off and root rot | Pythium ultimum Trow, 1901 | |
| Seed rot | Phytopythium spp. | |
| Bacterial blight | Pseudomonas syringae pv. glycinea (Coerper 1919) Young et al., 1978 | Bacterium |
| Bacterial pustule | Xanthomona campestris subsp. glycines (Nakano) Dye | |
| Bacterial tan spot | Curtobacterium flaccumfaciens pv. flaccumfaciens (Hedges 1922) Collins and Jones 1983 | |
| Bacterial wilt | Ralstonia solanacearum Yabuuchi et al., 1996 (Smith, 1896) | |
| Fasciation | Rhodococcus facians (Tilford 1936) Goodfellow 1984 | |
| Wildfire | Pseudomonas syringae pv. tabaci (Wolf and Foster, 1917) Young et al., 1978 | |
| Bean pod mottle | Bean pod mottle virus | Virus |
| Bud blight | Tobacco ringspot virus | |
| Mosaic | Soybean mosaic virus Gardner and Kendrick (1921) | |
| Soybean vein necrosis virus | Soybean vein necrosis virus | |
| Yellow mosaic | Bean yellow mosaic potyvirus |
| Phytophthora spp. | Effector Peptide | Host Target | Virulence Promotion Mechanism in Host | Reference |
|---|---|---|---|---|
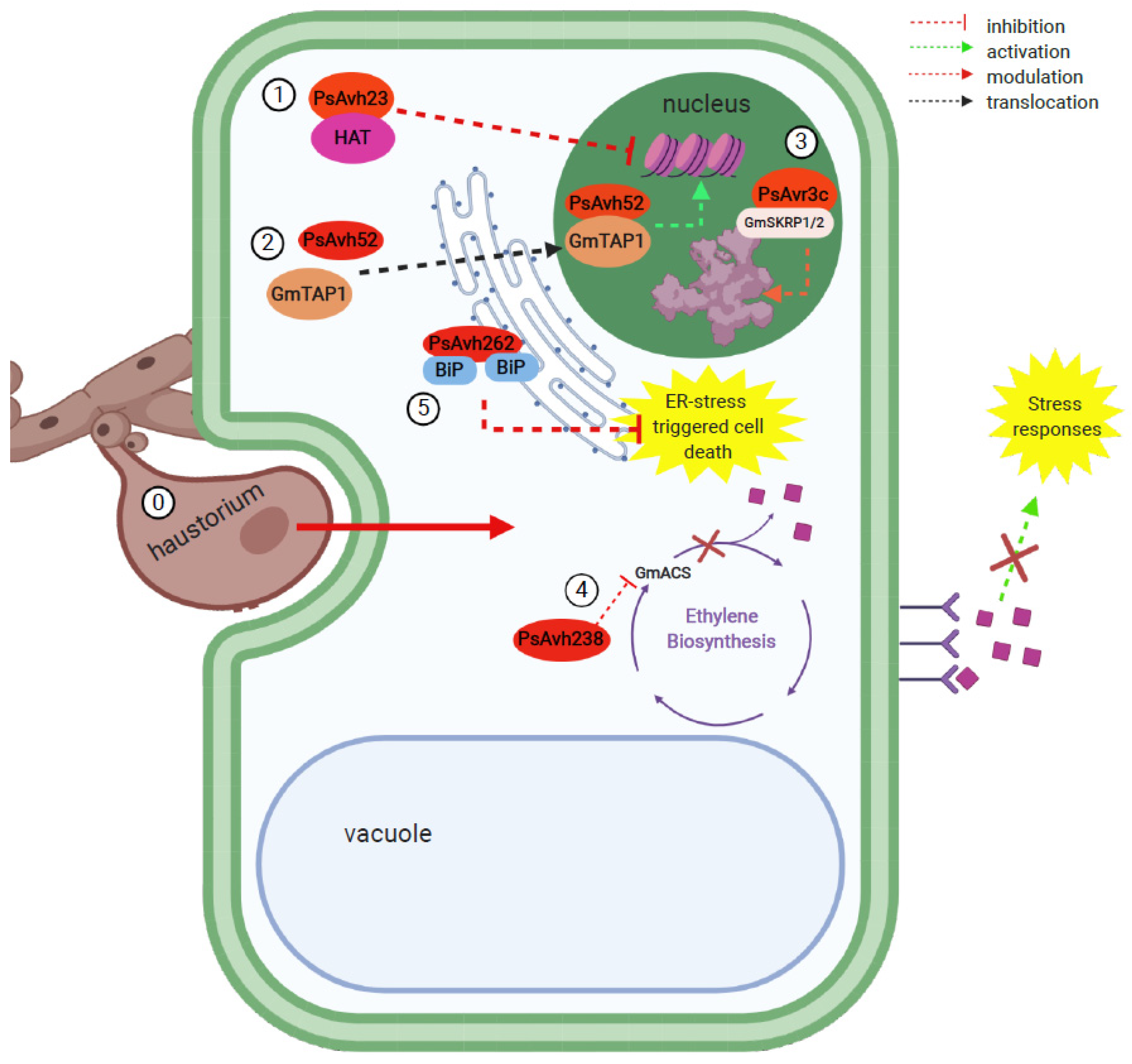
| P. sojae | PsAvh23 | ADA2 subunit of the ADA2/GCN5 module, part of the SAGA histone acetyltransferase (HAT) complex | Modulation of soybean H3K9 HAT by competitively binding to its regulatory subunit ADA2, preventing the association of catalytic subunit GCN5, thereby suppressing the activation of defense genes. | [15] |
| PsAvh52 | Putative transacetylase protein (GmTAP1) | Relocation of GmTAP1 to the nucleus, where it acetylates core histones to upregulate plant susceptibility genes. | [115] | |
| PsAvr3c | Serine/lysine/arginine-rich proteins (GmSKRP1/2) associated with spliceosome components | Stabilizes GmSKRP1, preventing its degradation. This leads to changes in host pre-mRNA splicing that ultimately lead to impaired plant immunity. | [117] | |
| PsAvh238 | Type 2 1-aminocyclopropane-1-carboxylate synthase (Type 2 GmACS) | Suppression of ethylene synthesis by interacting with key biosynthesis enzyme Type 2 GmACS to promote infection. | [120] | |
| PsAvh262 | Luminal binding immunoglobulin proteins (BiPs) | Stabilizes luminal binding BiPs of the endoplasmic reticulum (ER)-to suppress ER stress-triggered cell death and promote infection. | [121] | |
| P. infestans | PITG_22798 | Direct target still unknown | Transient expression in Nicotiana benthamiana showed nucleus localization and triggered cell death. The host avirulence effector 3b (AVR3b) suppressed PITG_22798-induced cell death. | [122] |
