Secretory Production of Functional Grouper Type I Interferon from Epinephelus septemfasciatus in Escherichia coli and Bacillus subtilis
Abstract
1. Introduction
2. Results and Discussion
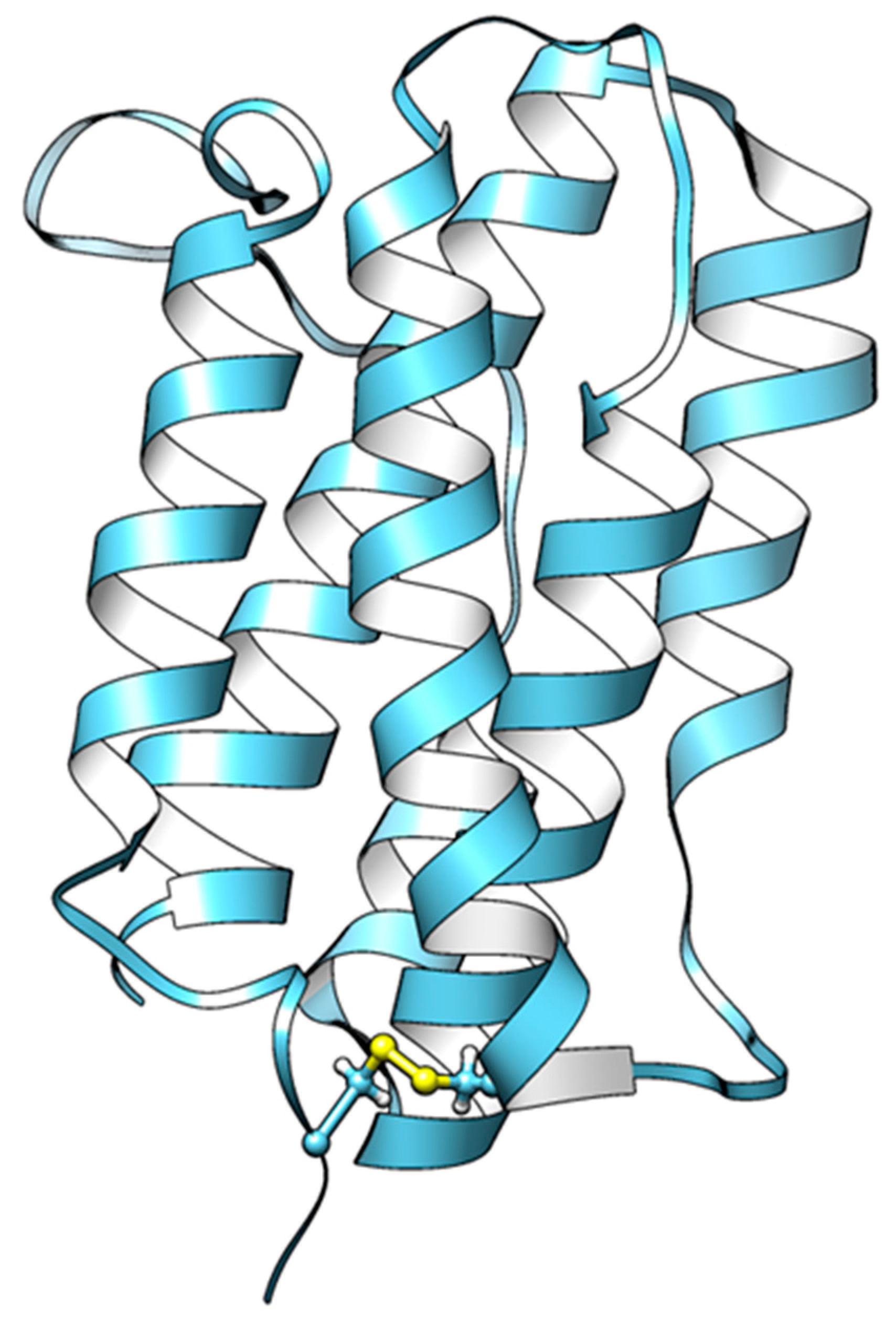
2.1. Multiple Sequence Alignment and Protein Modeling of Grouper IFN-α
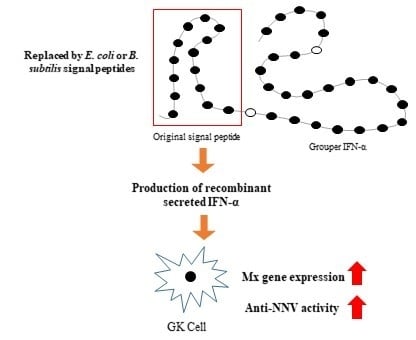
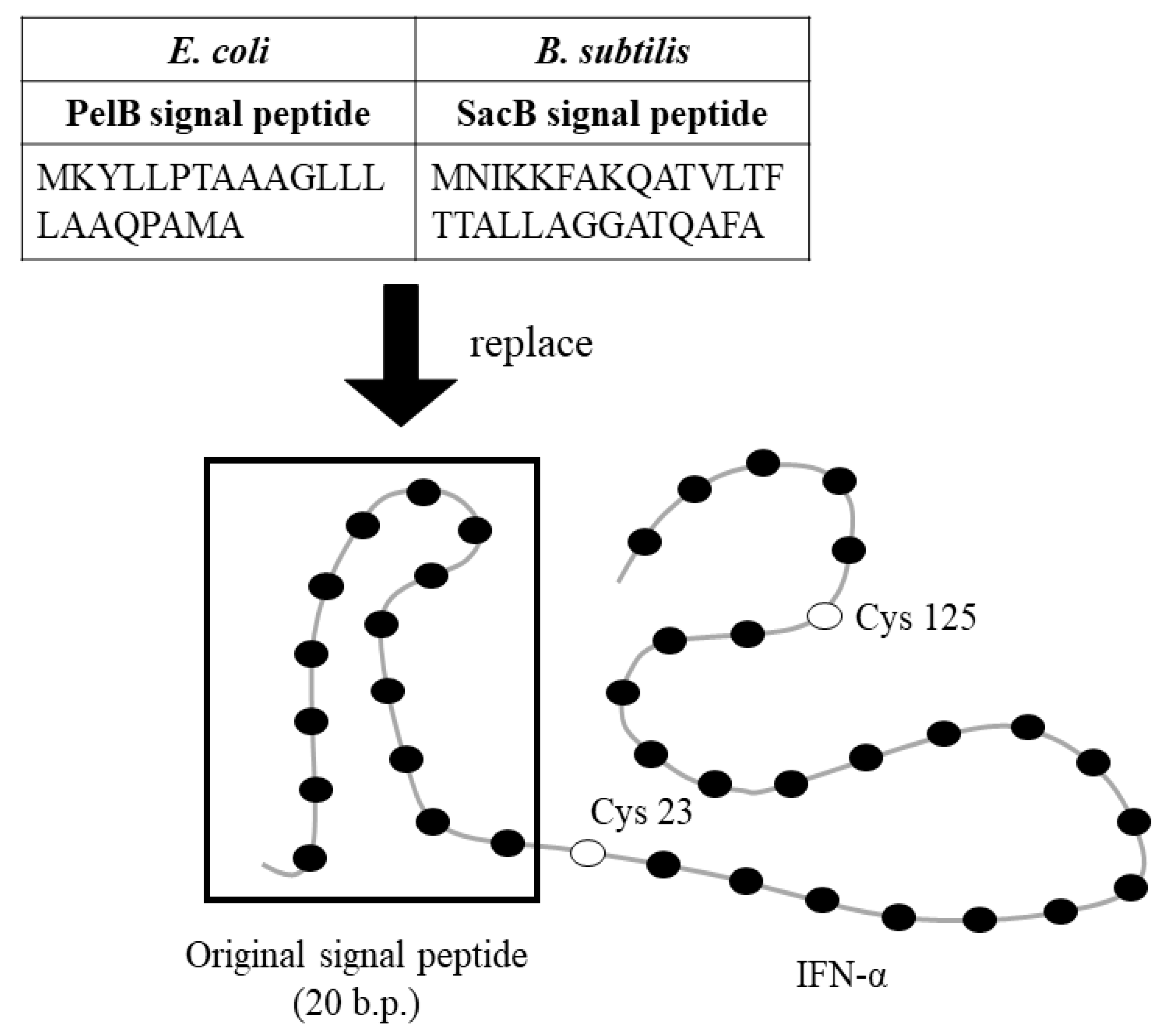
2.2. Strategy for Production of Secreted IFN-α in E. Coli and B. Subtilis
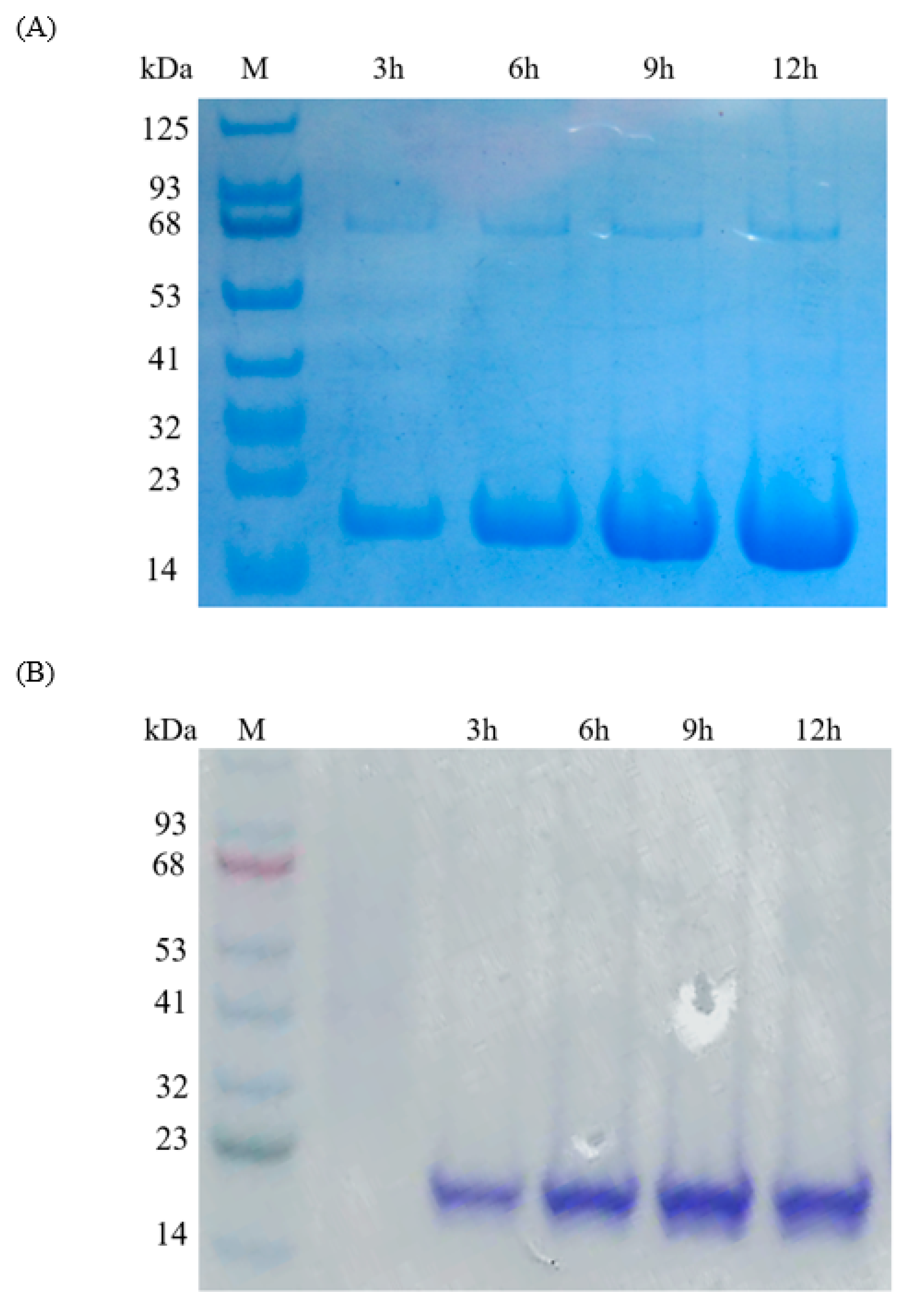
2.3. Effect of Inducing Time for IFN-α Overexpression
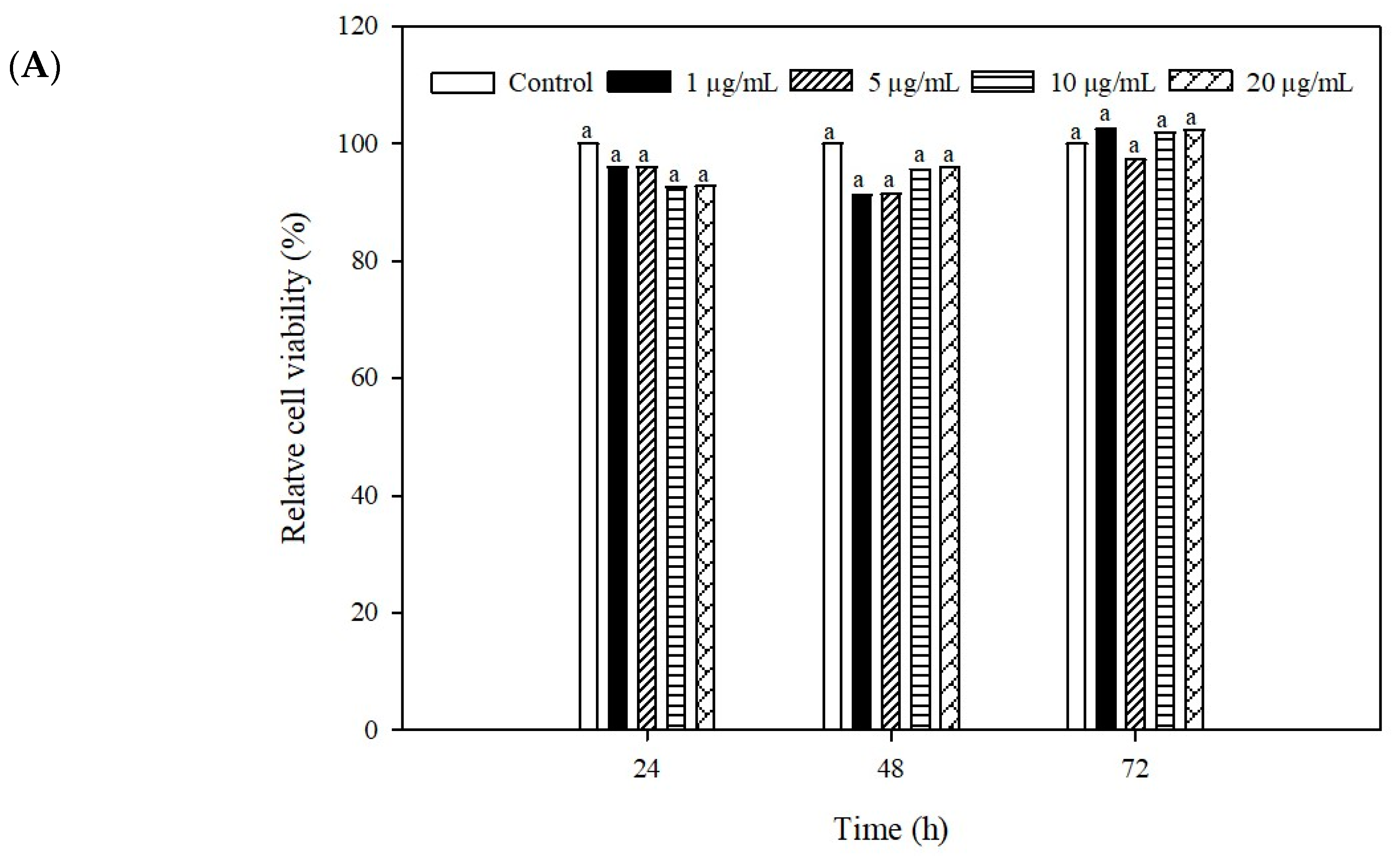
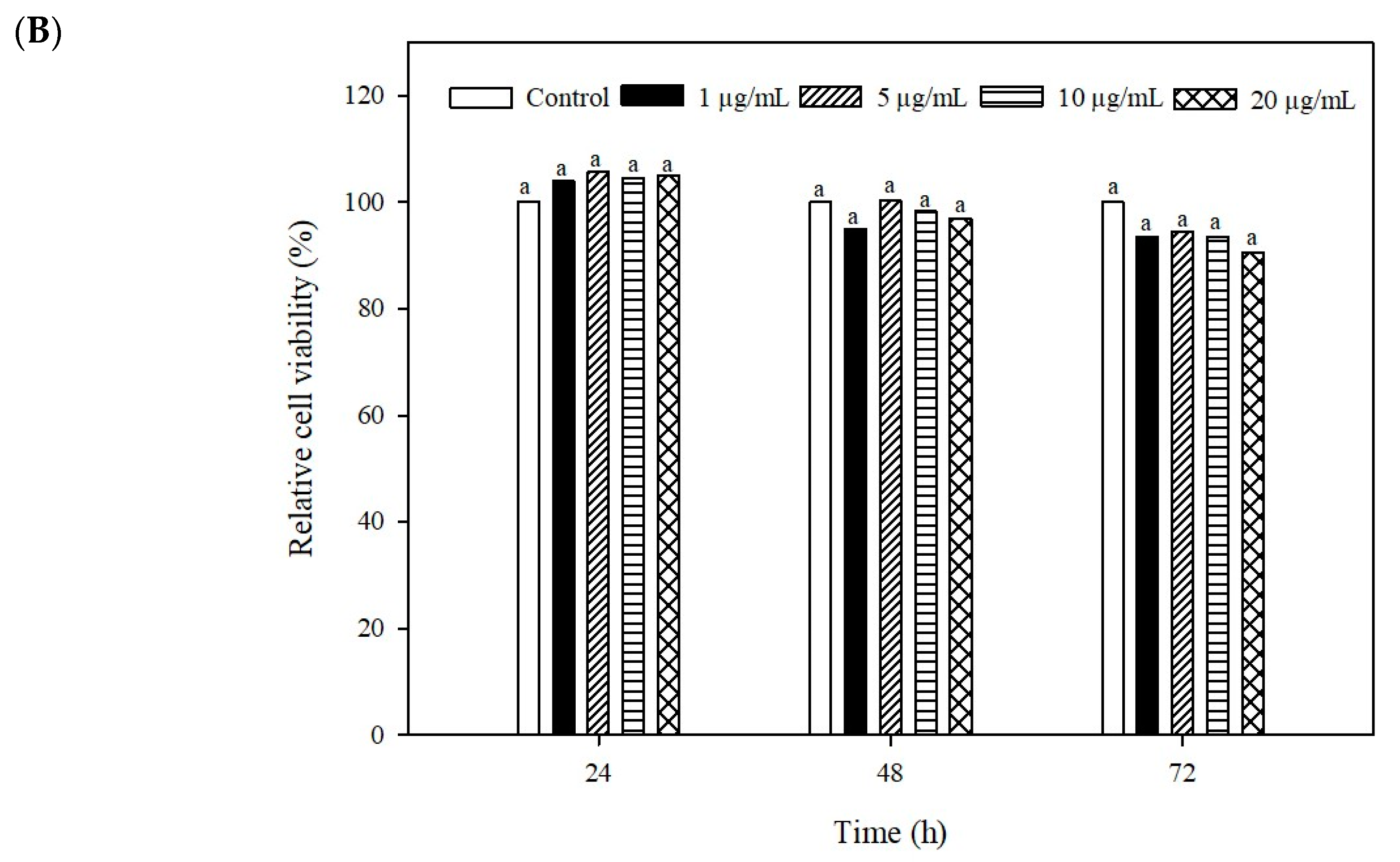
2.4. Cytotoxicity of the Recombinant IFN-α Against Grouper Head Kidney Cell
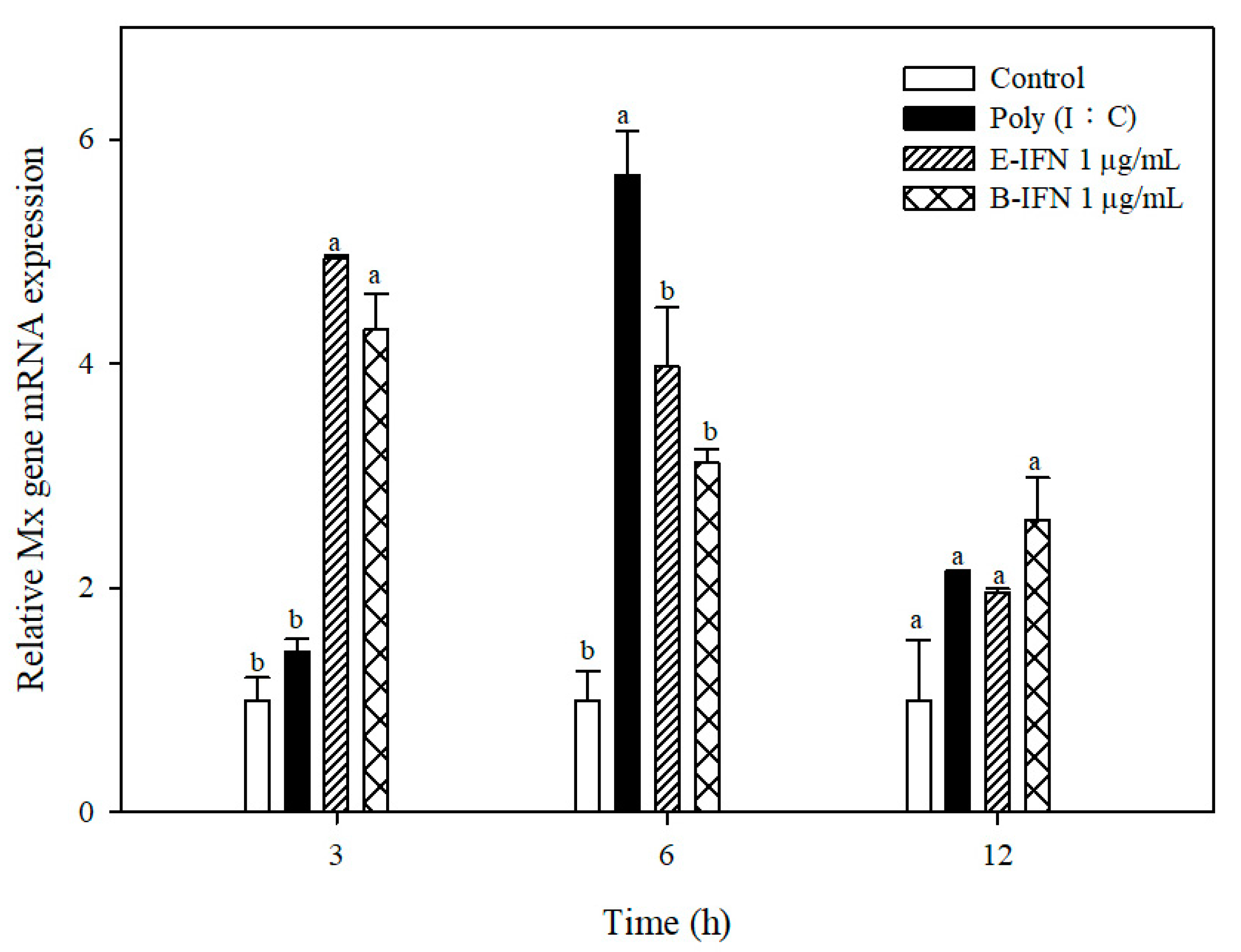
2.5. Recombinant IFN-α Stimulated Mx Gene Expression
2.6. Antiviral Effects of Recombinant IFN in GK Cells
3. Materials and Methods
3.1. Virus, Bacterial Strains, and Media
3.2. Cloning of ifn
3.3. IFN-α Overexpression and Purification from E. coli and B. Subtilis
3.4. Protein Quantification, Western Blot, and Peptide Mass Fingerprinting
3.5. Mx Gene Expression in the Cells
3.6. Cytotoxicity Test and Antiviral Effect of IFN
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hegde, A.; Chen, C.L.; Qin, Q.W.; Lam, T.J.; Sin, Y.M. Characterization, pathogenicity and neutralization studies of a nervous necrosis virus isolated from grouper, Epinephelus tauvina, in Singapore. Aquaculture 2002, 213, 55–72. [Google Scholar] [CrossRef]
