RNA-Targeting CRISPR–Cas Systems and Their Applications
Abstract
1. Introduction
2. Overview of RNA-Targeting CRISPR–Cas Systems
2.1. Type III (Cmr/Csm) Systems
2.2. Type VI (Cas13) Systems.
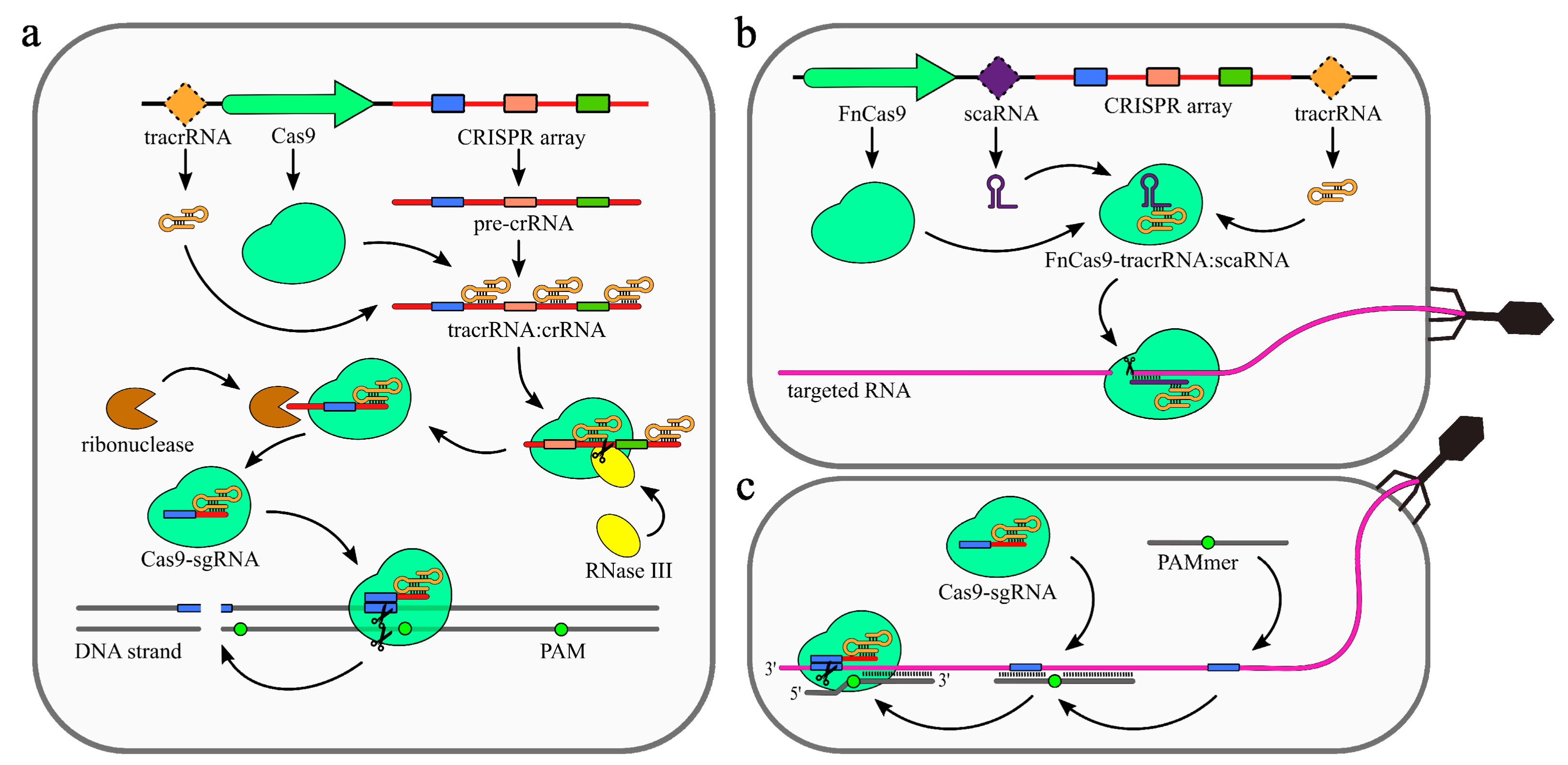
2.3. Type II (Cas9) Systems
3. Applications Based on RNA-Targeting CRISPR–Cas Systems
3.1. Type II(Cas9)-Based Applications
3.2. Type VI(Cas13)-Based Applications
3.3. Type III-Based Applications
4. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno-Associated Virus |
| ADAR | adenosine deaminase acting on RNA |
| CARF | CRISPR-associated Rossman fold |
| Cas | CRISPR-associated (protein) |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| crRNA | CRISPR RNA |
| dCas | catalytycally inactive Cas protein |
| HEPN | higher eukaryotes and prokaryotes nucleotide binding |
| KRAB | The Krüppel-associated box domain |
| MGEs | Mobile Genetic Elements |
| PAM | Protospacer-Associated Motif |
| PAMer | PAM-presenting oligonucleotide |
| PFS | Protospacer Flanking Sequence |
| REPAIR | RNA editing for programmable A-to-I replacement |
| RNP | ribonucleoprotein |
| scaRNA | small, CRISPR–Cas-associated RNA |
| sgRNA | single guide RNA |
| SHERLOCK | Specific High-Sensitivity Enzymatic Reporter unLOCKing |
| tracrRNA | Trans-activating crRNA |
References
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.; Keren, L.; Wurtzel, O.; Amitai, G.; Sorek, R. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet. Tig. 2010, 26, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Vercoe, R.B.; Chang, J.T.; Dy, R.L.; Taylor, C.; Gristwood, T.; Clulow, J.S.; Richter, C.; Przybilski, R.; Pitman, A.R.; Fineran, P.C. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013, 9, e1003454. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; van Embden, J.D.A.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012, 40, 5569–5576. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Reeks, J.; Naismith, J.H.; White, M.F. CRISPR interference: A structural perspective. Biochem. J. 2013, 453, 155–166. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Hale, C.R.; Majumdar, S.; Elmore, J.; Pfister, N.; Compton, M.; Olson, S.; Resch, A.M.; Glover, C.V.C.; Graveley, B.R.; Terns, R.M.; et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 2012, 45, 292–302. [Google Scholar] [CrossRef]
- Zhang, J.; Rouillon, C.; Kerou, M.; Reeks, J.; Brugger, K.; Graham, S.; Reimann, J.; Cannone, G.; Liu, H.; Albers, S.-V.; et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell 2012, 45, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Burmistrz, M.; Rodriguez Martinez, J.I.; Krochmal, D.; Staniec, D.; Pyrc, K. Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) RNAs in the Porphyromonas gingivalis CRISPR-Cas I-C System. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [PubMed]
- Semenova, E.; Jore, M.M.; Datsenko, K.A.; Semenova, A.; Westra, E.R.; Wanner, B.; van der Oost, J.; Brouns, S.J.J.; Severinov, K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. USA 2011, 108, 10098–10103. [Google Scholar] [CrossRef] [PubMed]
- Jore, M.M.; Lundgren, M.; van Duijn, E.; Bultema, J.B.; Westra, E.R.; Waghmare, S.P.; Wiedenheft, B.; Pul, U.; Wurm, R.; Wagner, R.; et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011, 18, 529–536. [Google Scholar] [CrossRef]
- Spilman, M.; Cocozaki, A.; Hale, C.; Shao, Y.; Ramia, N.; Terns, R.; Terns, M.; Li, H.; Stagg, S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol. Cell 2013, 52, 146–152. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Lander, G.C.; Zhou, K.; Jore, M.M.; Brouns, S.J.J.; van der Oost, J.; Doudna, J.A.; Nogales, E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 2011, 477, 486–489. [Google Scholar] [CrossRef]
