Abstract
Integrase strand-transfer inhibitors (INSTIs) are now included in preferred first-line antiretroviral therapy (ART) for HIV-infected adults. Studies of Western clade-B HIV-1 show increased resistance to INSTIs following mutations in integrase and nef 3′polypurine tract (3′-PPT). With anticipated shifts in Africa (where 25.6-million HIV-infected people resides) to INSTIs-based ART, it is critical to monitor patients in African countries for resistance-associated mutations (RAMs) affecting INSTIs efficacy. We analyzed HIV-1 integrase and 3′-PPT sequences in 345 clinical samples from INSTIs-naïve HIV-infected Cameroonians for polymorphisms and RAMs that affect INSTIs. Phylogeny showed high genetic diversity, with the predominance of HIV-1 CRF02_AG. Major INSTIs RAMs T66A and N155K were found in two (0.6%) samples. Integrase polymorphic and accessory RAMs found included T97A, E157Q, A128T, M50I, S119R, L74M, L74I, S230N, and E138D (0.3′23.5% of samples). Ten (3.2%) samples had both I72V+L74M, L74M+T97A, or I72V+T97A mutations; thirty-one (9.8%) had 3′-PPT mutations. The low frequency of major INSTIs RAMs shows that INSTIs-based ART can be successfully used in Cameroon. Several samples had ≥1 INSTIs accessory RAMs known to reduce INSTIs efficacy; thus, INSTIs-based ART would require genetic surveillance. The 3′-PPT mutations could also affect INSTIs. For patients failing INSTIs-based ART with no INSTIs RAMs, monitoring 3′-PPT sequences could reveal treatment failure etiology.
1. Introduction
During HIV replication, the integrase enzyme catalyzes the ligation of viral reverse-transcribed DNA into the chromosome of the host cells. This integration is a critical step for viral replication, enabling the proviral DNA to persist in the host cells and form a permanent viral template that can continuously initiate the production of new viruses and replicate with each cell division [1,2,3]. Integrase is a 32 kDa protein (288 amino acids (aa)) encoded by the C-terminal of HIV polymerase gene [4]. It has 3 functional domains: an N-terminal domain (aa 1-49) that facilitates protein multimerization; a catalytic core domain (aa 50-212) that contains the viral DNA binding site and is involved in the recognition of viral DNA substrate; and a C-terminal domain (aa 213-288) that help stabilize the integrase–viral DNA complex [4,5,6].
Considering its critical role in viral integration into the host genome and the formation of viral reservoirs, integrase has been a major HIV/AIDS therapeutic target. There are currently three FDA-approved integrase strand transfer inhibitors (INSTIs) used as part of antiretroviral therapy (ART) regimens [raltegravir (RAL), elvitegravir (EVG), and dolutegravir (DTG)], and a fourth INSTI, bictegravir, approved for use as a component of a fixed-dose combination ART [7]. Compared to other drug classes, these INSTIs have shown higher efficacy, superiority and higher genetic barrier to the development of resistance [8,9]. Thus, INSTIs are now recommended as part of initial or preferred ART regimens for most HIV-infected people in western countries [7,10] and are also recommended by the World Health Organization (WHO) as alternative 1st-line regimens for adults living with HIV/AIDS [11,12]. However, access to INSTIs in resource-limited countries has been restricted due to its high costs. Although WHO is now recommending DTG as alternative 1st-line regimen [11,12], its use is currently rare in resource-limited countries and it is estimated that only two countries in Sub-Saharan Africa (SSA), Kenya and Botswana, are currently providing DTG to HIV-infected persons as part of 1st-line ART regimens [13,14]. Nevertheless, with DTG license to generic pharmaceutical manufacturers through the Medicines Patent Pool [15], and recently negotiated reduced DTG prices through collective bargaining with many countries [15,16,17], INSTIs are expected to be increasingly available in several resource-limited countries. In fact, although global 2018 estimates show that DTG was used in only 4% of 1st-line ART regimens and in 6% of 2nd-line regimens, it is projected that by the year 2025, DTG, including single pills containing DTG and other antiretroviral drugs (ARVs), will be used by up to 57% of people living with HIV (PLWH) [16].
There is evidence of a worldwide increase in HIV resistance to many ARVs, including non-nucleoside reverse transcriptase inhibitors that form the backbone of many current WHO-recommended 1st- and 2nd-line regimens [11,12]; and many people failing these 1st- and 2nd-line regimens will require 3rd-line regimens and INSTIs as rescue therapies [11,12,18]. Resistance to ARVs is associated with mutations in the viral genome, including transmitted and acquired drug-resistance mutations (DRMs) [19]. These DRMs are associated with increased risk of treatment discontinuation, virological failure, and death [19]. It has also been shown that people with pretreatment DRMs are more likely to develop new DRMs after initiating treatment [19].