- Ransangan, J.; Manin, B.O. Genome analysis of Betanodavirus from cultured marine fish species in Malaysia. Vet. Microbiol. 2012, 156, 16–44. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.P.; Chung, C.L.; Hung, Y.F.; Lai, Y.S.; Chiou, P.P.; Lu, M.W.; Kong, Z.L. Comparison of the responses of different recombinant fish type I interferons against betanodavirus infection in grouper. Fish Shellfish Immunol. 2016, 49, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Ucko, M.; Colorni, A.; Diamant, A. Nodavirus infections in Israeli mariculture. J. Fish Dis. 2004, 27, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, T.; Furuhashi, M.; Nagai, T.; Nakai, T.; Muroga, K. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl. Environ. Microbiol. 1997, 63, 1633–1636. [Google Scholar] [CrossRef]
- Nishizawa, T.; Mori, K.; Furuhashi, M.; Nakai, T.; Furusawa, I.; Muroga, K. Comparison of the coat protein genes of five fish nodaviruses, the causative agents of viral nervous necrosis in marine fish. J. Gen. Virol. 1995, 76, 1563–1569. [Google Scholar] [CrossRef]
- Costa, J.Z.; Thompson, K.D. Understanding the interaction between Betanodavirus and its host for the development of prophylactic measures for viral encephalopathy and retinopathy. Fish Shellfish Immunol. 2016, 53, 35–49. [Google Scholar] [CrossRef]
- Chi, S.C.; Lo, B.J.; Lin, S.C. Characterization of grouper nervous necrosis virus (GNNV). J. Fish Dis. 2001, 24, 3–13. [Google Scholar] [CrossRef]
- Munday, B.L.; Kwang, J.; Moody, N. Betanodavirus infections of teleost fish: A review. J. Fish Dis. 2002, 25, 127–142. [Google Scholar] [CrossRef]
