An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae
Abstract
:1. Introduction
2. Defence-Related Proteins
3. Enzymes
4. Receptor-Like Proteins
5. Transcription Factors
6. Signal Transduction
7. Concluding Remarks
Funding
Conflicts of Interest
References
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity; and management of Verticillium Species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [Green Version]
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.K.; Wang, C.Y.; Xie, C.J.; Yang, X.Y. Advances in molecular mechanisms of Verticillium pathogenicity and plant resistance to Verticillium wilt. J. Henan. Agr. Sci. 2014, 43, 1–6. [Google Scholar]
- Zhang, J.; Fang, H.; Zhou, H.; Sanogo, S.; Ma, Z. Genetics; breeding; and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci. 2014, 54, 1289–1303. [Google Scholar] [CrossRef]
- Prieto, P.; Navarroraya, C.; Valverdecorredor, A.; Amyotte, S.G.; Dobinson, K.F.; Mercadoblanco, J.; Segura, A.; Preston, G.; Wit, P.D. Colonization process of olive tissues by Verticillium dahliae and its in planta interaction with the biocontrol root endophyte Pseudomonas fluorescens PICF7. Microb. Biotechnol. 2010, 2, 499–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Zhao, Y.L.; Jin, Y.; Zhang, T.; Guo, H.S. Colonization process of Arabidopsis thaliana roots by a green fluorescent protein-tagged isolate of Verticillium dahliae. Protein Cell 2014, 5, 94–98. [Google Scholar]
- Shaban, M.; Miao, Y.; Ullah, A.; Khan, A.H.; Ahmed, M.M.; Tabassum, M.A.; Zhu, L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 2018, 125, 193–204. [Google Scholar] [CrossRef]
- Temple, S.H.; DeVay, J.; Forrester, L.L. Temperature effects upon development and pathogenicity of defoliating and nondefoliating pathotypes of Verticillium dahliae in leaves of cotton plants. Phytopathology 1973, 63, 953–958. [Google Scholar] [CrossRef]
- Daayf, F. Verticillium wilts in crop plants: Pathogen invasion and host defence responses. Can. J. Plant Pathol. 2015, 37, 8–20. [Google Scholar] [CrossRef]
- Talboys, P.W. Association of tylosis and hyperplasia of the xylem with vascular invasion of the hop by Verticillium albo-atrum. Trans. Brit. Mycol. Soc. 1958, 41, 249–260. [Google Scholar] [CrossRef]
- Meyer, R.; Slater, V.; Dubery, I.A. A phytotoxic protein-lipopolysaccharide complex produced by Verticillium dahliae. Phytochemistry 1994, 35, 1449–1453. [Google Scholar] [CrossRef]
- Porter, C.; Green, R. Production of exotoxin in the genus Verticillium. Phytopathology 1952, 42, 472. [Google Scholar]
- Luo, X.; Xie, C.; Dong, J.; Yang, X.; Sui, A. Interactions between Verticillium dahliae and its host: Vegetative growth, pathogenicity, plant immunity. Appl. Microbiol. Biotechnol. 2014, 98, 6921–6932. [Google Scholar] [CrossRef] [PubMed]
- Belien, T.; Van, C.S.J.; Volckaert, G. Microbial endoxylanases, effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant-Microbe Interact. 2006, 19, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fradin, E.F.; Zhang, Z.; Rovenich, H.; Song, Y.; Liebrand, T.W.; Masini, L.; van den Berg, G.C.; Joosten, M.H.; Thomma, B.P. Functional analysis of the tomato immune receptor Ve1 through domain swaps with its non-functional homolog Ve2. PLoS ONE 2014, 9, e88208. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhang, Z.; Gao, Y.; Zhou, L.; Fang, L.; Chen, X.; Ning, Z.; Chen, T.; Guo, W.; Zhang, T. Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 2015, 5, 15048. [Google Scholar]
- Bu, B.; Qiu, D.; Zeng, H.; Guo, L.; Yuan, J.; Yang, X. A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 2014, 33, 461–470. [Google Scholar] [CrossRef]