- Van der Oost, J.; Westra, E.R.; Jackson, R.N.; Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. [Google Scholar] [CrossRef]
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61. [Google Scholar] [CrossRef]
- Burmistrz, M.; Pyrć, K. CRISPR-Cas Systems in Prokaryotes. Pol. J. Microbiol. 2015, 64, 193–202. [Google Scholar] [CrossRef]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant. Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Classification and Nomenclature of CRISPR-Cas Systems: Where from Here? Cris. J. 2018, 1, 325–336. [Google Scholar] [CrossRef]
- Brezgin, S.; Kostyusheva, A.; Kostyushev, D.; Chulanov, V. Dead Cas Systems: Types, Principles, and Applications. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Khosravi, S.; Ishii, T.; Dreissig, S.; Houben, A. Application and prospects of CRISPR/Cas9-based methods to trace defined genomic sequences in living and fixed plant cells. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Valenti, M.T.; Serena, M.; Carbonare, L.D.; Zipeto, D. CRISPR/Cas system: An emerging technology in stem cell research. World J. Stem Cells 2019, 11, 937–956. [Google Scholar] [CrossRef] [PubMed]
- Banan, M. Recent advances in CRISPR/Cas9-mediated knock-ins in mammalian cells. J. Biotechnol. 2019, 308, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Khanzadi, M.N.; Khan, A.A. CRISPR/Cas9: Nature’s gift to prokaryotes and an auspicious tool in genome editing. J. Basic Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Klompe, S.E.; Vlot, M.; van der Oost, J.; Staals, R.H.J. Shooting the messenger: RNA-targetting CRISPR-Cas systems. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Staals, R.H.J.; Zhu, Y.; Taylor, D.W.; Kornfeld, J.E.; Sharma, K.; Barendregt, A.; Koehorst, J.J.; Vlot, M.; Neupane, N.; Varossieau, K.; et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol. Cell 2014, 56, 518–530. [Google Scholar] [CrossRef]
- Benda, C.; Ebert, J.; Scheltema, R.A.; Schiller, H.B.; Baumgärtner, M.; Bonneau, F.; Mann, M.; Conti, E. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell 2014, 56, 43–54. [Google Scholar] [CrossRef]
- Nickel, L.; Ulbricht, A.; Alkhnbashi, O.S.; Förstner, K.U.; Cassidy, L.; Weidenbach, K.; Backofen, R.; Schmitz, R.A. Cross-cleavage activity of Cas6b in crRNA processing of two different CRISPR-Cas systems in Methanosarcina mazei Gö1. RNA Biol. 2018, 16, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.W.; Zhu, Y.; Staals, R.H.J.; Kornfeld, J.E.; Shinkai, A.; van der Oost, J.; Nogales, E.; Doudna, J.A. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 2015, 348, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Inanaga, H.; Sato, C.; Numata, T. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell 2015, 58, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.R.; Cocozaki, A.; Li, H.; Terns, R.M.; Terns, M.P. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014, 28, 2432–2443. [Google Scholar] [CrossRef]
- Tamulaitis, G.; Kazlauskiene, M.; Manakova, E.; Venclovas, Č.; Nwokeoji, A.O.; Dickman, M.J.; Horvath, P.; Siksnys, V. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell 2014, 56, 506–517. [Google Scholar] [CrossRef]
- Estrella, M.A.; Kuo, F.-T.; Bailey, S. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 2016, 30, 460–470. [Google Scholar] [CrossRef]