Despite their higher efficacy, evidence from HIV-infected people on INSTIs in western countries showed resistance to INSTIs, including DRMs resulting in treatment failures [20,21,22,23], and polymorphic mutations that contribute to the rescue of viral fitness and increased resistance to INSTIs [22,23]. All these evidences are from populations in developed countries, and there are major knowledge gaps on INSTIs resistance-associated mutations (RAMs) across diverse populations, especially in Sub-Saharan Africa (SSA) where over two-thirds of the current 38 million PLWH reside [24]. To address this scientific knowledge gap, WHO recently recommended that HIV drug resistance surveillance, including pretreatment DRMs, be implemented for INSTIs [18,19]. These recommendations are critically important considering the current shift of many ART programs to INSTIs-based regimens [7,10] and anticipated shifts in SSA and other resources-limited countries [16]. Therefore, the goal of our current study was to analyze integrase gene sequences in clinical samples to assess the occurrence of polymorphisms and INSTIs RAMs in PLWH in Cameroon, a SSA country that has not yet adopted INSTIs-based ART. Because it has been shown that mutations located outside the integrase gene, in the viral 3′ polypurine tract (PPT, located in the 3′ end of nef), can confer resistance to INSTIs [25,26,27], we also sequenced and analyzed nef and the PPT viral genome region in these samples. We further analyzed the HIV integrase gene and PPT sequences from 215 additional patient samples from Cameroon, obtained from the Los Alamos database [28], for INSTIs RAMs and natural polymorphisms that could affect INSTIs-based therapy.
2. Results
2.1. Patient Demographics
We collected samples from 235 HIV-infected subjects in Cameroon, of which we successfully amplified and sequenced the integrase and/or nef genes in 130 samples (Table 1). This cohort of 130 patients included 39 males [mean age: 37.28 ± 9.94 years] and 91 females (35.43 ± 8.75 years). The mean plasma viral load and CD4+ T-cells count at the time of specimen collection were, respectively, 4.64 ± 1.54 log copies/mL and 360.3 ± 320.4 cells /µL for males and 4.24 ± 1.50 log copies/mL and 311.2 ± 184.5 cells /µL for females. Of the samples with successfully amplified and sequenced integrase and nef genes, 33 (84.61%) males and 66 (72.52%) females were treatment-naïve, 5 (12.82%) males and 25 (27.47%) females were on ART (1st-line regimens), 1 female had discontinued treatment, and treatment information was not available for 1 male. None of the cohort patients had been exposed to INSTIs at the time of enrolment and specimen collection. We also analyzed 215 full-length Cameroon HIV-1 sequences (samples collected from HIV+ Cameroonians between 1991 and 2014, when INSTIs was not available in Cameroon and many other countries) previously deposited in the Los Alamos HIV sequence database (see Table S1). Most of the database sequences information did not include demographics, treatment status, CD4 levels, or viral loads data.

Table 1.
Demographic and clinical characteristics of cohort subjects by sex.
2.2. HIV-1 Subtyping
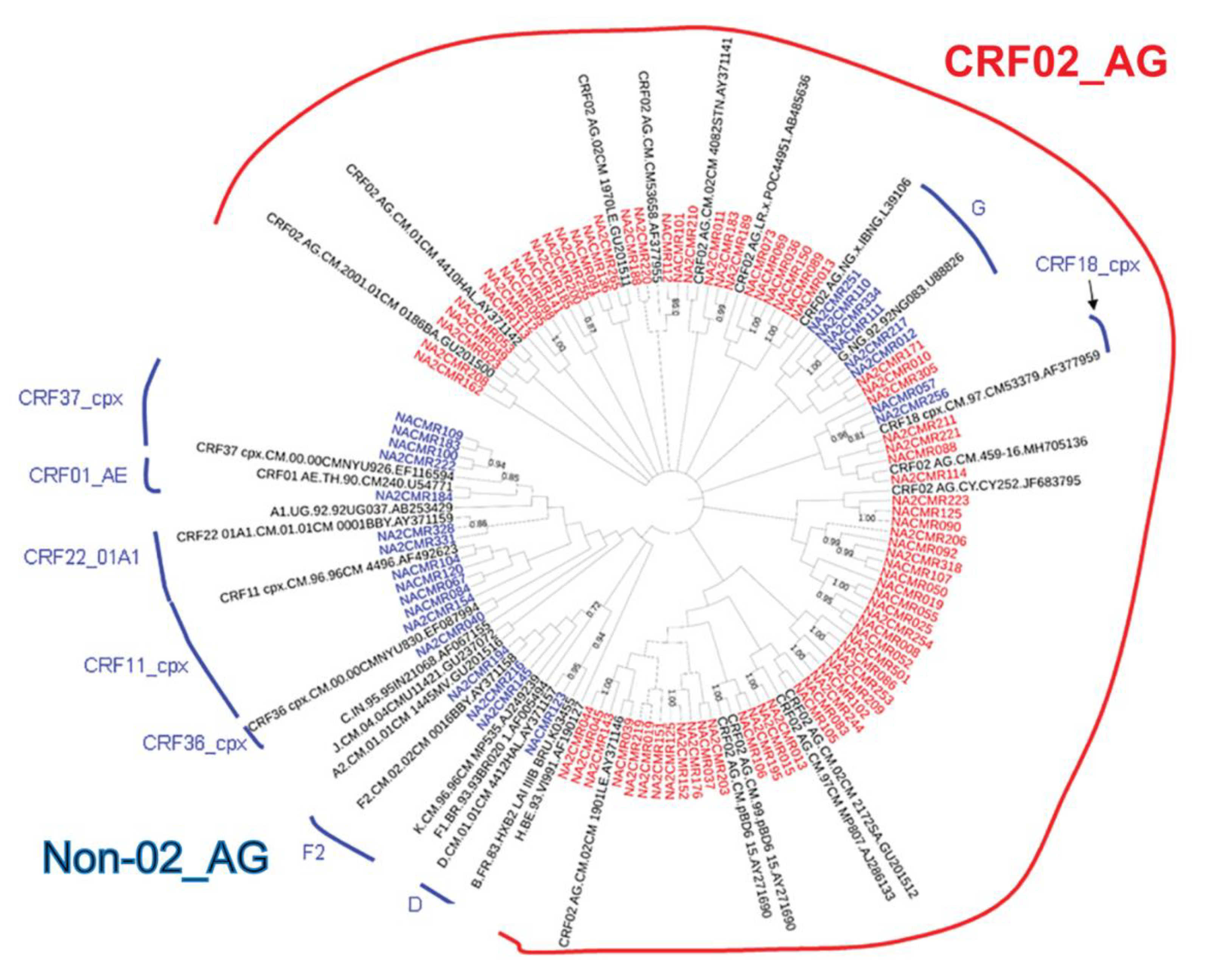
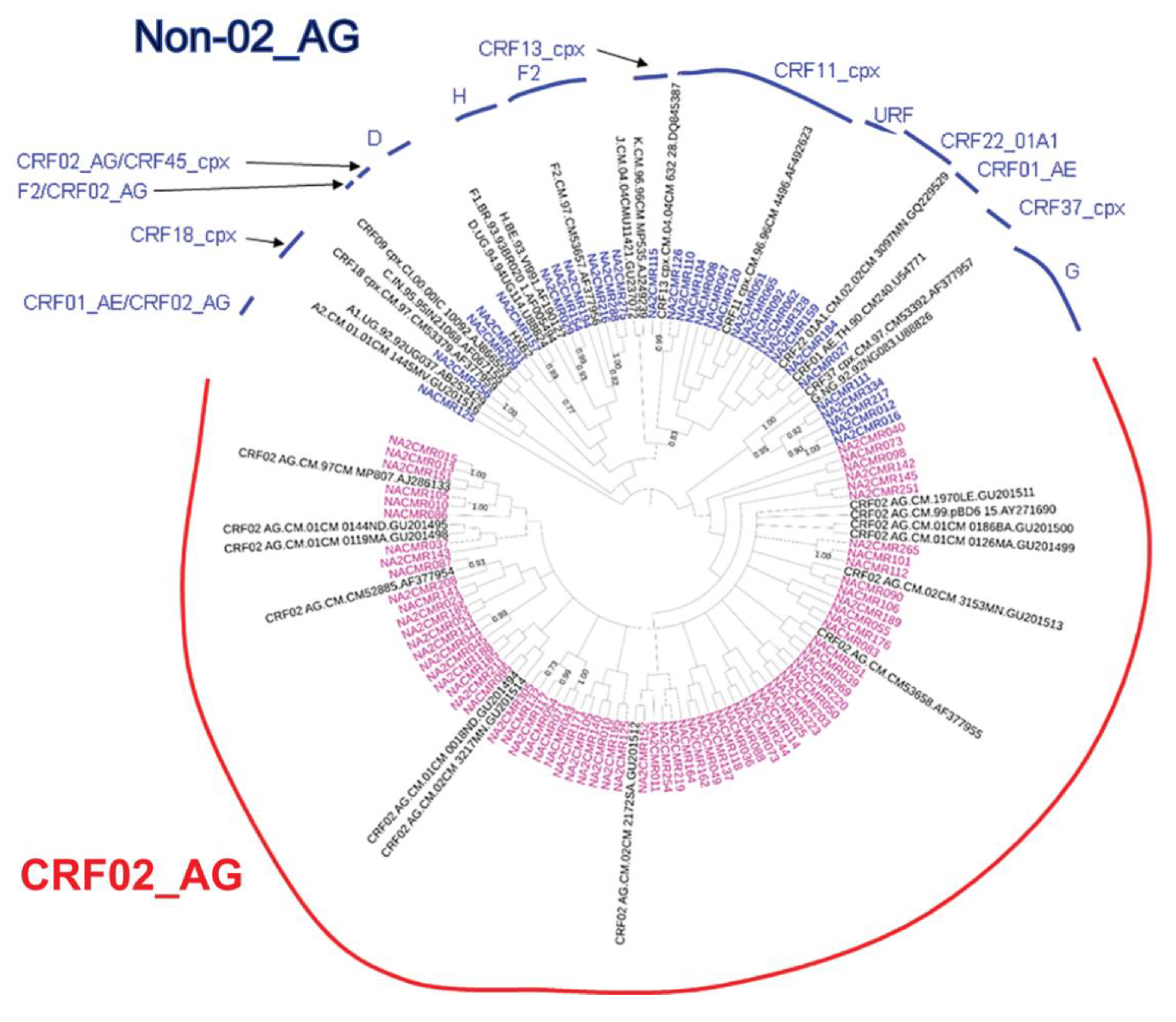
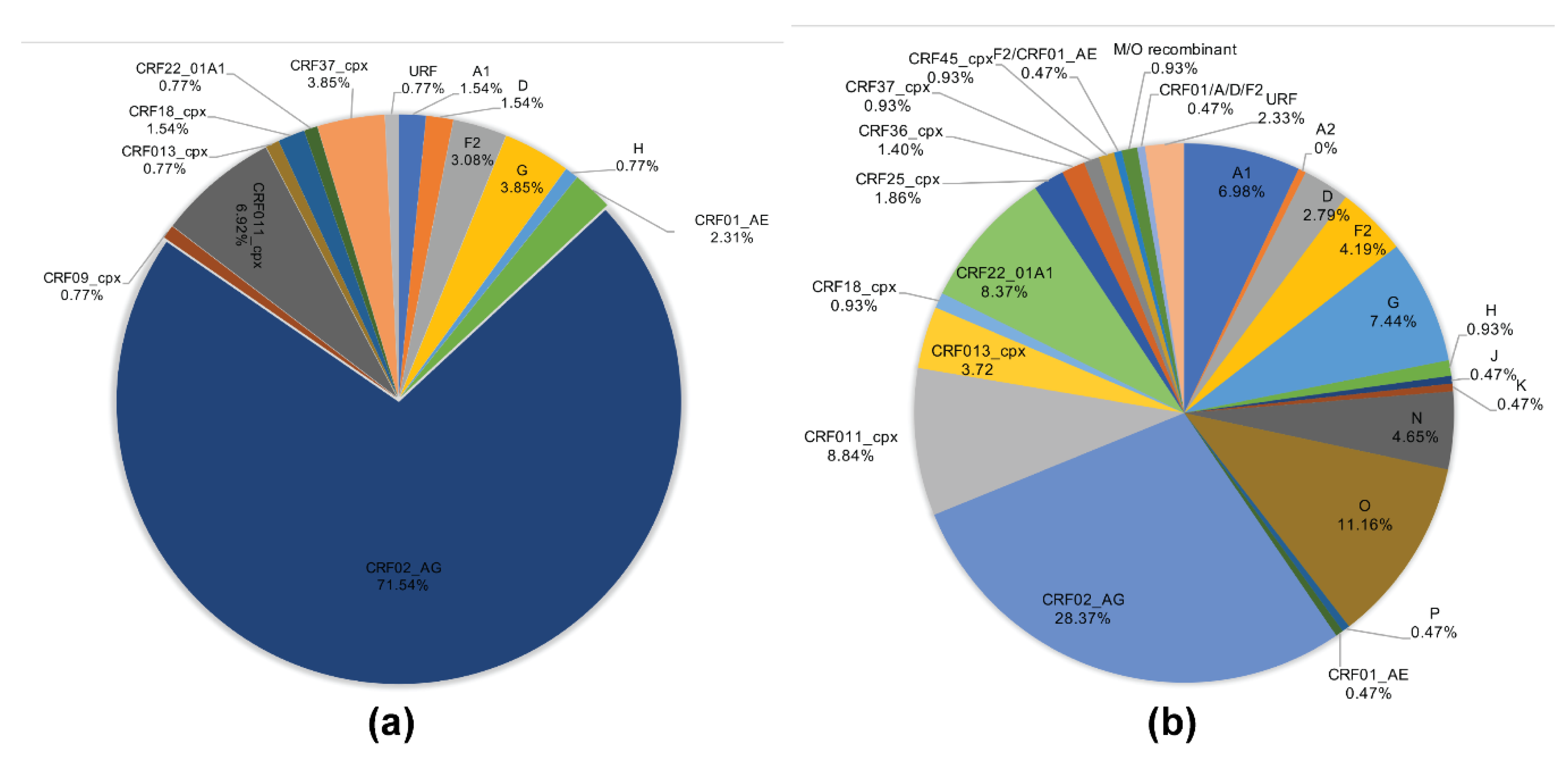
The viral subtypes of our study cohort were determined using the NCBI viral genotyping tool and phylogenetic analyses as described in Materials and Methods. Of the 100 samples successfully sequenced for the integrase gene, 75% were HIV-1 CRF02_AG (AG) and 25% were non-CRF02_AG (non-AG), including 6% subtype G, 4% each for CRF37_cpx and CRF11_cpx, 3% subtype F2, 2% each for CRF18_cpx and CRF22_01A1, and 1% each for subtype A1, D, CRF01_AE, CRF36_cpx (Figure 1). Of the 101 samples successfully sequenced for the nef gene, 69.31% were HIV-1 CRF02_AG and 30.69% were non-AG subtypes, including 8% CRF11_cpx, 5% subtype G, 4% subtype F2, 3% CRF22_01A1, 2% subtype H; 1% each for subtypes D, CRF01_AE, CRF13_cpx, CRF18_cpx, CRF37_cpx, CRF01_AE/CRF02_AG, CRF02_AG/CRF45_cpx, F2/CRF02_AG, and 1% unique recombinant form (URF) (Figure 2). Combined analyses based on both integrase and nef genes showed that 71.54% of samples were CRF02_AG and 28.46% were non-AG: 6.92% CRF11_cpx, 3.85% each subtypes G and CRF37_cpx, 3.08% subtype F2, 2.31% CRF01_AE, 1.54% each subtypes A1, D, and CRF18_cpx, 0.77% each subtypes H, CRF09_cpx, CRF13_cpx, CRF22_01A1, and a URF (0.77%) (Figure 3a).
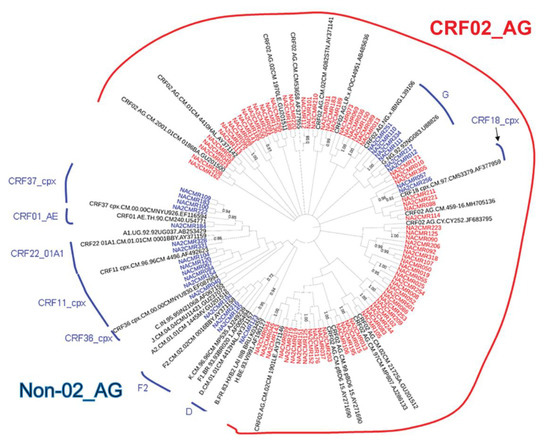
Figure 1.
Phylogenetic analysis of HIV-1 integrase gene sequences in cohort samples. The maximum likelihood phylogenetic tree was constructed using the MEGA 6 software package as described in Materials and Methods. The 31 reference sequences were from the Los Alamos database and included HIV-1 isolates from ten different countries [Belgium (1), Brazil (1), Cameroon (21), Cyprus (1), India (1), France (1), Liberia (1), Nigeria (2), Uganda (1), and United Arab Emirates(1)]. Bootstrap values of 1000 replicates (>70%) are shown at the corresponding nodes.
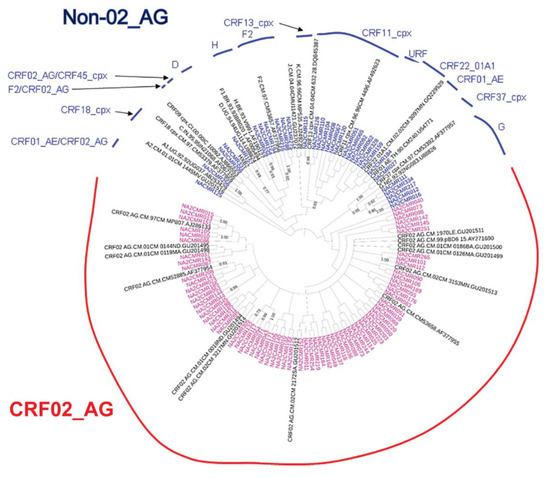
Figure 2.
Phylogenetic analysis of HIV-1 nef gene sequences in cohort samples. The maximum likelihood phylogenetic tree was constructed using the MEGA 6 software package as described in Materials and Methods. The 31 reference sequences were from the Los Alamos database and included HIV-1 isolates from nine different countries [Belgium (1), Brazil (1), Cameroon (22), Cote D’Ivoire (1), India (1), France (1), Nigeria (1), Uganda (2), and United Arab Emirates(1)]. Bootstrap values of 1000 replicates (>70%) are shown at the corresponding nodes.