- Liang, Y.; Cui, S.; Tang, X.; Zhang, Y.; Qiu, D.; Zeng, H.; Guo, L.; Yuan, J.; Yang, X. An asparagine-rich protein Nbnrp1 modulate Verticillium dahliae protein PevD1-induced cell death and disease resistance in Nicotiana benthamiana. Front. Plant Sci. 2018, 9, 303. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Khan, N.U.; Wang, N.; Yang, X.; Qiu, D. The protein elicitor PevD1 enhances resistance to pathogens and promotes growth in Arabidopsis. Int. J. Biol. Sci. 2016, 12, 931–943. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gao, Y.; Liang, Y.; Dong, Y.; Yang, X.; Qiu, D. Verticillium dahliae PevD1, an Alt a 1-like protein, targets cotton PR5-like protein and promotes fungal infection. J. Exp. Bot. 2019, 70, 613–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Zhu, T.; Han, L.; Liu, M.; Xu, M.; Liu, Y.; Han, D.; Qiu, D.; Gong, Q.; Liu, X. The asparagine-rich protein NRP interacts with the Verticillium effector PevD1 and regulates the subcellular localization of cryptochrome 2. J. Exp. Bot. 2017, 68, 3427–3440. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ma, X.; Yun, S.; Zhang, X.; Li, F.; Hou, Y. Necrotizing activity of Verticillium dahliae and Fusarium oxysporum f. sp. vasinfectum endopolygalacturonases in cotton. Plant Dis. 2017, 101, 1128–1138. [Google Scholar] [PubMed]
- Liu, N.; Zhang, X.; Sun, Y.; Wang, P.; Li, X.; Pei, Y.; Li, F.; Hou, Y. Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 2017, 7, 39840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazar, R.N.; Xu, X.; Kurosky, A.; Robb, J. Antagonistic function of the Ve R-genes in tomato. Plant Mol. Biol. 2018, 98, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, Y.M.; McKenna, J.A.; McGinness, B.S.; Hinch, J.; Poon, S.; Connelly, A.A.; Anderson, M.A.; Heath, R.L. Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J. Exp. Bot. 2014, 65, 1541–1550. [Google Scholar] [CrossRef]
- Li, Y.B.; Han, L.B.; Wang, H.Y.; Zhang, J.; Sun, S.T.; Feng, D.Q.; Yang, C.L.; Sun, Y.D.; Zhong, N.Q.; Xia, G.X. The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 2016, 170, 2392–2406. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y.; Wang, X.; Wang, W.; Li, Z.; Wu, J.; Wang, G.; Wu, L.; Zhang, G.; Ma, Z. HyPRP1 performs a role in negatively regulating cotton resistance to Verticillium dahliae via the thickening of cell walls and ROS accumulation. BMC Plant Boil. 2018, 18, 339. [Google Scholar]
- Pajerowska-Mukhtar, K.M.; Emerine, D.K.; Mukhtar, M.S. Tell me more: Roles of NPRs in plant immunity. Trends Plant Sci. 2013, 8, 402–411. [Google Scholar] [CrossRef]
- Parkhi, V.; Kumar, V.; Campbell, L.A.M.; Bell, A.A.; Rathore, K.S. Expression of Arabidopsis NPR1 in transgenic cotton confers resistance to non-defoliating isolates of Verticillium dahliae but not the defoliating isolates. J. Phytopathol. 2010, 158, 822–825. [Google Scholar] [CrossRef]
- Jue, D.; Liu, Y.; Shi, C.; Min, C.; Yang, Q. Cloning and characterization of a Solanum torvum NPR1 gene involved in regulating plant resistance to Verticillium dahliae. Acta Physiol. Plant. 2014, 36, 2999–3011. [Google Scholar]
- Yang, C.L.; Liang, S.; Wang, H.Y.; Han, L.B.; Wang, F.X.; Cheng, H.Q.; Wu, X.M.; Qu, Z.L.; Wu, J.H.; Xia, G.X. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol. Plant. 2015, 8, 399–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Li, C.; Zhou, X.; Liu, S.; Wu, F. Physiological response and sulfur metabolism of the V. dahliae-infected tomato plants in tomato/potato onion companion cropping. Sci. Rep. 2016, 6, 36445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munis, M.F.; Tu, L.; Deng, F.; Tan, J.; Xu, L.; Xu, S.; Long, L.; Zhang, X. A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem. Biophys. Res. Commun. 2010, 393, 38–44. [Google Scholar] [CrossRef]
- Li, F.; Shen, H.; Wang, M.; Fan, K.; Bibi, N.; Ni, M.; Yuan, S.; Wang, X. A synthetic antimicrobial peptide BTD-S expressed in Arabidopsis thaliana confers enhanced resistance to Verticillium dahliae. Mol. Genet. Genom. 2016, 291, 1647–1661. [Google Scholar] [CrossRef]
- Ni, M.; Zhao, Y.; Bibi, N.; Shao, M.; Yuan, S.; Fan, K.; Zhang, G.; Li, F.; Wang, X. A non-cyclic baboon θ-defensin derivative exhibiting antimicrobial activity against the phytopathogen Verticillium dahliae. Appl. Microbiol. Bio. 2013, 97, 2043–2052. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, C.; Mu, X.; Chao, L.; Ke, S.; Zhu, W.; Yang, Q. Cloning and expression of a wild eggplant cytochrome P450 gene, StoCYP77A2, involved in plant resistance to Verticillium dahliae. Plant Biotech. Rep. 2015, 9, 167–177. [Google Scholar]