- Maniv, I.; Jiang, W.; Bikard, D.; Marraffini, L.A. Impact of Different Target Sequences on Type III CRISPR-Cas Immunity. J. Bacteriol. 2016, 198, 941–950. [Google Scholar] [CrossRef]
- Kazlauskiene, M.; Tamulaitis, G.; Kostiuk, G.; Venclovas, Č.; Siksnys, V. Spatiotemporal Control of Type III-A CRISPR-Cas Immunity: Coupling DNA Degradation with the Target RNA Recognition. Mol. Cell 2016, 62, 295–306. [Google Scholar] [CrossRef]
- Niewoehner, O.; Garcia-Doval, C.; Rostøl, J.T.; Berk, C.; Schwede, F.; Bigler, L.; Hall, J.; Marraffini, L.A.; Jinek, M. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 2017, 548, 543–548. [Google Scholar] [CrossRef]
- Kazlauskiene, M.; Kostiuk, G.; Venclovas, Č.; Tamulaitis, G.; Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 2017, 357, 605–609. [Google Scholar] [CrossRef]
- Sheppard, N.F.; Glover, C.V.C.; Terns, R.M.; Terns, M.P. The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 2016, 22, 216–224. [Google Scholar] [PubMed]
- Hatoum-Aslan, A.; Samai, P.; Maniv, I.; Jiang, W.; Marraffini, L.A. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J. Biol. Chem. 2013, 288, 27888–27897. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Makarova, K.S.; Burroughs, A.M.; Koonin, E.V.; Aravind, L. Comprehensive analysis of the HEPN superfamily: Identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 2013, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Niewoehner, O.; Jinek, M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 2016, 22, 318–329. [Google Scholar]
- Goldberg, G.W.; McMillan, E.A.; Varble, A.; Modell, J.W.; Samai, P.; Jiang, W.; Marraffini, L.A. Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts. Nat. Commun. 2018, 9, 61. [Google Scholar] [CrossRef]
- Jiang, W.; Samai, P.; Marraffini, L.A. Degradation of Phage Transcripts by CRISPR-Associated RNases Enables Type III CRISPR-Cas Immunity. Cell 2016, 164, 710–721. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Burstein, D.; Knott, G.J.; Doudna, J.A. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol. Cell 2017, 66, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Ma, J.; Li, Z.; You, L.; Wang, J.; Wang, M.; Zhang, X.; Wang, Y. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 2017, 170, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Anantharaman, V.; Aravind, L.; Koonin, E.V. Live virus-free or die: Coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct 2012, 7, 40. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef]
- Sampson, T.R.; Saroj, S.D.; Llewellyn, A.C.; Tzeng, Y.-L.; Weiss, D.S. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 2013, 497, 254–257. [Google Scholar] [CrossRef]
- Dugar, G.; Leenay, R.T.; Eisenbart, S.K.; Bischler, T.; Aul, B.U.; Beisel, C.L.; Sharma, C.M. CRISPR RNA-Dependent Binding and Cleavage of Endogenous RNAs by the Campylobacter jejuni Cas9. Mol. Cell 2018, 69, 893–905. [Google Scholar] [CrossRef]
- Rousseau, B.A.; Hou, Z.; Gramelspacher, M.J.; Zhang, Y. Programmable RNA Cleavage and Recognition by a Natural CRISPR-Cas9 System from Neisseria meningitidis. Mol. Cell 2018, 69, 906–914. [Google Scholar] [CrossRef]
- Strutt, S.C.; Torrez, R.M.; Kaya, E.; Negrete, O.A.; Doudna, J.A. RNA-dependent RNA targeting by CRISPR-Cas9. Elife 2018, 7, e32724. [Google Scholar] [CrossRef]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; East-Seletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Zebec, Z.; Manica, A.; Zhang, J.; White, M.F.; Schleper, C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014, 42, 5280–5288. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, H.T.; Cooper, J.C.; Lo, L.; Potter, J.; Terns, R.M.; Terns, M.P. Programmable type III-A CRISPR-Cas DNA targeting modules. PLoS ONE 2017, 12, e0176221. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef]