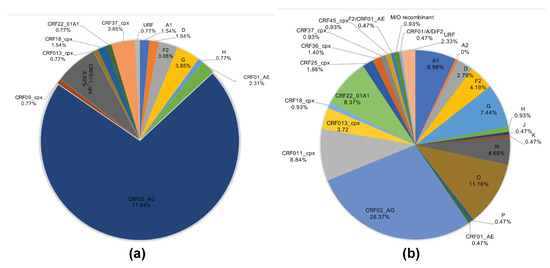
Figure 3.
HIV-1 genetic diversity in cohort and database samples. (a) Proportions of each HIV-1 subtype and CRFs identified in cohort samples based on NCBI viral genotyping tool (combined analysis of integrase and nef gene sequences). (b) Proportions of each HIV-1 subtype and CRFs identified using full-length sequence of database samples, based on NCBI viral genotyping tool. CRF: circulating recombinant form; URF: unique recombinant form.
Genotypes for the 215 database HIV samples as reported in their original sequences submissions included 180 (83.72%) group M, 24 (11.16%) group O, 10 (4.65%) group N, and 1 (0.47%) group P. The 180 HIV-1 group M samples included 61 (28.37%) CRF02_AG, 19 (8.84%) CRF09_cpx, 18 (8.37%) CRF22_01A1, 16 (7.44%) subtype G, 15 (6.98%) subtype A1, 9 (4.19%) subtype F2, 8 (3.72%) CRF13_cpx, 6 (2.79%) subtype D, 5 (2.33%) URFs, 4 (1.86%) CRF25_cpx, 3 (1.4%) CRF36_cpx, 2 (0.93%) each for subtypes H, CRF18_cpx, CRF37_cpx, CRF45_cpx, and M/O recombinant; 1 (0.47%) each for subtypes A2, J, K, CRF01_AE, F2/CRF02_AG, CRF01_AE/A/D/F2 (Figure 3b).
2.3. INSTI Resistance Mutations in Cohort Samples
No INSTIs major RAMs were present in our cohort samples. The INSTIs accessory RAMs E157Q and T97A were present, respectively, in 6 (all CRF02_AG) and 2 (1 CRF02_AG and 1 CRF09_cpx) samples (Table 2). Four other mutations were identified in cohort samples: M50I (in 6 samples), S119R (in 4 samples, all CRF02_AG), L74M (in 8 CRF02_AG and 1 CRF11_cpx samples), and L74I in 25 samples of which 22 (88%) were CRF02_AG (Table 2).

Table 2.
INSTIs resistance mutations in cohort samples.
2.4. INSTI Resistance Mutations in Database Samples
Two INSTIs major RAMs were identified in two database samples: T66A in 1 (0.47%) HIV-1 group M/O recombinant sample isolated in 1997 from a 29 years old ART-naïve Cameroonian female (GenBank accession number: AJ239083) [29]; and N155K in 1 (0.47%) CRF36_cpx sample isolated in 2000 from a 19 years old female in Cameroon (GenBank accession number: EF087995) [30] (Table 3). Three INSTIs accessory RAMs were identified: T97A (in 16 samples), A128T (in 1 M/O recombinant sample), and E157Q (in 3 samples) (Table 3). Other mutations in database samples included M50I, L74M, L74I, S119R, S230N, and E138D, identified in 67 (31%), 9 (4%), 49 (22.8%), 1 (0.47%), 2 (0.93%), and 1 (0.47%) of database samples (Table 3), respectively.

Table 3.
Profile of INSTIs drug resistance mutations of database samples.
2.5. INSTIs Resistance-Associated Mutations in Subjects Infected with HIV-1 CRF02_AG and Non-AG Viruses
The INSTIs accessory RAMs identified in both cohort and database samples included T97A, E157Q, M50I, L74M, L74I, and S119R. The INSTIs accessory RAMs identified in both HIV-1 CRF02_AG and non-AG cohort samples included T97A, M50I, L74M, and L74I (in 1.3% to 29.3% of AG and in 4% to 16% of non-AG cohort samples). E157Q and S119R mutations were observed only in cohort samples of AG subtype. There were no significant differences in the proportions of AG and non-AG cohort samples with INSTIs RAMs (Table 4).

Table 4.
Integrase gene mutations in cohort and database AG and non-AG samples.
The INSTIs accessory RAMs identified in both AG and AG database samples included T97A, E157Q, M50I, L74M, L74I, and S230N (in 3.5% to 60% of AG and in 1.4% to 86.5% of non-AG database samples). A significantly higher proportion of non-AG database samples (86.56%) had the M50I mutation compared to 17.6% in database AG samples (p < 0.000001, Table 4). A significantly higher proportion of AG database samples (21%) had the L74M mutation compared to 4.2% in database non-AG samples (p < 0.005, Table 4). The proportion of AG and non-AG database samples harboring other INSTIs RAMs was not significantly different and T66A, N155K, A128T, S119R, and S230N were observed only in a very small number (1.4%) of database samples of non-AG genotypes (Table 4).
2.6. Integrase Natural Polymorphisms in Subjects Infected with HIV-1 CRF02_AG and Non-AG Viruses
Similar polymorphisms (substitutions at similar aa positions) were observed in cohort and database samples, including a total of 27 natural polymorphisms (Table 5).

Table 5.
Integrase gene polymorphisms in cohort and database AG and non-AG samples.
Integrase polymorphisms in cohort samples: Comparative analyses of cohort subjects infected with CRF02_AG and non-AG subtypes showed that the G134N, I135V, K136T, and T206S polymorphisms were present in a significantly higher proportion of samples with AG subtype (81% to 97%) compared to samples with non-AG subtypes (28–52%) (p < 0.00001, Table 5). Similarly, T124A was present in a higher proportion of AG (90.67%) than non-AG (68%) samples (p = 0.031, Table 5), and R269K was only present in AG (30.67%) samples (p = 0.003, Table 5). For the cohort group, polymorphisms A21T, I72V, D167E, and D256E were significantly more prevalent in non-AG (24–48%) than in AG (5–20%) samples (p < 0.04, Table 5) and K136Q was only present in non-AG (44%) samples (p < 0.000001, Table 5).
Integrase polymorphisms in database samples: The L101I, T125A, G134N, I135V, K136T, T206S, T218I, and R269K polymorphisms were present in a significantly higher proportion of AG (44% to 95%) compared to non-AG (5–66%) database samples (p < 0.00002, Table 5). Similarly, K14R, V31I, T112V, and T214A polymorphisms were present in a significantly higher proportion of AG (75% to 95%) than non-AG (50% to 78%) database samples (Table 5). The E11D, A21T, G134D, K136Q, D167E, I208L, and D256E polymorphisms were significantly more prevalent in non-AG (14′51.3%) than in AG (3–26%) database samples (Table 5).
Integrase polymorphisms in both cohort and database samples: Overall, T124A, G134N, I135V, K136T, T206S, and R269K polymorphisms were significantly more prevalent in AG compared to non-AG samples, whereas A21T, K136Q, D167E, and D256E polymorphisms were significantly more prevalent in non-AG compared to AG samples.
2.7. Effects of ART and Immune Function on Integrase RAMs and Natural Polymorphisms
Additional analyses of cohort patients showed no significant differences in the occurrence of gene polymorphism or gene mutations among subjects who were treatment-naïve and those on ART. Similarly, there were no significant differences in the occurrence of gene polymorphism or INSTIs RAMs in cohort patients with CD4+ T-cell counts above 200 cells/μL (66%) and those with CD4+ T-cell counts below 200 cells/μL (34%). Integrase sequences for Cameroon HIV isolates downloaded from the Los Alamos database did not have information on patient’s treatment status or levels of CD4+ cells.