- Shi, H.; Liu, Z.; Zhu, L.; Zhang, C.; Chen, Y.; Zhou, Y.; Li, F.; Li, X. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Gao, E.; Shaban, M.; Wang, Y.; Wang, H.; Nie, X.; Zhu, L. GhUMC1, a blue copper-binding protein; regulates lignin synthesis and cotton immune response. Biochem. Bioph. Res. Commun. 2018, 504, 75–81. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.; Tian, L.; Wang, G.; Zhang, X.; Wang, X.; Guo, W. Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci. Rep. 2016, 6, 29022. [Google Scholar] [CrossRef]
- Cheng, X.X.; Zhao, L.H.; Klosterman, S.J.; Feng, H.J.; Feng, Z.L.; Feng, W.; Shi, Y.Q.; Li, Z.F.; Zhu, H.Q. The endochitinase VDECH from Verticillium dahliae inhibits spore germination and activates plant defense responses. Plant Sci. 2017, 259, 12. [Google Scholar] [CrossRef]
- Han, L.B.; Li, Y.B.; Wang, F.X.; Wang, W.Y.; Liu, J.; Wu, J.H.; Zhong, N.Q.; Wu, S.J.; Jiao, G.L.; Wang, H.Y.; et al. The cotton apoplastic protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen Verticillium dahliae. Plant Cell 2019, 31, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Zhu, L.; Tu, L.; Liu, L.; Yuan, D.; Jin, L.; Long, L.; Zhang, X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wu, L.; Chen, B.; Zhao, J.; Cui, J.; Li, Z.; Yang, J.; Wu, L.; Zhang, G.; Ma, Z. The cotton laccase gene GHLAC15 enhances Verticillium wilt via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 2018, 20, 309–322. [Google Scholar] [CrossRef] [Green Version]
- Duan, X.; Zhang, Z.; Wang, J.; Zuo, K. Characterization of a novel cotton subtilase gene GbSBT1 in response to extracellular stimulations and its role in Verticillium resistance. PLoS ONE 2016, 11, e0153988. [Google Scholar] [CrossRef] [Green Version]
- Mustafa, R.; Hamza, M.; Kamal, H.; Mansoor, S.; Scheffler, J.; Amin, I. tobacco rattle virus-based silencing of enoyl-CoA reductase gene and its role in resistance against cotton wilt disease. Mol. Biotechnol. 2017, 59, 241–250. [Google Scholar] [CrossRef]
- Long, L.; Zhao, J.R.; Xu, F.C.; Yang, W.W.; Liao, P.; Gao, Y.; Gao, W.; Song, C.P. Silencing of GbANS reduces cotton resistance to Verticillium dahliae through decreased ROS scavenging during the pathogen invasion process. Plant Cell Tiss. Org. 2018, 135, 213–221. [Google Scholar] [CrossRef]
- Qin, T.; Liu, S.; Zhang, Z.; Sun, L.; He, X.; Lindsey, K.; Zhu, L.; Zhang, X. GhCyP3 improves the resistance of cotton to Verticillium dahliae by inhibiting the E3 ubiquitin ligase activity of GhPUB17. Plant Mol. Biol. 2019, 99, 379–393. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Zhu, L.F.; Xu, L.; Gao, W.H.; Sun, L.Q.; Liu, L.L.; Zhang, X.L. Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell Proteomics 2013, 12, 3690–3703. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Nguyen, C.T.; Liang, Y.; Cao, Y.; Stacey, G. Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal Behav. 2013, 8, e22598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.; Tanaka, K.; Zhang, X.C.; Son, G.H.; Brechenmacher, L.; Nguyen, T.H.; Stacey, G. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012, 160, 396–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Wang, G.; Wang, J.; Li, Y.; Tian, L.; Wang, X.; Guo, W. The lysin motif-containing proteins, Lyp1, Lyk7 and LysMe3, play important roles in chitin perception and defense against Verticillium dahliae in cotton. BMC Plant Biol. 2017, 17, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Liu, T.; Ding, B.; Li, F.; Wang, Q.; Qian, S.; Ye, F.; Chen, T.; Yang, Y.; Wang, J.; et al. Two Lysin-motif receptor kinases; Gh-LYK1 and Gh-LYK2; contribute to resistance against Verticillium wilt in upland cotton. Front. Plant Sci. 2017, 8, 2133. [Google Scholar] [CrossRef] [Green Version]
- Nazar, R.N.; Castroverde, C.D.M.; Xu, X.; Kurosky, A.; Robb, J. Wounding induces tomato Ve1 R-gene expression. Planta 2019, 249, 1779–1797. [Google Scholar] [CrossRef]
- Kawchuk, L.M.; Hachey, J.; Lynch, D.R.; Kulcsar, F.; van Rooijen, G.; Waterer, D.R.; Robertson, A.; Kokko, E.; Byers, R.; Howard, R.J.; et al. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 6511–6515. [Google Scholar] [CrossRef] [Green Version]
- Castroverde, C.D.; Xu, X.; Blaya, F.J.; Nazar, R.N.; Robb, J. Epistatic influence in tomato Ve1-mediated resistance. Plant Biol. 2017, 19, 843–847. [Google Scholar] [CrossRef]