- Nelles, D.A.; Fang, M.Y.; O’Connell, M.R.; Xu, J.L.; Markmiller, S.J.; Doudna, J.A.; Yeo, G.W. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell 2016, 165, 488–496. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Price, A.A.; Sampson, T.R.; Ratner, H.K.; Grakoui, A.; Weiss, D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 6164–6169. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant. Biotechnol. J. 2018, 16, 1415–1423. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Programmable Inhibition and Detection of RNA Viruses Using Cas13. Mol. Cell 2019, 76, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; He, C.; Dickinson, B.C. Targeted m 6 A Reader Proteins to Study Epitranscriptomic Regulation of Single RNAs. J. Am. Chem. Soc. 2018, 140, 11974–11981. [Google Scholar] [CrossRef] [PubMed]
- Batra, R.; Nelles, D.A.; Pirie, E.; Blue, S.M.; Marina, R.J.; Wang, H.; Chaim, I.A.; Thomas, J.D.; Zhang, N.; Nguyen, V.; et al. Elimination of Toxic Microsatellite Repeat Expansion RNA by RNA-Targeting Cas9. Cell 2017, 170, 899.e10–912.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Konermann, S.; Brideau, N.J.; Lotfy, P.; Wu, X.; Novick, S.J.; Strutzenberg, T.; Griffin, P.R.; Hsu, P.D.; Lyumkis, D. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell 2018, 175, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.R.; Sheppard, N.F.; Ramia, N.; Deighan, T.; Li, H.; Terns, R.M.; Terns, M.P. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 2016, 30, 447–459. [Google Scholar] [CrossRef]
- Rouillon, C.; Zhou, M.; Zhang, J.; Politis, A.; Beilsten-Edmands, V.; Cannone, G.; Graham, S.; Robinson, C.V.; Spagnolo, L.; White, M.F. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell 2013, 52, 124–134. [Google Scholar] [CrossRef]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar] [CrossRef]
- Wang, F.; Wang, L.; Zou, X.; Duan, S.; Li, Z.; Deng, Z.; Luo, J.; Lee, S.Y.; Chen, S. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 2019, 37, 708–729. [Google Scholar] [CrossRef]


| Application | CRISPR–Cas Type (Method Name) | Key Features | Reference |
|---|---|---|---|
| RNA knockdown | Cas9 | programmable and specific RNA cleavage targeted RNA sequence does not require PAM motif as it is delivered by PAMer DNA corresponding to targeted RNA sequence is not affected | [61] |
| Cas13a/b/c | requires Cas13 variant and CRISPR array for activity multiple transcripts can be targeted simultaneously usually PFS is required in targeted RNA possible off target cleavage | [47,49,51,53,55] | |
| Csm/Cmr | utilizes three nuclease activities: specific RNA cleavage, non-specific ssDNA cleavage, non-specific RNA degradation complex structure of effector complex limits its application | [62,63] | |
| Cas13d (CasRx) | the smallest Cas13 variant known to date possible to be delivered by Adeno-Associated Virus (AAV) no PFS required good efficiency and specificity | [64] | |
| RNA imaging and tracking | dCas9 | dCas9 is fused to a fluorescent protein and nuclear localization signal enables recognition of endogenous, untagged, unmodified mRNA PAM is delivered by PAMers method does not disturb the functional mRNA and protein levels | [65] |
| dCas13a | catalytically inactive Cas13 is fused to GFP, zinc finger and The Krüppel-associated box domain (KRAB) domain enables recognition of endogenous, untagged, unmodified mRNA negative feedback system based on self-targeting zinc finger and repression of KRAB domain reduces background signal from unbound protein | [55] | |