- Oh, M.J.; Gye, H.J.; Nishizawa, T. Assessment of the sevenband grouper Epinephelus septemfasciatus with a live nervous necrosis virus (NNV) vaccine at natural seawater temperature. Vaccine 2013, 31, 2025–2027. [Google Scholar] [CrossRef]
- Tanaka, S.; Mori, K.; Arimoto, M.; Iwamoto, T.; Nakai, T. Protective immunity of sevenband grouper, Epinephelus septemfasciatus Thunberg, against experimental viral nervous necrosis. J. Fish Dis. 2001, 24, 15–22. [Google Scholar] [CrossRef]
- Liu, W.; Hsu, C.H.; Chang, C.Y.; Chen, H.H.; Lin, C.S. Immune response against grouper nervous necrosis virus by vaccination of virus-like particles. Vaccine 2006, 24, 6282–6287. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.H.; Wu, S.Y.; Lin, C.H. Oral immunization with cell-free self-assembly virus-like particles against orange-spotted grouper nervous necrosis virus in grouper larvae, Epinephelus coioides. Vet. Immunol. Immunopathol. 2018, 197, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Kung, C.W.; Chen, J.Y. Antiviral activity by fish antimicrobial peptides of epinecidin-1 and hepcidin 1-5 against nervous necrosis virus in medaka. Peptides 2010, 31, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T.; Ueda, Y.; Ito, K.; Miura, C.; Yamashita, H.; Miura, T.; Tozawa, Y. Anti-viral effects of interferon administration on sevenband grouper, Epinephelus septemfasciatus. Fish Shellfish Immunol. 2011, 30, 1064–1071. [Google Scholar] [CrossRef]
