Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family, and Interaction and Functional Analysis of TaWOX9 and TaWUS in Wheat
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of WOX Genes in Wheat
2.2. Multiple Sequence Alignment and Sequence Features of TaWOX Proteins
2.3. Expression Profile of TaWOX Genes in Wheat
2.4. Subcellular Localization and Transactivation Activity of TaWUS and TaWOX9
2.5. Identification of Proteins That Interact with TaWUS and TaWOX9
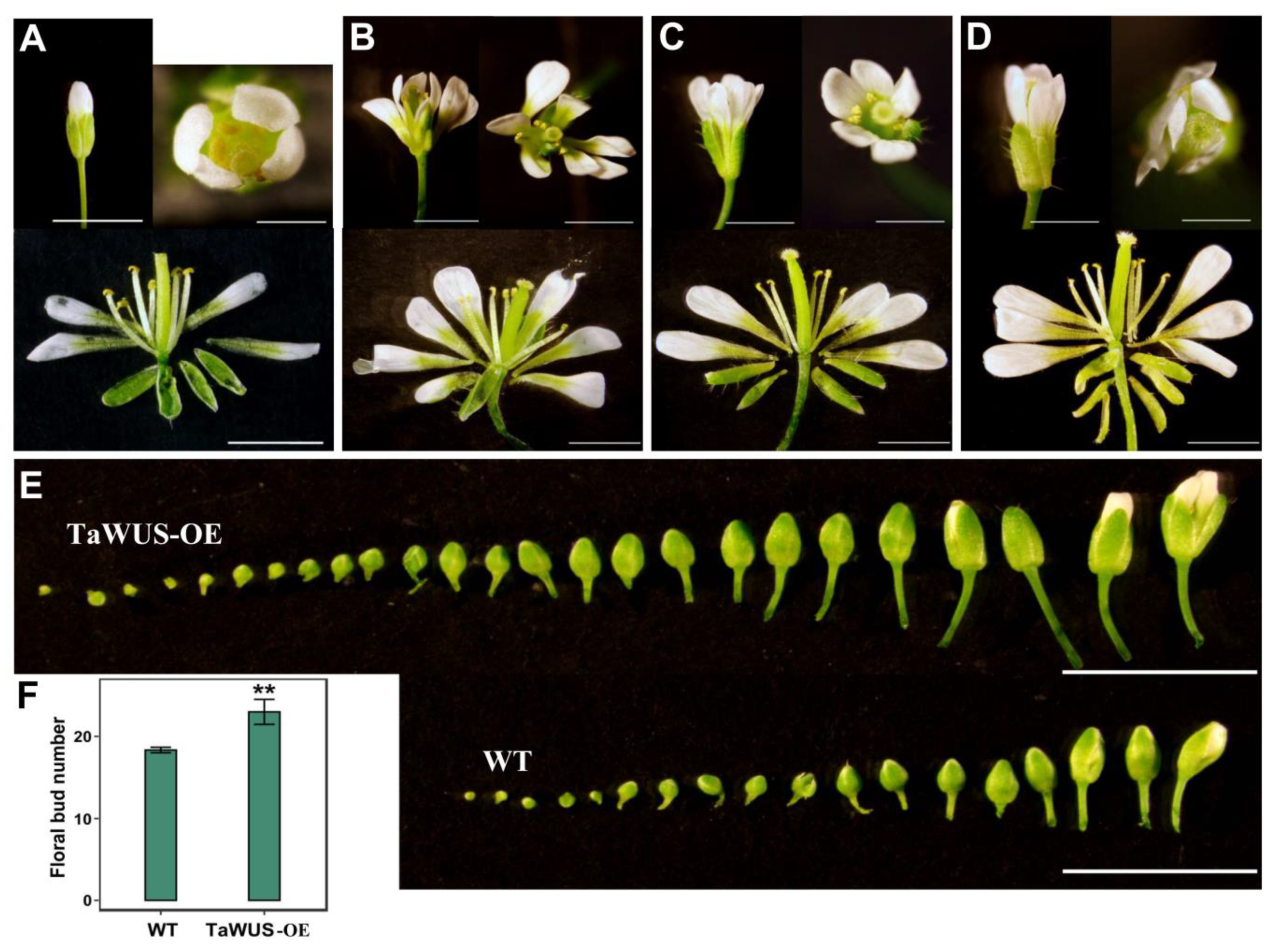
2.6. Overexpression Analysis of TaWUS and TaWOX9 in Arabidopsis
3. Discussion
3.1. Overview of the TaWOX Gene Family
3.2. Putative Functions of TaWOX Genes Based on Sequence Homology, Gene Expression Pattern and Heterologous Gene Expression
3.3. Identification of Interacting Genes of TaWUS and TaWOX9 Provides Clues for Further Study of Their Functions
4. Materials and Methods
4.1. Identification and Characterization Analysis of TaWOX Genes
4.2. Analysis of Conserved Protein Motifs
4.3. Planting and Collection of Plant Materials
4.4. Subcellular Localization and Transcriptional Activation in the Yeast GAL4 System
4.5. Yeast Two-Hybrid Screen and Assay
4.6. Bimolecular Fluorescence Complementation (BiFC) Assays
4.7. Quantitative Real Time Polymerase Chain Reaction (RT-qPCR) Analysis
4.8. Generation of Transgenic Arabidopsis
4.9. Phenotypic Analysis of Transgenic Arabidopsis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, L.; Zhang, J.; Li, J.; Zheng, H.; Chen, J.; Lu, M. WUSCHEL-related Homeobox genes in Populus tomentosa: Diversified expression patterns and a functional similarity in adventitious root formation. BMC Genom. 2014, 15, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef]
- Hao, Q.; Zhang, L.; Yang, Y.; Shan, Z.; Zhou, X.A. Genome-Wide Analysis of the WOX Gene Family and Function Exploration of GmWOX18 in Soybean. Plants 2019, 8, 215. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Niu, L.; McHale, N.A.; Ohme-Takagi, M.; Mysore, K.S.; Tadege, M. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc. Natl. Acad. Sci. USA 2013, 110, 366–371. [Google Scholar] [CrossRef] [Green Version]
- Laux, T.; Mayer, K.F.; Berger, J.; Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87. [Google Scholar]