2.8. Analysis of 3′-PPT and 5′ Terminal Nucleotides of 3′ Long Terminal Repeat
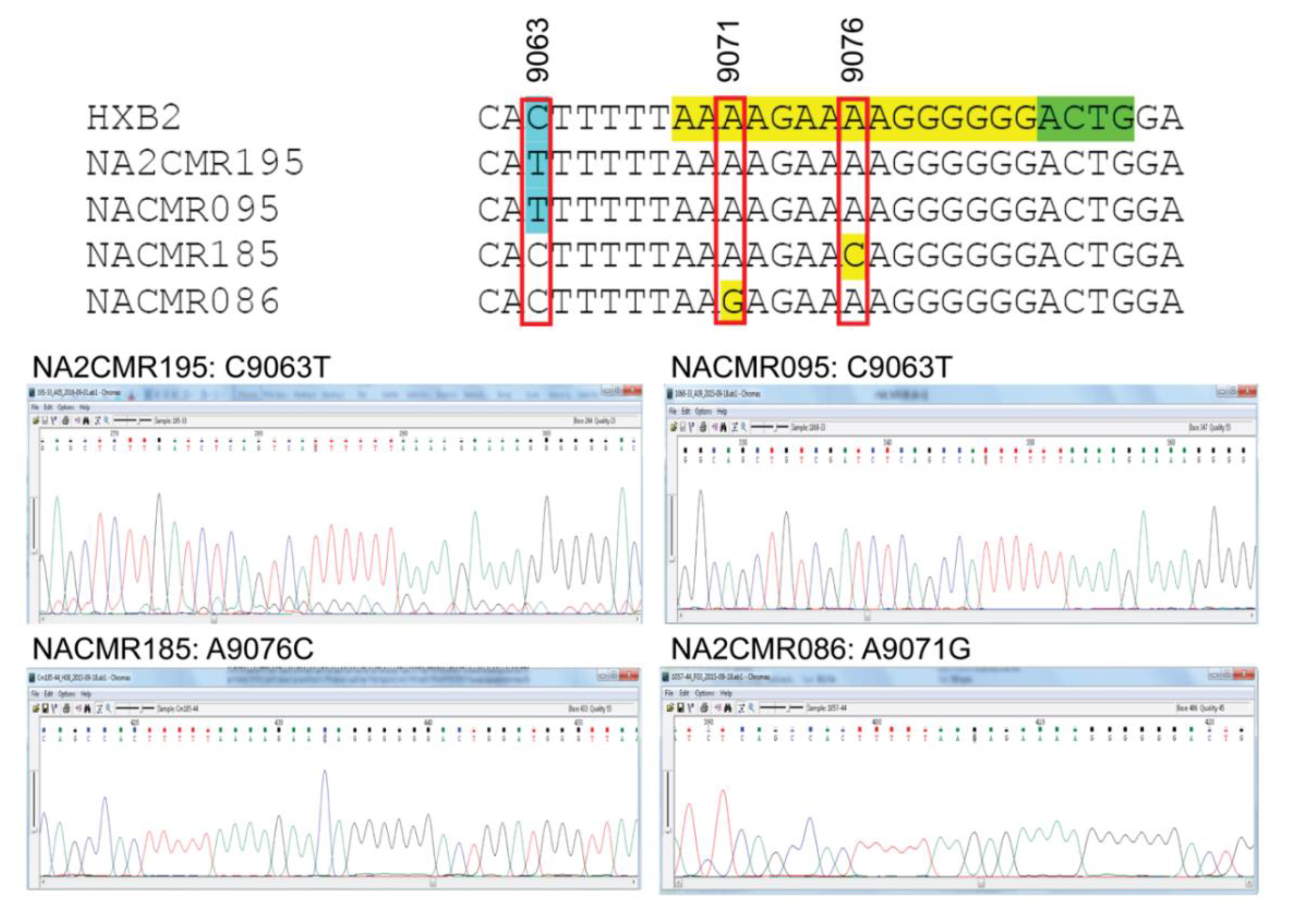
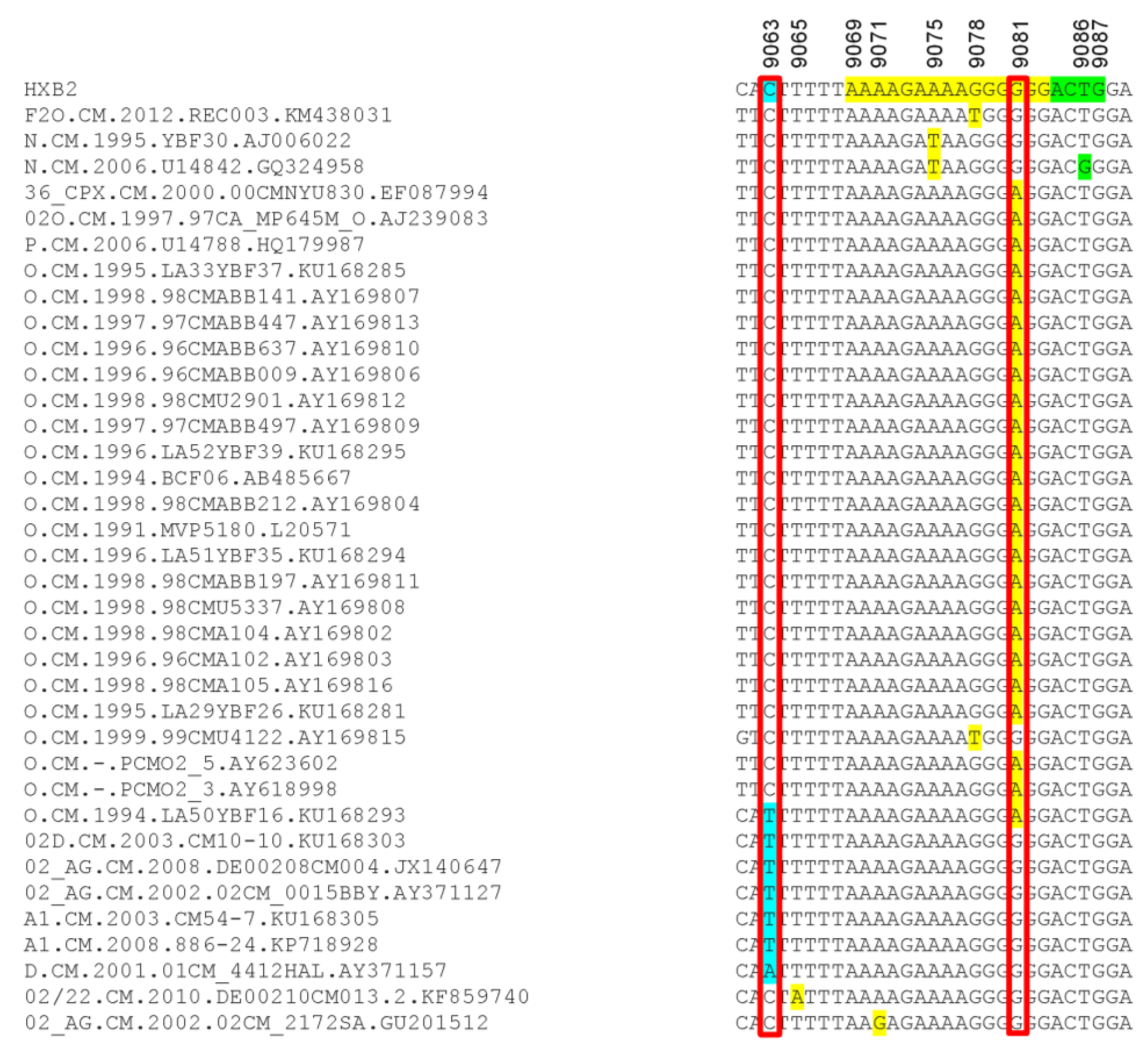
Because mutations in the HIV-1 nef /3′-PPT region can confer resistance to INSTIs [25,26,27], we also analyzed the sequences of the 3′-PPT viral genome, as well as the 8 nucleotides upstream of the 3′-PPT region (of nef), and the first four nucleotides downstream of the 3′-PPT [5′ terminal region of the 3′ long terminal repeat (LTR)], in comparison to the reference HXB2 genome sequence. Mutations within the 15 nucleotides 3′-PPT region (Figure 4, yellow highlight) was observed only in 2 cohort samples: A9076C mutation in subject NACMR185 and A9071G mutation in subject NACMR086 (Figure 4). Two other cohort samples (NA2CMR195 and NACMR095) had a C to T substitutions in the nef region upstream of the 3′-PPT region (nucleotide 9063). No mutation was observed downstream of the 3′-PPT (LTR) among cohort samples.
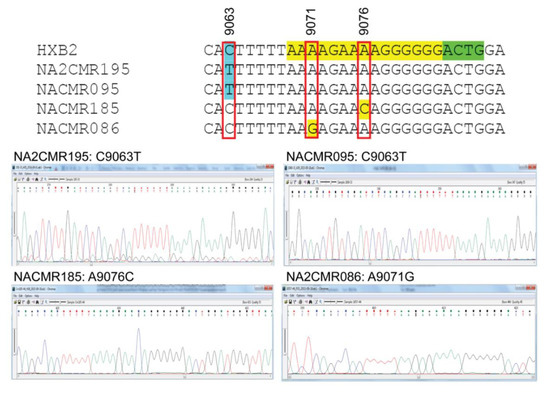
Figure 4.
Mutations detected in the 3′polypurine tract (3′-PPT) and its flanking regions for cohort samples. Nucleotide sequences alignments, using HXB2 as a reference sequence, are shown for the four study cohort samples in which mutations were detected. The 15 nucleotides of 3′-PPT sequences are shaded yellow. The 1st four nucleotides of 3′LTR are shaded green. Region with mutation upstream of the 3-PPT (nucleotide position 9063) is shaded in sky blue. Nucleotide numbers correspond to the numbering of the HXB2 reference sequence. Electropherograms showing data sequences and mutations are shown for the four cohort samples.
Analysis of the 215 database samples showed 29 samples with mutations in the viral 3′-PPT region [G9078T in one subtype F2 and one Group O; A9075T in two HIV-1 group N samples; G9081A in one HIV-1 CRF_36cpx, one HIV-1 group M/O recombinant, one HIV-1 group P, and 21 HIV-1 group O; A9071G mutation in one HIV-1 CRF02_AG sample] (Figure 5). Seven database samples had mutations upstream of the 3′-PPT region (C9063T in one Group O and 5 group M [3 CRF02_AG and 2 subtypes A1]; C9063A in one subtype D) (Figure 5). Mutation downstream of the 3′-PPT (LTR) was only observed in one database sample (T9086G in one Group N sample) (Figure 5).
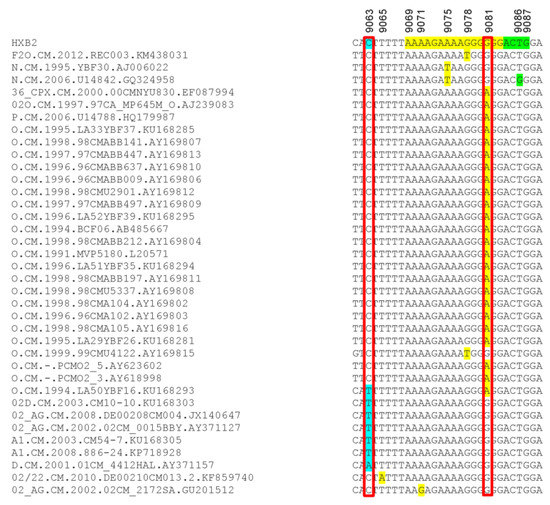
Figure 5.
Mutations detected in the 3′-PPT and its flanking regions for database samples. The nucleotides alignment, using HXB2 as a reference sequence, are shown for the database samples in which mutations were detected. The 15 nucleotides of 3′-PPT sequences are shaded yellow. The 1st four nucleotides of 3′LTR are shaded green. Region with mutation upstream of the 3-PPT (nucleotide position 9063) is shaded in sky blue. Nucleotide numbers correspond to the numbering of the HXB2 reference sequence.
3. Discussion
Despite INSTIs’ high efficacy and superiority compared to other ARV drug classes, studies in high-income countries show that resistance to INSTIs can occur, associated with transmitted and/or acquired DRMs, leading to decreased susceptibility to INSTIs and treatment failure [20,21]. The integrase mutations often associated with reduced susceptibility to INSTIs include both nonpolymorphic and polymorphic mutations [1,22,23]. In 2018, ART regimens including 2nd-generation INSTIs such as DTG constituted only 4% of 1st-line regimens and 6% of 2nd-line regimens worldwide [12,16]. However, INSTIs are now part of the WHO-recommended alternative 1st-line regimens for PLWH [7,11,12]. With ongoing efforts to expand INSTIs availability worldwide, it is projected that by the year 2025, up to 57% of PLWH will be on ART regimens containing DTG or other INSTIs [16]. Therefore, it is important to monitor PLWH for the presence of mutations in the integrase gene that can affect INSTIs efficacy and therapeutic outcomes. Furthermore, our current knowledge of INSTIs RAMs mostly comes from studies in western countries; patient-derived data on INSTIs RAMs in SSA are scarce and there is a major knowledge gap on INSTIs RAMs in SSA, whereas over two-thirds of the current 38 million PLWH reside in that region. Our current study contributes to filling that gap and addresses recent WHO recommendations that HIV drug resistance surveillance be implemented for INSTIs, including pretreatment DRMs [18,19]. We analyzed integrase gene sequences from Cameroon clinical isolates for the presence of mutations known to be associated with resistance or reduced susceptibility to INSTIs. Because mutations in the nef 3′-PPT region have been associated with resistance to INSTIs [25,26,27], we also analyzed nef gene sequences, their 3′-PPT regions, and sequences adjacent to the 3′-PPT regions.
Phylogeny of integrase and nef sequences showed high genetic diversity, with a predominance of AG infections (69–75% of cohort samples). This is similar to previous studies showing that over 60% of HIV+ Cameroonians harbored AG viruses [31,32,33]. The lower proportion of AG infections seen in database samples (28%) is likely due to overrepresentation of other genotypes such as HIV-1 groups O, N, and P from Cameroon in the database, from studies that specifically analyzed sequences from these subtypes [34], rather than sequences from randomly collected patient samples as in our cohort or other studies of Cameroon isolates [31,32,33].
No INSTIs major RAMs were present in cohort samples, and only 2 database samples had INSTIs major RAMs: T66A in a group M/O recombinant and N155K in a CRF36_cpx sample. Mutations in integrase aa 66 can significantly reduce viral susceptibility to INSTIs. T66A and T66I reduce EVG susceptibility, respectively, by 5-fold and 10-fold [35,36,37]. T66K reduced susceptibility to EVG by 40-fold, reduced susceptibility to RAL by 10-fold, and reduced susceptibility to DTG by 2- to 3-fold [21,36,38]. Mutations in integrase aa 155 have been associated with reduced susceptibility to RAL (by 10-fold) and EVG (by 30-fold) [35,39,40], and studies of patients failing DTG-based ART showed that patients with virologic failure had N155H substitution at baseline and also developed additional INSTIs-RAMs [41,42]. The fact that only 0.6% of samples analyzed had INSTIs major RAMs suggests that there will be a lesser likelihood of resistance to INSTIs in Cameroon and that INSTIs will be effective for HIV-infected Cameroonians. Similar trends were observed in other countries in Africa [13,34,43,44,45], Asia [46,47,48,49,50,51], Europe [52,53,54], and South America [55] where studies showed a low frequency of INSTIs major RAMs.