- Kitao, Y.; Kono, T.; Sakai, M. Characterization and expression analysis of type I interferon in common carp Cyprinus carpio L. Comp. Biochem. Physiol. A 2009, 154, S19. [Google Scholar] [CrossRef]
- Li, Z.S.; Xu, X.P.; Huang, L.C.; Wu, J.M.; Lu, Q.X.; Xiang, Z.M.; Liao, J.J.; Weng, S.P.; Yu, X.Q.; He, J.G. Administration of recombinant IFN1 protects zebrafish (Danio rerio) from ISKNV infection. Fish Shellfish Immunol. 2010, 29, 399–406. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Zhang, H.X.; Zhang, J.H.; Chen, W.H.; Ruan, X.F.; Xia, C. In vitro effects of recombinant zebrafish IFN on spring viremia of carp virus and infectious hematopoietic necrosis virus. J. Interferon Cytokine Res. 2006, 26, 256–259. [Google Scholar] [CrossRef]
- Samuel, C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef]
- Uze, G.; Monneron, D. IL-28 and IL-29: Newcomers to the interferon family. Biochimie 2007, 89, 729–734. [Google Scholar] [CrossRef]
- Lee, S.B.; Esteban, M. The interferon-induced double-stranded RNA-activated human p68 protein kinase inhibits the replication of vaccinia virus. Virology 1993, 193, 1037–1041. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Yu, H.P.; Chang, B.L.; Tang, W.C.; Liao, C.L.; Lin, Y.L. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J. Immunol. 2009, 183, 8035–8043. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Kochs, G. Interferon-induced mx proteins: Dynamin-like GTPases with antiviral activity. Traffic 2002, 3, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.W.; Chao, Y.M.; Guo, T.C.; Santi, N.; Evensen, O.; Kasani, S.K.; Hong, J.R.; Wu, J.L. The interferon response is involved in nervous necrosis virus acute and persistent infection in zebrafish infection model. Mol. Immunol. 2008, 45, 1146–1152. [Google Scholar] [CrossRef]