| RNA editing | Cas13b (REPAIR) | dCas13b is fused to ADAR2 domain enables to convert selected adenosine to inosine small size enables delivery via AAV | [66] |
| nucleic acid detection | Cas13a (SHERLOCKv1) | requires Cas13a and quenched fluorescent RNA reporter specific recognition of the target triggers nonspecific RNA cleavage that activates the fluorophore rapid, inexpensive RNA detection single-base mismatch specificity attomolar sensitivity can be utilized in detection of specific strains of pathogens, genotyping, and identification of mutations | [67] |
| Cas13 + Csm6 (SHERLOCKv2) | extended version of SHERLOCKv1 Csm6 strengthens the signal multiplex detection of up to four targets 3.5-fold increased sensitivity | [68] | |
| splicing alteration | dCas13d (dCasRx) | catalytically inactive CasRx is fused with negative splice factor hnRNPa1 exon skipping can be achieved by targeting cis-acting motifs like: intronic branch point, splice acceptor site, exon, splice donor site | [64] |
| resistance against RNA viruses | FnCas9 | PAM independent cytosol localization of FnCas9 is necessary to inhibit RNA viruses nuclear localization is needless, thus potential off-targets of FnCas9 are limited nuclease activity is not required production of RNA virus resistant plants possible multiplex targeting | [69,70] |
| Cas13a | provides stable immunity to viruses in plants codon optimization of Cas13a is required for proper protein functioning possible off-target activity multiplex targeting possible | [71,72] | |
| induction of apoptosis | Cas13a | programmed cell death is triggered by nonspecific RNA degradation in response to infection by cellular pathogen | [47] |
| regulation of gene expression | Cas13b | catalytically inactive Cas13 is fused with eukaryotic RNA-modifying enzyme N6-methyladenosine (m6A) variant fused to YTHDF1 protein can trigger assembly of translation machinery variant fused to YTHDF2 protein can induce RNA degradation | [73] |
| specific RNA isolation | dCas9 | catalytically inactive Cas9 is fused with biotin suitable for purification of tagless RNA transcripts affinity chromatography purification of biotinylated dCas9-gRNA-targetRNA on streptavidin resin RNA sequence of interest does not require PAM motif as it is delivered by PAMer | [61] |
| elimination of repetitive sequences | Cas9 | dCas9 is fused to the PIN RNA endonuclease no PAMer is required small size enables delivery via AAV potential application in treating genetic diseases connected with toxic microsatellite expansions | [74] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burmistrz, M.; Krakowski, K.; Krawczyk-Balska, A. RNA-Targeting CRISPR–Cas Systems and Their Applications. Int. J. Mol. Sci. 2020, 21, 1122. https://doi.org/10.3390/ijms21031122
Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-Targeting CRISPR–Cas Systems and Their Applications. International Journal of Molecular Sciences. 2020; 21(3):1122. https://doi.org/10.3390/ijms21031122
Chicago/Turabian StyleBurmistrz, Michal, Kamil Krakowski, and Agata Krawczyk-Balska. 2020. "RNA-Targeting CRISPR–Cas Systems and Their Applications" International Journal of Molecular Sciences 21, no. 3: 1122. https://doi.org/10.3390/ijms21031122
APA StyleBurmistrz, M., Krakowski, K., & Krawczyk-Balska, A. (2020). RNA-Targeting CRISPR–Cas Systems and Their Applications. International Journal of Molecular Sciences, 21(3), 1122. https://doi.org/10.3390/ijms21031122