| Pi17316 | A Yeast-2-Hybrid screen proposed interaction with the potato ortholog of the putative MAP3K VASCULAR HIGHWAY 1-interacting kinase (StVIK). | Pi17316 putatively acts in the StVIK signal transduction pathway to modulate plant immunity. More detailed studies are needed. | [123] | |
| P. capsici | PcAvh1 | Putatively interacts with the scaffolding subunit of protein phosphatase 2A (PP2Aa) | Interferes with pathways regulating plant immunity and growth. More detailed studies are needed. | [124] |
| P. parasitica | PPTG00121 (= PpE4) | Direct target still unknown | PpE4 is necessary for full virulence of P. parasitica, but further studies are needed to comprehend its mode of action. | [125] |
| PpRxLR2 | Direct target still unknown | Transient expression experiments in N. benthamiana showed the capacity of PpRxLR2 to suppress programmed cell death in cells challenged with the elicitin INF-1. | [126] |
| Peptide | Peptide Activities | Reference |
|---|---|---|
| Gm0025x00667(75–100) |
| [147] |
| Gm0026x00785(77–103) |
| [147] |
| GmOLPc |
| [156] |
| Gly m 4l |
| [159] |
| Gene ID # | Predicted Functions ^ | Expression Patterns ^ |
|---|---|---|
| Glyma.05G235200 | Stress response and antifungal | High expression in pods, seeds, and stems, relatively low in nodules |
| Glyma.08G042600 | Stress response and antifungal | High expression in stems, flowers, and leaves, relatively low in nodules |
| Glyma.09G223500 | Related to cell division | High expression in root hairs and shoot tips, relatively low in nodules |
| Glyma.10G133900 | Stress response and antifungal | High expression in roots and unopen flowers, relatively low in nodules |
| Glyma.13G094100 | Pathogenesis-related | High expression in nodules |
| Glyma.14G213600 | Stress response and antifungal | High expression in root hairs and nodules |
| Glyma.18G040800 | Stress response and antifungal | High expression in roots, stems, nodules |
| Glyma.19G168000 | Stress response and antifungal | High expression in nodules |
| Glyma.20G200200 | Stress response and antifungal | High expression in nodules |
| Association with Soybean Plant | Type of Microbe | Symbiotic Tissue | Strain | Target Microbe(s) | Reference(s) |
|---|---|---|---|---|---|
| Endophytic | Bacterium | Nodule | Paenibacillus sp. HKA-15 | Rhizoctonia bataticola | [21,22] |