- Guan, Y.X.; Pan, H.X.; Gao, Y.G.; Yao, S.J.; Cho, M.G. Refolding and purification of recombinant human interferon-gamma expressed as inclusion bodies in Escherichia coli using size exclusion chromatography. Biotechnol. Bioprocess Eng. 2005, 10, 122–127. [Google Scholar] [CrossRef]
- Reddy, P.K.; Reddy, S.G.; Narala, V.R.; Majee, S.S.; Konda, S.; Gunwar, S.; Reddy, R.C. Increased yield of high purity recombinant human interferon-gamma utilizing reversed phase column chromatography. Protein Expr. Purif. 2007, 52, 123–130. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Hamming, O.J.; Lutfalla, G.; Levraud, J.P.; Hartmann, R. Crystal structure of Zebrafish interferons I and II reveals conservation of type I interferon structure in vertebrates. J. Virol. 2011, 85, 8181–8187. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- Feige, M.J. Oxidative Folding of Proteins: Basic Principles, Cellular Regulation and Engineering; Royal Society of Chemistry: Croydon, UK, 2018. [Google Scholar]
- Morehead, H.; Johnston, P.D.; Wetzel, R. Roles of the 29-138 disulfide bond of subtype A of human alpha interferon in its antiviral activity and conformational stability. Biochemistry 1984, 23, 2500–2507. [Google Scholar] [CrossRef] [PubMed]