Several other integrase accessory RAMs and polymorphic mutations were observed in our current study, including the INSTIs accessory RAMs T97A (in 2% of cohort and 7.4% of database samples), E157Q (in 6% of cohort and 1.4% of database samples), A128T (in 0.47% of database samples); the polymorphic mutations M50I (in 6% of cohort and 31% of database samples), S119R (in 4% of cohort and 0.47% of database samples), L74M (in 9% of cohort and 4% of database samples), L74I (in 25% of cohort and 23% of database samples), E138D (in 0.47% database samples), and S230N (in 0.93% of database samples). Ten of the 11 INSTIs accessory RAMs observed were in the integrase central core domain. This integrase region contains the endonuclease and polynucleotidyl transferase site and is involved in DNA substrate recognition, binding, and chromosomal integration of the newly synthesized double-strand viral DNA into the host genomic DNA [56,57,58]. The other mutation (S230N) was in the C-terminal domain, a region that helps stabilize the integrase–viral DNA complex [4,5,6]. The location of these mutations in regions involved in recognition, binding, integration, and stabilization of HIV into the host DNA shows the potential for these mutations to affect the integrase function and response to INSTIs. In fact, mutations in integrase aa residue 230 reduce DTG susceptibility by 3-fold [59]. T97A mutation can reduce EVG susceptibility by 3-fold [36], and combination of T97A substitution with other INSTIs RAMs markedly reduce HIV susceptibility to RAL [60,61] and DTG [62,63]. Similarly, although L74M and E157Q individually have minimal effect on the susceptibility to INSTIs, a combination of L74M and E157Q with other INSTIs RAMs result in reduced susceptibility to INSTIs. In fact, patients harboring viruses with L74M [61] or E157Q [64] mutations in addition to other integrase RAMs showed reduced susceptibility to DTG.
Six of our subjects (3 cohort and 3 database samples) were infected with viruses that had both I72V and L74M mutations; one cohort sample had both L74M and T97A, and 3 database samples had both I72V and T97A mutations. These subjects could be susceptible to resistance to INSTIs, as there is evidence that the presence of multiple integrase accessory RAMs or polymorphic mutations can increase viral fitness and reduce susceptibility to INSTIs, even in the absence of INSTIs major RAMs [40,65]. Furthermore, the fact that most of the integrase mutations observed in our study occurred in the central core and C-terminal domains suggests that they could alter proviral DNA binding, integration, and stabilization and affect INSTIs efficacy. Thus, vigilance and surveillance for INSTIs DRMs would be required in Cameroon and other SSA countries when they do shift to current WHO-recommended DTG/INSTIs-based ART.
Previous studies showed that polymorphic differences among HIV subtypes can affect viral fitness and resistance pathways [66] and that different viral subtypes may support different mutational pathways and this could result in subtype-based differences in drug resistance [67,68,69]. There is also evidence that natural polymorphisms associated with integrase activity and occurrence of resistance to INSTIs are subtype-dependent and subtype-specific polymorphic mutations in the integrase gene affect integrase DNA binding affinity when additional mutations are present and can influence INSTIs efficacy [66,68,70,71,72]. Thus, we compared the occurrence of INSTIs RAMs among samples from subjects infected with HIV-1 CRF02_AG and subjects harboring non-AG subtypes. Our data showed polymorphisms and mutations in the integrase in both groups, similar to previous studies showing that mutations and resistance to INSTIs can occur in both clades-B, CRF02_AG and non-AG subtypes [70,71,73]. However, the frequencies of such mutations can vary based on viral genotype [74]. The aa substitutions E92Q, S119R, E138A, Y143R, G148H/R, and S230R/N are more prevalent in subtype-B than in non-B subtypes [71,74,75,76], whereas mutations such as L74I/M, T97A, L101I, E157Q, T214A, and V201I are more prevalent in non-B subtypes compared to HIV-1 subtype B [71,74,77]. Because the vast majority of PLWH on INSTIs are people infected with HIV-1 subtype B, most INSTIs resistance-conferring mutations that have been characterized pertain to HIV-1 subtype B. It is likely that several mutations prevalent in non-B subtypes could affect the genetic barrier and INSTIs efficacy. In fact, computational modeling of INSTIs resistance development across different HIV-1 subtypes showed that compared to subtype B, the presence of M50I in subtypes A and C, L74I in subtypes A and CRF02_AG, G163R in CRF01_AE, and V165I in subtypes F and CRF01_AE would be associated with lower genetic barrier to resistance in these non-B clades [74]. With increased use of INSTIs by individuals infected with non-B viruses, it is important to monitor these subjects for viral escape in the context of selection pressure [74] and identify the RAM and polymorphic mutations that alter the genetic barrier to resistance and INSTIs efficacy and the role of viral genotype.
The genetic barrier is measured by evolutionary time to viral escape in the context of a selection pressure [74] and previous studies showed that development of HIV resistance to nucleoside reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, and protease inhibitors resulted in part to subtype-associated preferential codon usages and that different amino acid substitutions influenced the likelihood of development of drug resistance [78,79,80]. INSTIs are the newest class of anti-HIV drugs approved for use in HIV/AIDS treatment, and modeling of HIV-1 integrase sequences showed that viral subtypes and codon substitution would differentially affect the genetic barrier to the development of INSTIs resistance [74]. Most mutational pathways linked to higher or lower genetic barrier to resistance to INSTIs come from subtype B infected subjects in resource-rich countries. The role of baseline genetic diversity and subtype-specific polymorphism among non-B subtypes in the development of resistance to INSTIs has not been systematically investigated. With the increasing use of INSTIs worldwide, including in low- and middle-income countries where non-B subtypes predominate, monitoring of nucleotide substitutions that evolves into INSTIs-resistant viruses will help better understand the factors underlying the development of resistance to INSTIs across more diverse HIV subtypes.
Recent studies of patients failing DTG therapy showed that some patients with viral failure had no DRM in the viral integrase but instead had mutations in the nef 3′-PPT region [25,26,27]. These results suggested that mutations outside the integrase, in the 3′-PPT, can confer resistance to DTG and other INSTIs. Furthermore, the 3′-PPT is closely associated with the 5′ terminal of the 3′ LTR, and mutations within the six guanine residues (G-track) of the 3′-PPT can alter RNase H-mediated cleavage at the PPT 3′ terminus, and integrase activity [81,82]. This suggests that mutations in the 3′-PPT G-track might confer an alternative pathway to resistance to DTG and other INSTIs. Thirty-one (9.8%) subjects in our study were infected with viruses that had mutations in the 3′-PPT, including 26 with mutations in the 3′-PPT G-track. The potential effects of these 3′-PPT mutations on integrase function and susceptibility to INSTIs are not known. If DTG/INSTIs-based ART is adopted in Cameroon, studies of patients failing INSTIs-based ART should include both analyses of these 3′-PPT mutations and integrase DRMs to elucidate the potential role of these mutations on the susceptibility to DTG and other INSTIs.
In summary, the current study of integrase and nef viral sequences from 345 HIV-infected Cameroonians showed only 2 subjects with INSTIs major RAMs, but several subjects with INSTIs accessory RAMs and polymorphic mutations, including 10 subjects harboring viruses that simultaneously had two different INSTIs accessory RAMs. Individually, INSTIs accessory RAMs do not have major effects on susceptibility to INSTIs, but the simultaneous presence of several accessory mutations or their presence in combination with other mutations has been associated with reduced susceptibility to INSTIs, increased viral fitness and virologic failure [40,60,61,62,63,64,65]. With the current worldwide push to expand the use of DTG/INSTIs-based ART, it is inevitable that some patients on these regimens could experience virologic failure at some point during the course of their treatment. Genetic surveillance for management of such cases should include screening for the presence of INSTIs major RAMs, accessory and polymorphic mutations, and potential combinations of these, as well as mutations in the 3′-PPT region.
4. Materials and Methods
4.1. Ethics Statement
The study cohort samples were collected as part of an ongoing project aimed at analyzing the influence of HIV genetic diversity on viral neuropathogenesis in Cameroon. This study was conducted in accordance with the Declaration of Helsinki and the protocol was approved by the Cameroon National Ethics Committee (Ethical Clearance Authorization #146/CNE/SE/2012, approved on 13 June 2006 and re-approved on 2 May 2012), as well as the Institutional Review Board of the University of Nebraska Medical Center (UNMC) (IRB #307-06-FB, approved on 26 March 2007 and re-approved annually until 2018). Written informed consent was obtained from all study participants and data were processed using unique identifiers to ensure confidentiality.
4.2. Specimen Collection, HIV Serology, CD4 Cell Counts, and Viral Loads
Sample collection, serology, CD4 cell counts, and viral loads analyses were performed in the Hematology laboratory of the Yaoundé University Teaching Hospital or the International Reference Center “Chantal Biya”, Cameroon, between 2008 and 2016. Venous blood samples were collected and stored at room temperature in the outpatient clinic and analyses performed in the Hematology laboratory within 6 h of blood collection. The HIV status of each participant was determined using the rapid immunochromatographic HIV-1/2 test (Abbott Diagnostics, Chicago, IL, USA) and the Murex HIV antigen/antibody Combination ELISA (Abbott Diagnostics), according to the manufacturer’s instructions. A participant was considered HIV positive if he/she tested positive for the two tests, HIV negative if non-reactive for both tests, and discordant if reactive for only one test. No discordant result was observed in our study population.
CD4 T-lymphocyte counts were quantified by flow cytometry, using a Fluorescence-Activated Cell Sorting (FACS) Count Instrumentation System and the BD FACSCount CD4 reagent kit (BD Biosciences, San Jose, CA, USA), according to the manufacturer’s instructions. The FACS instrument was calibrated and quality control tested before each experiment. For viral loads quantification, HIV plasma viral load was quantified by reverse transcription polymerase chain reaction (RT-PCR), using Amplicor HIV-1 Monitor Test (Roche Diagnostic Systems, Pleasanton, CA, USA), according to the manufacturer’s protocol. The assay detection limit was 40 viral RNA copies/mL. For additional molecular analyses, plasma samples were stored in 1mL aliquots at −80 °C until further use.
4.3. RNA Extraction and PCR Amplification
Plasma samples were shipped on dry ice (−70 ℃) to UNMC, where sequencing and analyses of the integrase and nef (viral 3′-PPT region) genes were performed. HIV-1 RNA was extracted from plasma using QIAamp Viral RNA mini kit (Qiagen Inc., Germantown, MD, USA) per manufacturer’s protocol. cDNA synthesis and 1st round of PCR amplification of integrase and nef genes was performed using SuperScriptTM III One-Step RT-PCR system (Life Technologies, Grand Island, NY, USA). The 50 µL reaction volume contained 500 ng of purified RNA, 25 µL of 2X reaction buffer, 10 pMol of forward and reverse primers (Table 6) and 1 µL reverse transcriptase /Platinum Taq DNA polymerase mix. Reverse transcription was carried out at 50 ℃ for 1 h followed by PCR consisting of an initial denaturation at 94 ℃ for 2 min; followed by 40 cycles of 94 ℃, 15 s; 55 ℃, 30 s; 68 ℃, 1 min; and a final extension step at 72 ℃, 10 min. The nested PCR amplification of integrase and nef genes was carried out in a total volume of 50 µL containing 5 µL of the 1st-round PCR amplicon, 25 µL of 2X KAPA HiFi HotStart ready Mix (KAPA Biosystems, Wilmington, MA, USA), 10 pMol of forward and reverse primers. The nested PCR thermal cycling condition were 95 ℃, 5 min; followed by 35 cycles of 94 ℃, 10s, 60 C, 30s; 72 ℃, 1 min; and a final extension step at 72 ℃, 10 min. Amplicons were detected by agarose gel electrophoresis (1% agarose), visualized by ethidium bromide (0.5 µg/mL) staining under ultraviolet light (260 nm), and images captured using an automated gel documentation system (Syngene, Frederick, MD). Sequences of the primers used for PCR amplifications are detailed in Table 6.