- De Jonge, R.; van Esse, H.P.; Maruthachalam, K.; Bolton, M.D.; Santhanam, P.; Saber, M.K.; Zhang, Z.; Usami, T.; Lievens, B.; Subbarao, K.V.; et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 5110–5115. [Google Scholar] [CrossRef] [Green Version]
- Castroverde, C.D.; Xu, X.; Nazar, R.N.; Robb, J. Biotic factors that induce the tomato Ve1 R-gene. Plant Sci. 2017, 265, 61–69. [Google Scholar] [CrossRef]
- Gayoso, C.; Pomar, F.; Novo-Uzal, E.; Merino, F.; de Ilarduya, O.M. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 2010, 10, 232. [Google Scholar] [CrossRef] [Green Version]
- Fradin, E.F.; Abd-El-Haliem, A.; Masini, L.; van den Berg, G.C.; Joosten, M.H.; Thomma, B.P. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 2011, 156, 2255–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fradin, E.F.; Zhang, Z.; Juarez-Ayala, J.C.; Castroverde, C.D.; Nazar, R.N.; Robb, J.; Liu, C.M.; Thomma, B.P. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009, 150, 320–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Liu, L.; Wang, Y.; Valkenburg, D.J.; Zhang, X.; Zhu, L.; Thomma, B. Transfer of tomato immune receptor Ve1 confers Ave1-dependent Verticillium resistance in tobacco and cotton. Plant Biotechnol. J. 2018, 16, 638–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Fradin, E.; de Jonge, R.; van Esse, H.P.; Smit, P.; Liu, C.M.; Thomma, B.P. Optimized agroinfiltration and virus-induced gene silencing to study Ve1-mediated Verticillium resistance in tobacco. Mol. Plant Microbe. Interact. 2013, 26, 182–190. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; van Esse, H.P.; van Damme, M.; Fradin, E.F.; Liu, C.M.; Thomma, B.P. Ve1-mediated resistance against Verticillium does not involve a hypersensitive response in Arabidopsis. Mol. Plant Pathol. 2013, 14, 719–727. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Z.; Seidl, M.F.; Majer, A.; Jakse, J.; Javornik, B.; Thomma, B.P. Broad taxonomic characterization of Verticillium wilt resistance genes reveals an ancient origin of the tomato Ve1 immune receptor. Mol. Plant Pathol. 2017, 18, 195–209. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, Y.; Chen, T.; Yu, W.; Liu, T.; Li, H.; Fan, X.; Ren, Y.; Shen, D.; Liu, L. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS ONE 2012, 7, e51091. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Chen, T.; Ling, X.; Ma, Z. Gbvdr6, a gene encoding a receptor-like protein of cotton (Gossypium barbadense), confers resistance to Verticillium wilt in Arabidopsis and upland cotton. Front. Plant Sci. 2017, 8, 2272. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.J.; Zhang, H.P.; Chen, Q.Z.; Tang, N.; Wang, L.K.; Wang, R.F.; Zhang, B.L. Molecular cloning and analysis of a receptor-like promoter of Gbvdr3 gene in sea island cotton. Genet. Mol. Res. 2016, 15, gmr.15028636. [Google Scholar] [CrossRef]
- Chen, T.; Kan, J.; Yang, Y.; Ling, X.; Chang, Y.; Zhang, B. A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae. Plant Physiol. Biochem. 2006, 98, 101–111. [Google Scholar] [CrossRef]
- Tang, J.; Lin, J.; Yang, Y.; Chen, T.; Ling, X.; Zhang, B.; Chang, Y. Ectopic expression of a Ve homolog VvVe gene from Vitis vinifera enhances defense response to Verticillium dahliae infection in tobacco. Gene 2016, 576, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhu, Y.; Xie, C.; Jue, D.; Hong, Y.; Chen, M.; Hubdar, A.K.; Yang, Q. Transgenic potato plants expressing StoVe1 exhibit enhanced resistance to Verticillium dahliae. Plant Mol. Biol. Rep. 2012, 30, 1032–1039. [Google Scholar] [CrossRef]
- Chai, Y.; Zhao, L.; Liao, Z.; Sun, X.; Zuo, K.; Zhang, L.; Wang, S.; Tang, K. Molecular cloning of a potential Verticillium dahliae resistance gene SlVe1 with multi-site polyadenylation from Solanum licopersicoides. DNA Seq. 2003, 14, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Chai, Y.; Wang, J.; Lin, J.; Sun, X.; Sun, C.; Zuo, K.; Tang, K. cDNA cloning and characterization of the Ve homologue gene StVe from Solanum torvum Swartz. DNA Seq. 2004, 15, 88–95. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets; including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Ellendorff, U.; Fradin, E.F.; Thomma, B.P.; De Jonge, R. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 2008, 60, 591–602. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Sun, Q.; Jiang, H.; Zhu, X.; Mo, J.; Long, L.; Xiang, L.; Xie, Y.; Shi, Y.; Yuan, Y. Identification of novel microRNAs in the Verticillium wilt-resistant upland cotton variety KV-1 by high-throughput sequencing. SpringerPlus 2014, 3, 564. [Google Scholar] [CrossRef] [Green Version]