- Hastings, K.T.; Cresswell, P. Disulfide eeduction in the endocytic pathway: Immunological functions of gamma-interferon-inducible lysosomal thiol reductase. Antioxid. Redox Signal. 2011, 15, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Kongton, K.; Phongdara, A.; Tonganunt-Srithaworn, M.; Wanna, W. Molecular cloning and expression analysis of the interferon-gamma-inducible lysosomal thiol reductase gene from the shrimp Penaeus monodon. Mol. Biol. Rep. 2011, 38, 3463–3470. [Google Scholar] [CrossRef]
- Braun, A.; Kwee, L.; Labow, M.A.; Alsenz, J. Protein aggregates seem to play a key role among the parameters influencing the antigenicity of interferon alpha (IFN-alpha) in normal and transgenic mice. Pharmaceut. Res. 1997, 14, 1472–1478. [Google Scholar] [CrossRef]
- Berkmen, M. Production of disulfide-bonded proteins in Escherichia coli. Protein Expr. Purif. 2012, 82, 240–251. [Google Scholar] [CrossRef]
- Diress, A.; Lorbetskie, B.; Larocque, L.; Li, X.G.; Alteen, M.; Isbrucker, R.; Girard, M. Study of aggregation, denaturation and reduction of interferon alpha-2 products by size-exclusion high-performance liquid chromatography with fluorescence detection and biological assays. J. Chromatogr. A 2010, 1217, 3297–3306. [Google Scholar] [CrossRef]
- Meima, R.; Eschevins, C.; Fillinger, S.; Bolhuis, A.; Hamoen, L.W.; Dorenbos, R.; Quax, W.J.; van Dijl, J.M.; Provvedi, R.; Chen, I.; et al. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J. Biol. Chem. 2002, 277, 6994–7001. [Google Scholar] [CrossRef]