Table 6.
Primers used for amplification and sequencing of integrase and nef genes [83,84].
4.4. Gene Sequencing
PCR products were purified using PureLink Quick PCR Purification Kit (Invitrogen, Carlsbad, CA, USA) and subjected to double-strand DNA sequencing to cover the entire amplicon using a set of sequencing primers (Table 6). The sequencing reactions were carried out using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) per manufacturer’s instructions, followed by capillary electrophoresis performed on an Applied Biosystems PRISM 3730 Genetic Analyzer at the UNMC DNA Sequencing Core facility. Sequences of the primers used for DNA sequencing are described in Table 6.
4.5. Sequence Analysis of the Study Cohort Samples
Raw sequence data were manually edited, spliced, and assembled by Sequencher v4.9 to generate the final contig. Multiple sequence alignment of full-length HIV-1 integrase gene was performed with all known HIV-1 group M reference sequences, using Clustal W [85]. All the reference sequences were obtained from the Los Alamos HIV Sequence Database. Phylogenetic trees were constructed using the neighbor-joining method, as well as the Maximum Likelihood method and General Time Reversible model, with 1000 bootstrap replication tests, using MEGA.v.6.0 software [86]. Samples’ HIV subtypes were determined using the NCBI viral genotyping tool. The INSTIs drug resistance associated mutation screening was done using the Stanford University HIV Drug resistance Database v.8.7. [87]. The 3′-PPT and its flanking nucleotide sequences were curated from the full-length nef gene sequences, and their sequence alignments performed using HXB2 as a reference sequence and the Clustal W program [85].
4.6. Sequence Analysis of the Database Samples
We analyzed additional Cameroon HIV-1 sequences (integrase and nef/3′-PPT sequences) using full-length HIV-1 sequences from Cameroon previously submitted to the Los Alamos HIV sequence database [28]. We downloaded all Cameroon sequences available in the database and after eliminating duplicate sequences from the same patient, a total of 215 full-length sequences were included in the analysis. Full-length integrase gene sequences for these 215 samples were used to screen for INSTIs RAMs, using the Stanford University HIV Drug resistance Database v.8.7. [87]. The 3′-PPT and its flanking nucleotide sequences were curated from the full length HIV-1 genomic sequences, and their sequence alignments performed using HXB2 as reference sequence and the Clustal W program [85].
4.7. Statistical Analyses
Comparative analyses of males’ and females’ demographic data were performed using the Student’s t-tests (for continuous variables) and Fisher’s exact test (for binary variables). Descriptive statistics including counts and percentages were used to summarize gene polymorphism or mutation occurrences for both cohort and database samples. Fisher’s exact tests were used to compare the proportion of gene polymorphism or mutation occurrences between groups for each gene. False discovery rate (FDR) was controlled to be no more than 0.05 to account for multiple comparisons [88]. All analyses were done using SAS version 9.4 or Prims version 7.0d.
4.8. Data Availability
The full-length HIV-1 integrase and nef gene sequences generated from this study are available in the NCBI database with GenBank accession numbers: MK327828-MK327927 (integrase sequences) and MK333810-MK333910 (nef [including 3′-PPT] sequences).
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/5/1553/s1.
Author Contributions
Conceptualization, A.A. and G.D.K.; investigation, A.A., C.T.T., E.N., and L.K.; data curation, G.D.K., J.Y.F., C.T.T., and D.M.; formal analysis, A.A., M.J., and G.D.K.; writing–original draft preparation, A.A. and G.D.K.; writing–review and editing, G.D.K., A.A., and A.K.N.; project administration, G.D.K., D.M., and A.K.N.; supervision, G.D.K.; funding acquisition, G.D.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Institute of Mental Health MH094160 and the Fogarty International Center.
Acknowledgments
We thank all volunteers who participated in this study. We thank the University of Nebraska Medical Center High-Throughput DNA Sequencing and Genotyping Core Facility for assistance with gene sequencing.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| HIV | Human immunodeficiency virus |
| INSTIs | Integrase strand-transfer inhibitors |
| ART | Antiretroviral therapy |
| 3′-PPT | 3′polypurine tract |
| RAMs | Resistance-associated mutations |
| CRF | Circulating recombinant form |
| T | Threonine |
| A | Alanine |
| N | Asparagine |
| K | Lysine |
| E | Glutamic Acid |
| Q | Glutamine |
| M | Methionine |
| I | Isoleucine |
| S | Serine |
| R | Arginine |
| L | Leucine |
| D | Aspartic Acid |
| V | Valine |
| G | Glycine |
| C | Cysteine |
| DNA | Deoxyribonucleic acid |
| kDa | Kilodalton |
| Aa | Amino acids |
| AIDS | Acquired immunodeficiency syndrome |
| FDA | United States Food and Drug Administration |
| RAL | Raltegravir |
| EVG | Elvitegravir |
| DTG | Dolutegravir |
| WHO | World Health Organization |
| SSA | Sub-Saharan Africa |
| PLWH | People living with HIV |
| DRMs | Drug-resistance mutations |
| URF | Unique recombinant form |
| LTR | Long terminal repeats |
| UNMC | University of Nebraska Medical Center |
| ELISA | Enzyme-linked immunosorbent assay |
| CD4 | cluster of differentiation 4 |
| FACS | Fluorescence-Activated Cell Sorting |
| RT-PCR | Reverse transcription polymerase chain reaction |
| RNA | Ribonucleic acid |
| cDNA | complementary DNA |
| PCR | Polymerase chain reaction |
| NCBI | National Center for Biotechnology Information |
| FDR | False discovery rate |
References
- Ceccherini-Silberstein, F.; Malet, I.; D’Arrigo, R.; Antinori, A.; Marcelin, A.G.; Perno, C.F. Characterization and structural analysis of HIV-1 integrase conservation. Aids Rev. 2009, 11, 17–29. [Google Scholar] [PubMed]
- Li, X.; Krishnan, L.; Cherepanov, P.; Engelman, A. Structural biology of retroviral DNA integration. Virology 2011, 411, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Lesbats, P.; Engelman, A.N.; Cherepanov, P. Retroviral DNA Integration. Chem. Rev. 2016, 116, 12730–12757. [Google Scholar] [CrossRef]
- Asante-Appiah, E.; Skalka, A.M. HIV-1 integrase: Structural organization, conformational changes, and catalysis. Adv. Virus. Res. 1999, 52, 351–369. [Google Scholar]
- Zheng, R.; Jenkins, T.M.; Craigie, R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 1996, 93, 13659–13664. [Google Scholar] [CrossRef]
- Cai, M.; Zheng, R.; Caffrey, M.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997, 4, 567–577. [Google Scholar] [CrossRef]
- DHHS. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. AIDSinfo 2019. Available online: https://aidsinfo.nih.gov/guidelines/html/1/adult-and-adolescent-arv/0 (accessed on 22 June 2019).
- Wong, E.; Trustman, N.; Yalong, A. HIV pharmacotherapy: A review of integrase inhibitors. JAAPA 2016, 29, 36–40. [Google Scholar] [CrossRef]
- Psichogiou, M.; Poulakou, G.; Basoulis, D.; Paraskevis, D.; Markogiannakis, A.; Daikos, G.L. Recent Advances in Antiretroviral Agents: Potent Integrase Inhibitors. Curr Pharm Des. 2017, 23, 2552–2567. [Google Scholar] [CrossRef]
- EACS. Guidelines. European AIDS Clinical Society, 2019. Available online: https://www.eacsociety.org/files/2019_guidelines-10.0_final.pdf (accessed on 21 June 2019).
- WHO. Consolidated Guidelines On THE USE OF ANTIRETROVIRAL DRUGS FOR TREATING AND PREVENTING HIV INFECTION. Recommendations for a Public Health Approach. Second Edition. World Health Organization, 2016. Available online: https://apps.who.int/iris/bitstream/handle/10665/208825/9789241549684_eng.pdf?sequence=1 (accessed on 21 June 2019).
- WHO. Update of Recommendations on First- and Second-Line Antiretroviral Regimens. World Health Organization, 2019. Available online: https://www.who.int/hiv/pub/arv/arv-update-2019-policy/en/ (accessed on 21 February 2020).
- Inzaule, S.C.; Hamers, R.L.; Noguera-Julian, M.; Casadella, M.; Parera, M.; Rinke de Wit, T.F.; Paredes, R. Primary resistance to integrase strand transfer inhibitors in patients infected with diverse HIV-1 subtypes in sub-Saharan Africa. J. Antimicrob. Chemother. 2018, 73, 1167–1172. [Google Scholar] [CrossRef]
- WHO. Guidelines on THE PUBLIC HEALTH RESPONSE TO PRETREATMENT HIV DRUG RESISTANCE World Health Organization 2017. Available online: https://apps.who.int/iris/bitstream/handle/10665/255880/9789241550055-eng.pdf;jsessionid=E4F7C6DA3BC67323033C2666046BBC99?sequence=1 (accessed on 21 June 2019).
- MPP. Update on Progress of Sublicensees. Medicines Patent Pool, 2019. Available online: https://medicinespatentpool.org/what-we-do/global-licence-overview/update-on-progress-of-mpp-sublicensees/ (accessed on 4 February 2019).
- Gupta, A.; Juneja, S.; Vitoria, M.; Habiyambere, V.; Nguimfack, B.D.; Doherty, M.; Low-Beer, D. Projected Uptake of New Antiretroviral (ARV) Medicines in Adults in Low- and Middle-Income Countries: A Forecast Analysis 2015-2025. PLoS ONE 2016, 11, e0164619. [Google Scholar] [CrossRef]
- MPP. Five Years on, 3.9 Million People in the Developing World Have Access to HIV Treatment Dolutegravir, Thanks to Access-oriented Voluntary Licensing Agreements. Medicines Patent Pool, 2019. Available online: https://medicinespatentpool.org/mpp-media-post/five-years-on-3-9-million-people-in-the-developing-world-have-access-to-hiv-treatment-dolutegravir-thanks-to-access-oriented-voluntary-licensing-agreements/ (accessed on 4 February 2019).
- WHO. GLOBAL ACTION PLAN ON HIV DRUG RESISTANCE 2017–2021. World Health Organization, 2017. Available online: https://apps.who.int/iris/bitstream/handle/10665/255883/9789241512848-eng.pdf?sequence=1 (accessed on 21 June 2019).
- WHO. HIV Drug Resistance Report 2019. World Health Organization, 2019. Available online: https://www.who.int/hiv/pub/drugresistance/hivdr-report-2019/en/ (accessed on 21 February 2020).
- Lepik, K.J.; Harrigan, P.R.; Yip, B.; Wang, L.; Robbins, M.A.; Zhang, W.W.; Toy, J.; Akagi, L.; Lima, V.D.; Guillemi, S.; et al. Emergent drug resistance with integrase strand transfer inhibitor-based regimens. AIDS 2017, 31, 1425–1434. [Google Scholar] [CrossRef]
- Hurt, C.B.; Sebastian, J.; Hicks, C.B.; Eron, J.J. Resistance to HIV integrase strand transfer inhibitors among clinical specimens in the United States, 2009–2012. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2014, 58, 423–431. [Google Scholar] [CrossRef]
- Anstett, K.; Brenner, B.; Mesplede, T.; Wainberg, M.A. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 2017, 14, 36. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Liu, T.F.; Kiuchi, M.; Zioni, R.; Gifford, R.J.; Holmes, S.P.; Shafer, R.W. Natural variation of HIV-1 group M integrase: Implications for a new class of antiretroviral inhibitors. Retrovirology 2008, 5, 74. [Google Scholar] [CrossRef]
- UNAIDS. Global HIV & AIDS statistics—2019 fact sheet. HIV Epidemic Update. 2019. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 21 February 2019).