- He, X.H.; Shi, M.; Sun, Q.; Cai, Y.F. Expression profile analysis and target gene prediction of three conserved MicroRNAs in resistant Gossypium hirsutum cv. KV-1 responding to Verticillium dahlia. In Advanced Materials Research; Trans Tech Publications: Zurich, Switzerland, 2014; pp. 441–445. [Google Scholar]
- Yang, L.; Mu, X.; Liu, C.; Cai, J.; Shi, K.; Zhu, W.; Yang, Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integ. Plant. Boil. 2015, 57, 1078–1088. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, M.; Li, N.; Wang, H.; Qiu, P.; Pei, L.; Xu, Z.; Wang, T.; Gao, E.; Liu, J. Long noncoding RNA s involve in resistance to Verticillium dahliae; a fungal disease in cotton. Plant Biotechnol. J. 2018, 16, 1172–1185. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.Q.; Han, L.B.; Yang, C.L.; Wu, X.M.; Zhong, N.Q.; Wu, J.H.; Wang, F.X.; Wang, H.Y.; Xia, G.X. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 2016, 67, erw016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Wang, T.; Zhu, W.; Wang, Y.; Zhu, L. GhHB12, a HD-ZIP I transcription factor, negatively regulates the cotton resistance to Verticillium dahliae. Int. J. Mol. Sci. 2018, 19, 3997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Wang, K.; Sun, L.; Xing, H.; Wang, S.; Li, L.; Chen, S.; Guo, H.S.; Zhang, J. The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. eLife 2018, 7, e34902. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Xu, L.; Lindsey, K.; Zhang, X.; Zhu, L. Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J. Integr. Plant Biol. 2016, 58, 503–513. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Sun, L.; Wassan, G.M.; He, X.; Shaban, M.; Zhang, L.; Zhu, L.; Zhang, X. GbSOBIR1 confers Verticillium wilt resistance by phosphorylating the transcriptional factor GbbHLH171 in Gossypium barbadense. Plant Biotechnol. J. 2019, 17, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Gong, Q.; Yang, Z.; Wang, X.; Butt, H.I.; Chen, E.; He, S.; Zhang, C.; Zhang, X.; Li, F. Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 2017, 17, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, Q.; Yang, Z.; Chen, E.; Sun, G.; He, S.; Butt, H.I.; Zhang, C.; Zhang, X.; Yang, Z.; Du, X.; et al. A Phi-class glutathione S-transferase gene for Verticillium wilt resistance in Gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 2018, 59, 275–289. [Google Scholar] [CrossRef]
- Mo, H.J.; Sun, Y.X.; Zhu, X.L.; Wang, X.F.; Zhang, Y.; Yang, J.; Yan, G.J.; Ma, Z.Y. Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 2016, 243, 1023–1039. [Google Scholar] [CrossRef]
- Mo, H.; Wang, X.; Zhang, Y.; Yang, J.; Ma, Z. Cotton ACAULIS5 is involved in stem elongation and the plant defense response to Verticillium dahliae through thermospermine alteration. Plant Cell Rep. 2015, 34, 1975–1985. [Google Scholar] [CrossRef]
- Mo, H.; Wang, X.; Zhang, Y.; Zhang, G.; Zhang, J.; Ma, Z. Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 2015, 83, 962–975. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Pei, Y.; Sun, Y.; Liu, N.; Wang, P.; Liu, D.; Ge, X.; Li, F.; Hou, Y. A cotton cyclin-dependent kinase E confers resistance to Verticillium dahliae mediated by jasmonate-responsive pathway. Front. Plant Sci. 2018, 9, 642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; He, X.; Luo, X.; Xu, L.; Liu, L.; Min, L.; Jin, L.; Zhu, L.; Zhang, X. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 2014, 166, 2179–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Zhu, L.; Zhang, X.; Guan, Q.; Xiao, S.; Min, L.; Zhang, X. GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol. 2018, 178, 876–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.X.; Ma, Y.P.; Yang, C.L.; Zhao, P.M.; Yao, Y.; Jian, G.L.; Luo, Y.M.; Xia, G.X. Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 2011, 11, 4296–4309. [Google Scholar] [CrossRef]