- Chen, Y.M.; Kuo, C.E.; Chen, G.R.; Kao, Y.T.; Zou, J.; Secombes, C.J.; Chen, T.Y. Functional analysis of an orange-spotted grouper (Epinephelus coioides) interferon gene and characterisation of its expression in response to nodavirus infection. Dev. Comp. Immunol. 2014, 46, 117–128. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Beacham, I.R. Periplasmic enzymes in gram-negative bacteria. Int. J. Biochem. 1979, 10, 877–883. [Google Scholar] [CrossRef]
- Rathore, A.S.; Bilbrey, R.E.; Steinmeyer, D.E. Optimization of an osmotic shock procedure for isolation of a protein product expressed in E. coli. Biotechnol. Prog. 2003, 19, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Kakeshita, H.; Kageyama, Y.; Ara, K.; Ozaki, K.; Nakamura, K. Enhanced extracellular production of heterologous proteins in Bacillus subtilis by deleting the C-terminal region of the SecA secretory machinery. Mol. Biotechnol. 2010, 46, 320. [Google Scholar] [CrossRef][Green Version]
- Yoon, S.O.; Hirata, R.D.C.; da Silva, A.C.R.; Nguyen, N.Y.; Hirata, M.H. Cloning and expression of soluble recombinant protein comprising the extracellular domain of the human type I interferon receptor 2c subunit (IFNAR-2c) in E. coli. Biotechnol. Lett. 2002, 24, 1443–1448. [Google Scholar] [CrossRef]
- Zhu, F.F.; Wang, Q.; Pu, H.F.; Gu, S.S.; Luo, L.; Yin, Z.M. Optimization of soluble human interferon-gamma production in Escherichia coli using SUMO fusion partner. World J. Microb. Biot. 2013, 29, 319–325. [Google Scholar] [CrossRef]
- ISO 10993-5:2009—Biological Evaluation of Medical Devices—Part 5: Tests for in vitro Cytotoxicity. Available online: https://www.iso.org/standard/36406.html (accessed on 18 September 2019).