- Malet, I.; Subra, F.; Charpentier, C.; Collin, G.; Descamps, D.; Calvez, V.; Marcelin, A.G.; Delelis, O. Mutations Located outside the Integrase Gene Can Confer Resistance to HIV-1 Integrase Strand Transfer Inhibitors. mBio 2017, 8. [Google Scholar] [CrossRef]
- Wijting, I.E.A.; Lungu, C.; Rijnders, B.J.A.; van der Ende, M.E.; Pham, H.T.; Mesplede, T.; Pas, S.D.; Voermans, J.J.C.; Schuurman, R.; van de Vijver, D.; et al. HIV-1 Resistance Dynamics in Patients With Virologic Failure to Dolutegravir Maintenance Monotherapy. J. Infect. Dis. 2018, 218, 688–697. [Google Scholar] [CrossRef]
- Das, A.T.; Berkhout, B. How Polypurine Tract Changes in the HIV-1 RNA Genome Can Cause Resistance against the Integrase Inhibitor Dolutegravir. mBio 2018, 9. [Google Scholar] [CrossRef]
- LANL. HIV Reference Sequence Database; Los Alamos National Library: Los Alamos, NM, USA, 2019. Available online: https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html (accessed on 21 February 2020).
- Peeters, M.; Liegeois, F.; Torimiro, N.; Bourgeois, A.; Mpoudi, E.; Vergne, L.; Saman, E.; Delaporte, E.; Saragosti, S. Characterization of a highly replicative intergroup M/O human immunodeficiency virus type 1 recombinant isolated from a Cameroonian patient. J. Virol. 1999, 73, 7368–7375. [Google Scholar] [CrossRef]
- Powell, R.L.; Zhao, J.; Konings, F.A.; Tang, S.; Nanfack, A.; Burda, S.; Urbanski, M.M.; Saa, D.R.; Hewlett, I.; Nyambi, P.N. Identification of a novel circulating recombinant form (CRF) 36_cpx in Cameroon that combines two CRFs (01_AE and 02_AG) with ancestral lineages of subtypes A and G. Aids Res. Hum. Retrovir. 2007, 23, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Bodelle, P.; Coffey, R.; Devare, S.G.; Golden, A.; Hackett, J., Jr.; Harris, B.; Holzmayer, V.; Luk, K.C.; Schochetman, G.; et al. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J. Acquir. Immune. Defic. Syndr. 2008, 49, 432–439. [Google Scholar] [CrossRef]
- Teto, G.; Fonsah, J.Y.; Tagny, C.T.; Mbanya, D.; Nchindap, E.; Kenmogne, L.; Fokam, J.; Njamnshi, D.M.; Kouanfack, C.; Njamnshi, A.K.; et al. Molecular and Genetic Characterization of HIV-1 Tat Exon-1 Gene from Cameroon Shows Conserved Tat HLA-Binding Epitopes: Functional Implications. Viruses 2016, 8, 196. [Google Scholar] [CrossRef]
- Teto, G.; Tagny, C.T.; Mbanya, D.; Fonsah, J.Y.; Fokam, J.; Nchindap, E.; Kenmogne, L.; Njamnshi, A.K.; Kanmogne, G.D. Gag P2/NC and pol genetic diversity, polymorphism, and drug resistance mutations in HIV-1 CRF02_AG- and non-CRF02_AG-infected patients in Yaounde, Cameroon. Sci. Rep. 2017, 7, 14136. [Google Scholar] [CrossRef]
- Villabona-Arenas, C.J.; Domyeum, J.; Mouacha, F.; Butel, C.; Delaporte, E.; Peeters, M.; Mpoudi-Ngole, E.; Aghokeng, A.F. HIV-1 group O infection in Cameroon from 2006 to 2013: Prevalence, genetic diversity, evolution and public health challenges. Infection, genetics and evolution. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 36, 210–216. [Google Scholar] [CrossRef]
- Margot, N.A.; Hluhanich, R.M.; Jones, G.S.; Andreatta, K.N.; Tsiang, M.; McColl, D.J.; White, K.L.; Miller, M.D. In vitro resistance selections using elvitegravir, raltegravir, and two metabolites of elvitegravir M1 and M4. Antivir. Res. 2012, 93, 288–296. [Google Scholar] [CrossRef]
- Abram, M.E.; Hluhanich, R.M.; Goodman, D.D.; Andreatta, K.N.; Margot, N.A.; Ye, L.; Niedziela-Majka, A.; Barnes, T.L.; Novikov, N.; Chen, X.; et al. Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness. Antimicrob. Agents Chemother. 2013, 57, 2654–2663. [Google Scholar] [CrossRef]
- Van der Borght, K.; Verheyen, A.; Feyaerts, M.; Van Wesenbeeck, L.; Verlinden, Y.; Van Craenenbroeck, E.; van Vlijmen, H. Quantitative prediction of integrase inhibitor resistance from genotype through consensus linear regression modeling. Virol. J. 2013, 10, 8. [Google Scholar] [CrossRef]
- Winters, M.A.; Lloyd, R.M., Jr.; Shafer, R.W.; Kozal, M.J.; Miller, M.D.; Holodniy, M. Development of elvitegravir resistance and linkage of integrase inhibitor mutations with protease and reverse transcriptase resistance mutations. PLoS ONE 2012, 7, e40514. [Google Scholar] [CrossRef]
- Van Wesenbeeck, L.; Rondelez, E.; Feyaerts, M.; Verheyen, A.; Van der Borght, K.; Smits, V.; Cleybergh, C.; De Wolf, H.; Van Baelen, K.; Stuyver, L.J. Cross-resistance profile determination of two second-generation HIV-1 integrase inhibitors using a panel of recombinant viruses derived from raltegravir-treated clinical isolates. Antimicrob. Agents Chemother. 2011, 55, 321–325. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fransen, S.; Gupta, S.; Frantzell, A.; Petropoulos, C.J.; Huang, W. Substitutions at amino acid positions 143, 148, and 155 of HIV-1 integrase define distinct genetic barriers to raltegravir resistance in vivo. J. Virol. 2012, 86, 7249–7255. [Google Scholar] [CrossRef]
- Carganico, A.; Dupke, S.; Ehret, R.; Berg, T.; Baumgarten, A.; Obermeier, M.; Walter, H. New dolutegravir resistance pattern identified in a patient failing antiretroviral therapy. J. Int. AIDS Soc. 2014, 17, 19749. [Google Scholar] [CrossRef]
- Hardy, I.; Brenner, B.; Quashie, P.; Thomas, R.; Petropoulos, C.; Huang, W.; Moisi, D.; Wainberg, M.A.; Roger, M. Evolution of a novel pathway leading to dolutegravir resistance in a patient harbouring N155H and multiclass drug resistance. J. Antimicrob. Chemother. 2015, 70, 405–411. [Google Scholar] [CrossRef]
- Nyamache, A.K.; Muigai, A.W.; Nganga, Z.; Khamadi, S.A. HIV Type 1 genetic diversity and naturally occurring polymorphisms in HIV type 1 Kenyan isolates: Implications for integrase inhibitors. Aids Res. Hum. Retrovir. 2012, 28, 933–936. [Google Scholar] [CrossRef]
- Brado, D.; Obasa, A.E.; Ikomey, G.M.; Cloete, R.; Singh, K.; Engelbrecht, S.; Neogi, U.; Jacobs, G.B. Analyses of HIV-1 integrase sequences prior to South African national HIV-treatment program and available of integrase inhibitors in Cape Town, South Africa. Sci. Rep. 2018, 8, 4709. [Google Scholar] [CrossRef]
- Ndashimye, E.; Avino, M.; Kyeyune, F.; Nankya, I.; Gibson, R.M.; Nabulime, E.; Poon, A.F.Y.; Kityo, C.; Mugyenyi, P.; Quinones-Mateu, M.E.; et al. Absence of HIV-1 Drug Resistance Mutations Supports the Use of Dolutegravir in Uganda. Aids Res. Hum. Retrovir. 2018, 34, 404–414. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, E.J.; Choi, J.Y.; Kwon, O.K.; Kim, G.J.; Choi, S.Y.; Kim, S.S. Genetic variation of the HIV-1 integrase region in newly diagnosed anti-retroviral drug-naive patients with HIV/AIDS in Korea. Clinical microbiology and infection. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2011, 17, 1155–1159. [Google Scholar] [CrossRef]
- Nouhin, J.; Donchai, T.; Hoang, K.T.; Ken, S.; Kamkorn, J.; Tran, T.; Ayouba, A.; Peeters, M.; Chaix, M.L.; Lien, T.X.; et al. Natural polymorphisms of HIV-1 CRF01_AE integrase coding region in ARV-naive individuals in Cambodia, Thailand and Vietnam: An ANRS AC12 working group study. Infection, genetics and evolution. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2011, 11, 38–43. [Google Scholar] [CrossRef]
- Kotaki, T.; Khairunisa, S.Q.; Sukartiningrum, S.D.; Witaningrum, A.M.; Rusli, M.; Diansyah, M.N.; Arfijanto, M.V.; Rahayu, R.P.; Nasronudin; Kameoka, M. Detection of drug resistance-associated mutations in human immunodeficiency virus type 1 integrase derived from drug-naive individuals in Surabaya, Indonesia. Aids Res. Hum. Retrovir. 2014, 30, 489–492. [Google Scholar] [CrossRef]
- Kim, Y.; Chin, B.S.; Kim, G.; Shin, H.S. Integrase Strand Transfer Inhibitor Resistance Mutations in Antiretroviral Treatment-naive Patients in Korea: A Prospective, Observational Study. J. Korean. Med. Sci. 2018, 33, e173. [Google Scholar] [CrossRef]
- Tsai, H.C.; Chen, I.T.; Wu, K.S.; Tseng, Y.T.; Sy, C.L.; Chen, J.K.; Lee, S.S.; Chen, Y.S. HIV-1 integrase strand-transfer inhibitor resistance in southern Taiwan. Oncotarget 2018, 9, 24927–24935. [Google Scholar] [CrossRef][Green Version]
- Chang, S.-Y.; Lin, P.-H.; Cheng, C.-L.; Chen, M.-Y.; Sun, H.-Y.; Hsieh, S.-M.; Sheng, W.-H.; Su, Y.-C.; Su, L.-H.; Chang, S.-F.; et al. Prevalence of Integrase Strand Transfer Inhibitors (INSTI) Resistance Mutations in Taiwan. Sci. Rep. 2016, 6, 35779. [Google Scholar] [CrossRef]
- Meixenberger, K.; Yousef, K.P.; Smith, M.R.; Somogyi, S.; Fiedler, S.; Bartmeyer, B.; Hamouda, O.; Bannert, N.; von Kleist, M.; Kucherer, C. Molecular evolution of HIV-1 integrase during the 20 years prior to the first approval of integrase inhibitors. Virol. J. 2017, 14, 223. [Google Scholar] [CrossRef]
- Avi, R.; Huik, K.; Sadam, M.; Karki, T.; Krispin, T.; Ainsalu, K.; Paap, P.; Schmidt, J.; Nikitina, N.; Lutsar, I. Characterization of integrase region polymorphisms in HIV type 1 CRF06_cpx viruses in treatment-naive patients in Estonia. Aids Res. Hum. Retrovir. 2010, 26, 1109–1113. [Google Scholar] [CrossRef]
- Tostevin, A.; White, E.; Dunn, D.; Croxford, S.; Delpech, V.; Williams, I.; Asboe, D.; Pozniak, A.; Churchill, D.; Geretti, A.M.; et al. Recent trends and patterns in HIV-1 transmitted drug resistance in the United Kingdom. Hiv Med. 2017, 18, 204–213. [Google Scholar] [CrossRef]