- Pantelides, I.S.; Tjamos, S.E.; Paplomatas, E.J. Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae. Mol. Plant Pathol. 2010, 11, 191–202. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Zhao, G.; Zhao, J.; Du, H.; He, X.; Zhang, H. A novel Gossypium barbadense ERF transcription factor, GbERFb, regulation host response and resistance to Verticillium dahliae in tobacco. Physiol. Mol. Biol. Plant. 2017, 23, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Jin, L.; Miao, Y.; He, X.; Hu, Q.; Guo, K.; Zhu, L.; Zhang, X. An ethylene response-related factor; GbERF1-like; from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef]
- Liu, Y.; Xin, L.; Lu, L.; Wang, W.; Quan, S.; Bo, L.; Wang, C.; Cheng, J.; Zhang, Y.; Xie, Y. GbABR1 is associated with Verticillium wilt resistance in cotton. Biologia. 2018, 73, 449–457. [Google Scholar] [CrossRef]
- Li, T.; Ma, X.; Li, N.; Zhou, L.; Liu, Z.; Han, H.; Gui, Y.; Bao, Y.; Chen, J.; Dai, X. Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L). Plant Biotechnol. J. 2017, 15, 1520–1532. [Google Scholar] [CrossRef] [Green Version]
- Li, N.Y.; Ma, X.F.; Short, D.P.G.; Li, T.G.; Zhou, L.; Gui, Y.J.; Kong, Z.Q.; Zhang, D.D.; Zhang, W.Q.; Li, J.J.; et al. The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 2018, 19, 1466–1479. [Google Scholar] [CrossRef] [Green Version]
- Li, N.Y.; Zhou, L.; Zhang, D.D.; Klosterman, S.J.; Li, T.G.; Gui, Y.J.; Kong, Z.Q.; Ma, X.F.; Short, D.P.G.; Zhang, W.Q.; et al. Heterologous expression of the cotton NBS-LRR gene GbaNA1 enhances Verticillium wilt resistance in Arabidopsis. Front. Plant Sci. 2018, 9, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, X.; Li, Y.; Wu, L.; Zhou, H.; Zhang, G.; Ma, Z. Ectopic expression of a novel Ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis. Plant Cell Rep. 2013, 32, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Gao, H.; Zhai, W.; Shi, J.; Zhang, M.; Zhang, W.; Jian, G.; Zhang, M.; Qi, F. Subtle regulation of cotton resistance to Verticillium wilt mediated by MAPKK family members. Plant Sci. 2018, 272, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wheeler, T.; Li, Z.; Kenerley, C.M.; He, P.; Shan, L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
 , phosphorylation; CRY2, cryptochrome 2; ET, ethylene; GCC-box, ethylene-responsive element binding factor associated amphiphilic repression domain; GSH, phi-class glutathione; HDTF1: homeodomain transcription factor gene 1; JA, jasmonic acid; JAZ1, Jasmonate Zim-domain1; NBS-LRR, nucleotide-binding site leucine-rich repeat; PevD1, an elicitor from V. dahliae; SARD1, the Arabidopsis master immune regulator; SA: salicylic acid; Spd, spermidine; Spm, spermine; ROS, reactive oxygen species; SA, salicylic acid; VdSCP41, a secretory protein from V. dahliae [7,16,17,18,19,20,21,22]. Red lines represent negative regulation and green lines represent positive regulation.
, phosphorylation; CRY2, cryptochrome 2; ET, ethylene; GCC-box, ethylene-responsive element binding factor associated amphiphilic repression domain; GSH, phi-class glutathione; HDTF1: homeodomain transcription factor gene 1; JA, jasmonic acid; JAZ1, Jasmonate Zim-domain1; NBS-LRR, nucleotide-binding site leucine-rich repeat; PevD1, an elicitor from V. dahliae; SARD1, the Arabidopsis master immune regulator; SA: salicylic acid; Spd, spermidine; Spm, spermine; ROS, reactive oxygen species; SA, salicylic acid; VdSCP41, a secretory protein from V. dahliae [7,16,17,18,19,20,21,22]. Red lines represent negative regulation and green lines represent positive regulation.
 , phosphorylation; CRY2, cryptochrome 2; ET, ethylene; GCC-box, ethylene-responsive element binding factor associated amphiphilic repression domain; GSH, phi-class glutathione; HDTF1: homeodomain transcription factor gene 1; JA, jasmonic acid; JAZ1, Jasmonate Zim-domain1; NBS-LRR, nucleotide-binding site leucine-rich repeat; PevD1, an elicitor from V. dahliae; SARD1, the Arabidopsis master immune regulator; SA: salicylic acid; Spd, spermidine; Spm, spermine; ROS, reactive oxygen species; SA, salicylic acid; VdSCP41, a secretory protein from V. dahliae [7,16,17,18,19,20,21,22]. Red lines represent negative regulation and green lines represent positive regulation.