| Root | Enterobacter ludwigii (ID 226) | Sclerotinia sclerotiorum, 61Xag | [23] | ||
| Root | Enterobacter sp. (ID 231) | Sclerotinia sclerotiorum | |||
| Root | Enterobacter sp. (ID 219) | Sclerotinia sclerotiorum | |||
| Stem | Agrobacterium tumefaciens/Rhizobium sp. (ID 179) | Sclerotinia sclerotiorum | |||
| Leaf | Kosakonia cowardii (ID 79) | Sclerotinia sclerotiorum | |||
| Root | Variovorax sp. (ID 41) | Sclerotinia sclerotiorum | |||
| Stem | Bacillus sp. (ID 152) | Sclerotinia sclerotiorum | |||
| Root | Burkholderia sp. (ID 137) | Sclerotinia sclerotiorum, Pseudomonas sojae, Rhizoctonia solani | |||
| Root | Burkholderia sp. (ID 130) | Sclerotinia sclerotiorum, Rhizoctonia solani | |||
| Root | Burkholderia sp. (ID 243) | Sclerotinia sclerotiorum, Pseudomonas sojae | |||
| Leaf | Pantoea vagans (ID 106) | Sclerotinia sclerotiorum | |||
| Leaf | Serratia marcescens (ID 245) | Sclerotinia sclerotiorum | |||
| Root | Enterobacter sp. (ID 110) | Sclerotinia sclerotiorum | |||
| Rhizospheric | Bacillus amyloliquefaciens BNM340 |
| [24] | ||
| Paenibacillus polymyxa BRF-1 | Rhizoctonia solani | [27] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, Y.-S.; Cheng, S.-S.; Gerhardt, A.; Cheung, M.-Y.; Contador, C.A.; Poon, L.-Y.W.; Lam, H.-M. Secretory Peptides as Bullets: Effector Peptides from Pathogens against Antimicrobial Peptides from Soybean. Int. J. Mol. Sci. 2020, 21, 9294. https://doi.org/10.3390/ijms21239294
Ku Y-S, Cheng S-S, Gerhardt A, Cheung M-Y, Contador CA, Poon L-YW, Lam H-M. Secretory Peptides as Bullets: Effector Peptides from Pathogens against Antimicrobial Peptides from Soybean. International Journal of Molecular Sciences. 2020; 21(23):9294. https://doi.org/10.3390/ijms21239294
Chicago/Turabian StyleKu, Yee-Shan, Sau-Shan Cheng, Aisha Gerhardt, Ming-Yan Cheung, Carolina A. Contador, Lok-Yiu Winnie Poon, and Hon-Ming Lam. 2020. "Secretory Peptides as Bullets: Effector Peptides from Pathogens against Antimicrobial Peptides from Soybean" International Journal of Molecular Sciences 21, no. 23: 9294. https://doi.org/10.3390/ijms21239294
APA StyleKu, Y.-S., Cheng, S.-S., Gerhardt, A., Cheung, M.-Y., Contador, C. A., Poon, L.-Y. W., & Lam, H.-M. (2020). Secretory Peptides as Bullets: Effector Peptides from Pathogens against Antimicrobial Peptides from Soybean. International Journal of Molecular Sciences, 21(23), 9294. https://doi.org/10.3390/ijms21239294