- Passaes, C.B.; Guimaraes, M.L.; Fernandez, S.L.; Lorete Rdos, S.; Teixeira, S.L.; Fernandez, J.C.; Morgado, M.G. Lack of primary mutations associated with integrase inhibitors among HIV-1 subtypes B, C, and F circulating in Brazil. J. Acquir. Immune. Defic. Syndr. 2009, 51, 7–12. [Google Scholar] [CrossRef]
- Mouscadet, J.F.; Delelis, O.; Marcelin, A.G.; Tchertanov, L. Resistance to HIV-1 integrase inhibitors: A structural perspective. Drug Resist. Updates: Rev. Comment. Antimicrob. Anticancer Chemother. 2010, 13, 139–150. [Google Scholar] [CrossRef]
- Li, Y.; Xuan, S.; Feng, Y.; Yan, A. Targeting HIV-1 integrase with strand transfer inhibitors. Drug Discov. Today 2015, 20, 435–449. [Google Scholar] [CrossRef]
- Mulu, A.; Maier, M.; Liebert, U.G. Lack of integrase inhibitors associated resistance mutations among HIV-1C isolates. J. Transl. Med. 2015, 13, 377. [Google Scholar] [CrossRef]
- Pham, H.T.; Labrie, L.; Wijting, I.E.A.; Hassounah, S.; Lok, K.Y.; Portna, I.; Goring, M.E.; Han, Y.; Lungu, C.; van der Ende, M.E.; et al. The S230R Integrase Substitution Associated With Virus Load Rebound During Dolutegravir Monotherapy Confers Low-Level Resistance to Integrase Strand-Transfer Inhibitors. J. Infect. Dis. 2018, 218, 698–706. [Google Scholar] [CrossRef]
- Fransen, S.; Gupta, S.; Danovich, R.; Hazuda, D.; Miller, M.; Witmer, M.; Petropoulos, C.J.; Huang, W. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J. Virol. 2009, 83, 11440–11446. [Google Scholar] [CrossRef]
- Eron, J.J.; Clotet, B.; Durant, J.; Katlama, C.; Kumar, P.; Lazzarin, A.; Poizot-Martin, I.; Richmond, G.; Soriano, V.; Ait-Khaled, M.; et al. Safety and efficacy of dolutegravir in treatment-experienced subjects with raltegravir-resistant HIV type 1 infection: 24-week results of the VIKING Study. J. Infect. Dis. 2013, 207, 740–748. [Google Scholar] [CrossRef]
- Naeger, L.K.; Harrington, P.; Komatsu, T.; Deming, D. Effect of dolutegravir functional monotherapy on HIV-1 virological response in integrase strand transfer inhibitor resistant patients. Antivir. Ther. 2016, 21, 481–488. [Google Scholar] [CrossRef]
- George, J.M.; Kuriakose, S.S.; Dee, N.; Stoll, P.; Lalani, T.; Dewar, R.; Khan, M.A.; Rehman, M.T.; Grossman, Z.; Maldarelli, F.; et al. Rapid Development of High-Level Resistance to Dolutegravir With Emergence of T97A Mutation in 2 Treatment-Experienced Individuals With Baseline Partial Sensitivity to Dolutegravir. Open Forum. Infect. Dis. 2018, 5, ofy221. [Google Scholar] [CrossRef]
- Charpentier, C.; Malet, I.; Andre-Garnier, E.; Storto, A.; Bocket, L.; Amiel, C.; Morand-Joubert, L.; Tumiotto, C.; Nguyen, T.; Maillard, A.; et al. Phenotypic analysis of HIV-1 E157Q integrase polymorphism and impact on virological outcome in patients initiating an integrase inhibitor-based regimen. J. Antimicrob. Chemother. 2018, 73, 1039–1044. [Google Scholar] [CrossRef]
- Hombrouck, A.; Voet, A.; Van Remoortel, B.; Desadeleer, C.; De Maeyer, M.; Debyser, Z.; Witvrouw, M. Mutations in human immunodeficiency virus type 1 integrase confer resistance to the naphthyridine L-870,810 and cross-resistance to the clinical trial drug GS-9137. Antimicrob. Agents Chemother. 2008, 52, 2069–2078. [Google Scholar] [CrossRef]
- Quashie, P.K.; Oliviera, M.; Veres, T.; Osman, N.; Han, Y.S.; Hassounah, S.; Lie, Y.; Huang, W.; Mesplede, T.; Wainberg, M.A. Differential effects of the G118R, H51Y, and E138K resistance substitutions in different subtypes of HIV integrase. J. Virol. 2015, 89, 3163–3175. [Google Scholar] [CrossRef]
- Bar-Magen, T.; Sloan, R.D.; Faltenbacher, V.H.; Donahue, D.A.; Kuhl, B.D.; Oliveira, M.; Xu, H.; Wainberg, M.A. Comparative biochemical analysis of HIV-1 subtype B and C integrase enzymes. Retrovirology 2009, 6, 103. [Google Scholar] [CrossRef]
- Bar-Magen, T.; Donahue, D.A.; McDonough, E.I.; Kuhl, B.D.; Faltenbacher, V.H.; Xu, H.; Michaud, V.; Sloan, R.D.; Wainberg, M.A. HIV-1 subtype B and C integrase enzymes exhibit differential patterns of resistance to integrase inhibitors in biochemical assays. AIDS 2010, 24, 2171–2179. [Google Scholar] [CrossRef]
- Fish, M.Q.; Hewer, R.; Wallis, C.L.; Venter, W.D.; Stevens, W.S.; Papathanasopoulos, M.A. Natural polymorphisms of integrase among HIV type 1-infected South African patients. Aids Res. Hum. Retrovir. 2010, 26, 489–493. [Google Scholar] [CrossRef]
- Maiga, A.I.; Malet, I.; Soulie, C.; Derache, A.; Koita, V.; Amellal, B.; Tchertanov, L.; Delelis, O.; Morand-Joubert, L.; Mouscadet, J.F.; et al. Genetic barriers for integrase inhibitor drug resistance in HIV type-1 B and CRF02_AG subtypes. Antivir. Ther. 2009, 14, 123–129. [Google Scholar]
- Han, Y.S.; Mesplede, T.; Wainberg, M.A. Differences among HIV-1 subtypes in drug resistance against integrase inhibitors. Infection, genetics and evolution. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 46, 286–291. [Google Scholar] [CrossRef]
- Hill, K.J.; Rogers, L.C.; Njenda, D.T.; Burke, D.H.; Sarafianos, S.G.; Sonnerborg, A.; Neogi, U.; Singh, K. Strain-specific effect on biphasic DNA binding by HIV-1 integrase. AIDS 2019, 33, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Cahn, P.; Pozniak, A.L.; Mingrone, H.; Shuldyakov, A.; Brites, C.; Andrade-Villanueva, J.F.; Richmond, G.; Buendia, C.B.; Fourie, J.; Ramgopal, M.; et al. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: Week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 2013, 382, 700–708. [Google Scholar] [CrossRef]
- Theys, K.; Libin, P.J.K.; Van Laethem, K.; Abecasis, A.B. An Evolutionary Model-Based Approach To Quantify the Genetic Barrier to Drug Resistance in Fast-Evolving Viruses and Its Application to HIV-1 Subtypes and Integrase Inhibitors. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Soriano, V.; Geretti, A.M.; Zahonero, N.; Garcia, S.; Booth, C.; Gutierrez, F.; Viciana, I.; de Mendoza, C. Resistance associated mutations to dolutegravir (S/GSK1349572) in HIV-infected patients--impact of HIV subtypes and prior raltegravir experience. Antivir. Res. 2011, 90, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Modica, S.; Rossetti, B.; Lombardi, F.; Lagi, F.; Maffeo, M.; D’Autilia, R.; Pecorari, M.; Vicenti, I.; Bruzzone, B.; Magnani, G.; et al. Prevalence and determinants of resistance mutations in HIV-1-infected patients exposed to integrase inhibitors in a large Italian cohort. Hiv Med. 2019, 20, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Saladini, F.; Meini, G.; Bianco, C.; Monno, L.; Punzi, G.; Pecorari, M.; Borghi, V.; Di Pietro, M.; Filice, G.; Gismondo, M.R.; et al. Prevalence of HIV-1 integrase mutations related to resistance to dolutegravir in raltegravir naive and pretreated patients. Clin. Microbiol. Infect 2012, 18, E428–E430. [Google Scholar] [CrossRef][Green Version]
- Abecasis, A.B.; Deforche, K.; Bacheler, L.T.; McKenna, P.; Carvalho, A.P.; Gomes, P.; Vandamme, A.M.; Camacho, R.J. Investigation of baseline susceptibility to protease inhibitors in HIV-1 subtypes C, F, G and CRF02_AG. Antivir. Ther. 2006, 11, 581–589. [Google Scholar]
- Theys, K.; Vercauteren, J.; Snoeck, J.; Zazzi, M.; Camacho, R.J.; Torti, C.; Schulter, E.; Clotet, B.; Sonnerborg, A.; De Luca, A.; et al. HIV-1 subtype is an independent predictor of reverse transcriptase mutation K65R in HIV-1 patients treated with combination antiretroviral therapy including tenofovir. Antimicrob. Agents Chemother. 2013, 57, 1053–1056. [Google Scholar] [CrossRef]
- Turner, D.; Brenner, B.; Moisi, D.; Detorio, M.; Cesaire, R.; Kurimura, T.; Mori, H.; Essex, M.; Maayan, S.; Wainberg, M.A. Nucleotide and amino acid polymorphisms at drug resistance sites in non-B-subtype variants of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2004, 48, 2993–2998. [Google Scholar] [CrossRef]
- Rausch, J.W.; Le Grice, S.F. ‘Binding, bending and bonding’: Polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int. J. Biochem. Cell Biol. 2004, 36, 1752–1766. [Google Scholar] [CrossRef]
- Julias, J.G.; McWilliams, M.J.; Sarafianos, S.G.; Alvord, W.G.; Arnold, E.; Hughes, S.H. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J. Virol. 2004, 78, 13315–13324. [Google Scholar] [CrossRef]
- Malet, I.; Delelis, O.; Valantin, M.A.; Montes, B.; Soulie, C.; Wirden, M.; Tchertanov, L.; Peytavin, G.; Reynes, J.; Mouscadet, J.F.; et al. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 2008, 52, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Jubier-Maurin, V.; Saragosti, S.; Perret, J.L.; Mpoudi, E.; Esu-Williams, E.; Mulanga, C.; Liegeois, F.; Ekwalanga, M.; Delaporte, E.; Peeters, M. Genetic characterization of the nef gene from human immunodeficiency virus type 1 group M strains representing genetic subtypes A, B, C, E, F, G, and H. Aids Res. Hum. Retrovir. 1999, 15, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Aiyar, A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 2000, 132, 221–241. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Integrase Strand Transfer Inhibitors Resistance Notes. HIV Drug Resistance Database 2019, Stanford University. HIVdb version 8.9-1. Available online: https://hivdb.stanford.edu/dr-summary/resistance-notes/INSTI/ (accessed on 21 February 2020).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).