, phosphorylation; CRY2, cryptochrome 2; ET, ethylene; GCC-box, ethylene-responsive element binding factor associated amphiphilic repression domain; GSH, phi-class glutathione; HDTF1: homeodomain transcription factor gene 1; JA, jasmonic acid; JAZ1, Jasmonate Zim-domain1; NBS-LRR, nucleotide-binding site leucine-rich repeat; PevD1, an elicitor from V. dahliae; SARD1, the Arabidopsis master immune regulator; SA: salicylic acid; Spd, spermidine; Spm, spermine; ROS, reactive oxygen species; SA, salicylic acid; VdSCP41, a secretory protein from V. dahliae [7,16,17,18,19,20,21,22]. Red lines represent negative regulation and green lines represent positive regulation.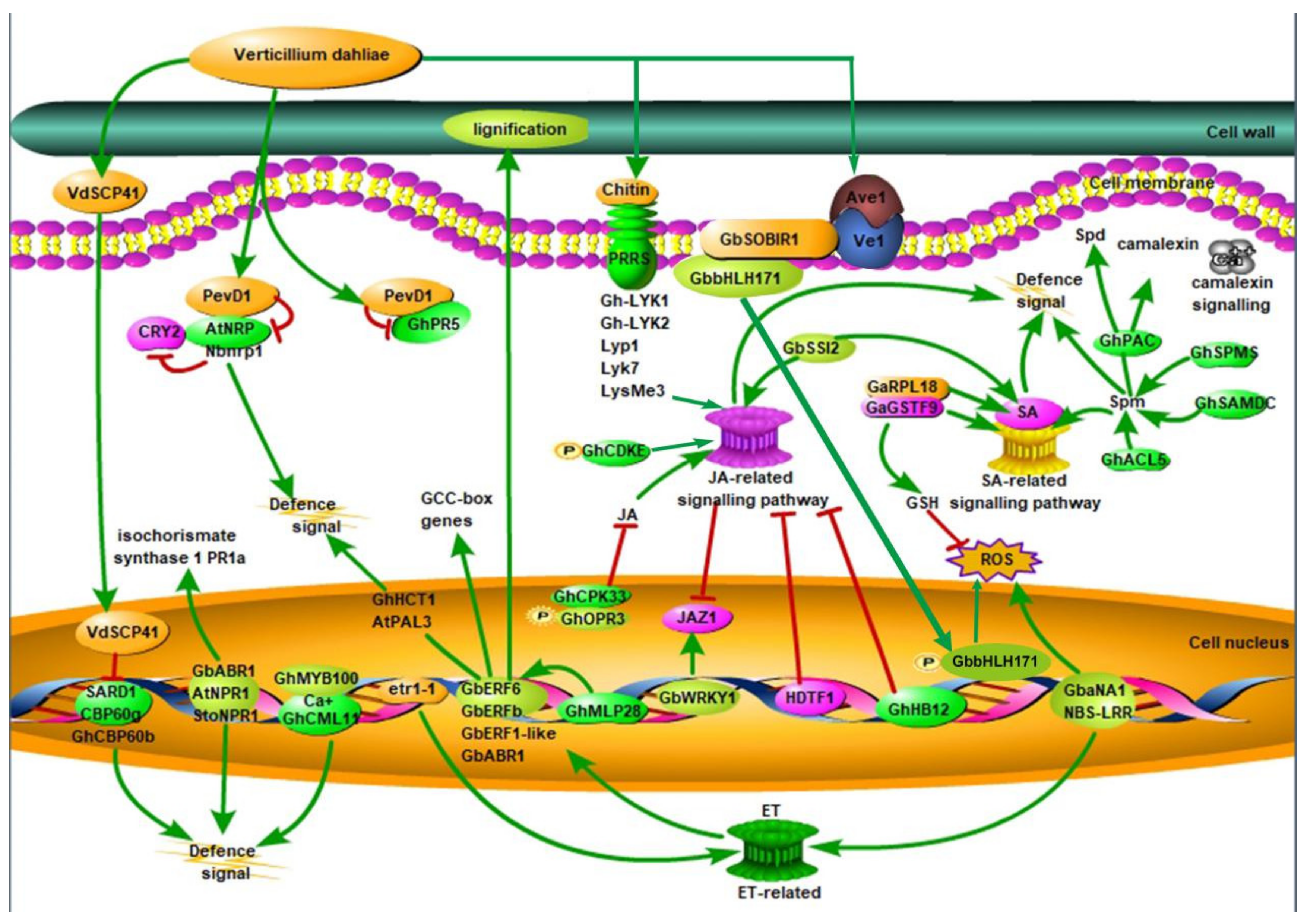
| Classification | Protein (Gene) Name | Annotation | Host | Resistance Mechanism | References |
|---|---|---|---|---|---|
| Defence-related proteins | PGIP | plant defence protein | Ck, Gh | inhibit fungal polygalacturonase activity | [24] |
| NaD1 | plant defensin | Na | antifungal activity | [26] | |
| GbNRX1 | apoplastic thioredoxin protein | Gb | apoplastic immune response and scavenge ROS | [27] | |
| GbHyPRP1 | proline-rich protein | Gb | thickening cell walls and ROS accumulation | [28] | |
| AtNPR1 | non-expressor of Pr1 | At | upregulating expression of ICS1 and PR1a | [30] | |
| GhMLP28 | defence-related major latex protein | St | enhance GhERF6 activity | [32] | |
| GbTLP1 | thaumatin-like protein | Gb | secondary cell wall development | [34] | |
| BTD-S | synthetic defensin derivative | Synthetic | antifungal activity | [35,36] | |
| StoCYP77A2 | cytochrome P450 | Nt | synthesis of antimicrobial compounds | [37] | |
| Enzymes | Chi28 | class IV chitinase subfamily | Gh, Gb | degrade the fungal cell wall | [42] |
| GhPMEI3 | pectin methylesterases | Gh | degrade the fungal cell wall | [43] | |
| GhLAC15 | laccase | Gh | lignification of the cell wall | [45] | |
| GbSBT1 | a subtilase gene | Gb | activating defence-related genes expressionn | [46] | |
| GhECR | enoyl-CoA reductase | Gh | production of very long chain fatty acids | [47] | |
| GbANS | anthocyanidin synthase | Gb | regulating biosynthesis of anthocyanins | [48] | |
| GhPUB17 | U-box E3 ubiquitin ligase | Gh | negatively regulating immunity | [49] | |
| Receptor-like proteins | GhDIR1 | putative dirigent protein | Gh | lignification of the cell wall | [38] |
| GhUMC1 | umecyanin-like protein | Gh | [39] | ||
| Lyp1, Lyk7,LysMe3 | lysin-motif receptor kinases | Gb | recognize chitin, receptor-mediated endocytosis-like signals and leucine zipper, enhance the expression of the JA/ET signalling pathway-related genes, increase the expressions of defence-related genes | [53] | |