- Haller, O.; Stertz, S.; Kochs, G. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect 2007, 9, 1636–1643. [Google Scholar] [CrossRef]
- Ooi, E.L.; Verjan, N.; Hirono, I.; Nochi, T.; Kondo, H.; Aoki, T.; Kiyono, H.; Yuki, Y. Biological characterisation of a recombinant Atlantic salmon type I interferon synthesized in Escherichia coli. Fish Shellfish Immunol. 2008, 24, 506–513. [Google Scholar] [CrossRef]
- Miroux, B.; Walker, J.E. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996, 260, 289–298. [Google Scholar] [CrossRef]
- Lai, Y.S.; John, J.A.; Lin, C.H.; Guo, I.C.; Chen, S.C.; Fang, K.; Lin, C.H.; Chang, C.Y. Establishment of cell lines from a tropical grouper, Epinephelus awoara (Temminck & Schlegel), and their susceptibility to grouper irido- and nodaviruses. J. Fish Dis. 2003, 26, 31–42. [Google Scholar]
- Chi, S.C.; Lin, S.C.; Su, H.M.; Hu, W.W. Temperature effect on nervous necrosis virus infection in grouper cell line and in grouper larvae. Virus Res. 1999, 63, 107–114. [Google Scholar] [CrossRef]
- Tjalsma, H.; Bolhuis, A.; Jongbloed, J.D.; Bron, S.; van Dijl, J.M. Signal peptide-dependent protein transport in Bacillus subtilis: A genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 2000, 64, 515–547. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]


| IFNs | Source | Signal Peptides | Expression Host | Recombinant IFN Location | Yield a (mg/L Culture) | References |
|---|---|---|---|---|---|---|
| IFN | D. rerio | none | E. coli | Cytosolic | Insoluble | [18] |
| IFN-α | E. septemfasciatus | none | E. coli | Cytosolic | Soluble | [15] |
| IFN-α | E. septemfasciatus | none | E. coli | Cytosolic | Insoluble | [3] |
| IFN-α | E. septemfasciatus | SacB | B. subtilis | Extracellular | 1.3 | This study |
| IFN-α | E. septemfasciatus | PelB | E. coli | Periplasmic | 6.0 | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-J.; Xiao Joe, J.T.; Lu, W.-J.; Huang, M.-Y.; Sun, T.-H.; Lin, S.-P.; Li, Y.-C.; Tsui, Y.-C.; Lu, M.-W.; Victor Lin, H.-T. Secretory Production of Functional Grouper Type I Interferon from Epinephelus septemfasciatus in Escherichia coli and Bacillus subtilis. Int. J. Mol. Sci. 2020, 21, 1465. https://doi.org/10.3390/ijms21041465
Lin H-J, Xiao Joe JT, Lu W-J, Huang M-Y, Sun T-H, Lin S-P, Li Y-C, Tsui Y-C, Lu M-W, Victor Lin H-T. Secretory Production of Functional Grouper Type I Interferon from Epinephelus septemfasciatus in Escherichia coli and Bacillus subtilis. International Journal of Molecular Sciences. 2020; 21(4):1465. https://doi.org/10.3390/ijms21041465
Chicago/Turabian StyleLin, Hsuan-Ju, Joan Tang Xiao Joe, Wen-Jung Lu, Mei-Ying Huang, Ting-Hsuan Sun, Sheng-Pao Lin, Yi-Chuan Li, Ya-Chin Tsui, Ming-Wei Lu, and Hong-Ting Victor Lin. 2020. "Secretory Production of Functional Grouper Type I Interferon from Epinephelus septemfasciatus in Escherichia coli and Bacillus subtilis" International Journal of Molecular Sciences 21, no. 4: 1465. https://doi.org/10.3390/ijms21041465
APA StyleLin, H.-J., Xiao Joe, J. T., Lu, W.-J., Huang, M.-Y., Sun, T.-H., Lin, S.-P., Li, Y.-C., Tsui, Y.-C., Lu, M.-W., & Victor Lin, H.-T. (2020). Secretory Production of Functional Grouper Type I Interferon from Epinephelus septemfasciatus in Escherichia coli and Bacillus subtilis. International Journal of Molecular Sciences, 21(4), 1465. https://doi.org/10.3390/ijms21041465