| Gh-LYK1,Gh-LYK2 | Gh | [54] | |||
| Ve1 and Ve2 | cell-surface glycoproteins | Sl | [16,25] | ||
| GbSOBIR1 | defence-related receptor-like kinases | Gb | [85] | ||
| Gbvdr3, Gbvdr6 Gbve1, VvVe, StVe StoVe1,SlVe1, GbRLK | Ve1 homologues | Gb, Vv St, Sl | [17,64,68,69,70,71,72,73,74] | ||
| miR482e | miR482 superfamily | St | target disease-resistance proteins with NBS and LRR motifs | [79] | |
| Transcription factors | GhHB12 | HD-ZIP I transcription factor | Gh | suppressing JA-response genes | [82] |
| GhMYB108 | plant MYB transcription factors | Gh | enhance defence signalling molecules | [81] | |
| CBP60g and SARD1 | plant-specific transcription factors | At | regulating SA signalling | [83] | |
| Signal transduction | GaRPL18 | ribosomal protein L18 | Ga | mediate resistance by SA-signalling | [86] |
| GaGSTF9 | phi-class glutathione S-transferase | Ga | regulating ROS via catalytic reduction of glutathione | [87] | |
| GhSAMDC,GhSPMS | S-adenosylmethionine decarboxylase | Gh | regulating Spm biosynthesis by SA-signalling | [88] | |
| GhPAO | polyamine oxidase | Gh | regulating Spm and camalexin signalling | [90] | |
| GhCDKE | cyclin-dependent kinase | Gh | enhance plant resistance by JA pathway | [91] | |
| HDTF1 | homeodomain transcription factor | Gh | activation of JA-mediated signalling | [84] | |
| GbWRKY1 | regulator mediating | Gb | activating JAZ1 expression | [92] | |
| GbSSI2,GbCAD1 | regulating signal pathways | Gb | activating JA-mediated signalling | [50] | |
| GbaNA1 | NBS-LRR protein | Gb | regulating ROS and ET signalling pathway | [100,101] | |
| ETR1 | ET receptor | At | activation and increased accumulation of defence proteins | [95] | |
| GbERF1-like | ET response-related factor | Gb | positive regulator in lignin synthesis | [97] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, R.; Li, J.; Xie, C.; Jian, W.; Yang, X. An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae. Int. J. Mol. Sci. 2020, 21, 1120. https://doi.org/10.3390/ijms21031120
Song R, Li J, Xie C, Jian W, Yang X. An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae. International Journal of Molecular Sciences. 2020; 21(3):1120. https://doi.org/10.3390/ijms21031120
Chicago/Turabian StyleSong, Ranran, Junpeng Li, Chenjian Xie, Wei Jian, and Xingyong Yang. 2020. "An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae" International Journal of Molecular Sciences 21, no. 3: 1120. https://doi.org/10.3390/ijms21031120
APA StyleSong, R., Li, J., Xie, C., Jian, W., & Yang, X. (2020). An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae. International Journal of Molecular Sciences, 21(3), 1120. https://doi.org/10.3390/ijms21031120