- Mayer, K.F.X.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in Regulating Stem Cell Fate in the Arabidopsis Shoot Meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef] [Green Version]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Haecker, A.; Groß-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, R.; Ji, J.; Kelsey, E.; Ohtsu, K.; Schnable, P.S.; Scanlon, M.J. Tissue specificity and evolution of meristematic WOX3 function. Plant Physiol. 2009, 149, 841–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, X.; Lu, S.; Tian, H.; Ding, Z. WOX5 is Shining in the Root Stem Cell Niche. Trends Plant Sci. 2015, 20, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Park, S.O.; Zheng, Z.; Oppenheimer, D.G.; Hauser, B.A. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005, 132, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Dabi, T.; Weigel, D. Requirement of Homeobox Gene STIMPY/WOX9 for Arabidopsis Meristem Growth and Maintenance. Curr. Biol. 2005, 15, 436–440. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an Ortholog of WUSCHEL in Rice, Is Required for Tiller Bud Formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef]
- Mjomba, F.M.; Zheng, Y.; Liu, H.; Tang, W.; Hong, Z.; Wang, F.; Wu, W. Homeobox Is Pivotal for OsWUS Controlling Tiller Development and Female Fertility in Rice. G3 Genes Genomes Genet. 2016, 6, 2013. [Google Scholar] [CrossRef] [Green Version]
- Shao, G.; Lu, Z.; Xiong, J.; Wang, B.; Jing, Y.; Meng, X.; Liu, G.; Ma, H.; Liang, Y.; Chen, F.; et al. Tiller Bud Formation Regulators MOC1 and MOC3 Cooperatively Promote Tiller Bud Outgrowth by Activating FON1 Expression in Rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.H.; Yoo, S.C.; Zhang, H.; Pandeya, D.; Koh, H.J.; Hwang, J.Y.; Kim, G.T.; Paek, N.C. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 2013, 198, 1071–1084. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Tanaka, S.Y.; Masumoto, Y.; Nobori, N.; Ishii, H.; Hibara, K.; Itoh, J.; Tanisaka, T.; Taketa, S. Barley NARROW LEAFED DWARF1 encoding a WUSCHEL-RELATED HOMEOBOX 3 (WOX3) regulates the marginal development of lateral organs. Breed. Sci. 2016, 66, 416–424. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A Core Regulatory Pathway Controlling Rice Tiller Angle Mediated by the LAZY1 Dependent Asymmetric Distribution of Auxin. Plant Cell 2018, 30, 1461. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Hu, Y.; Dai, M.; Huang, L.; Zhou, D.X. The WUSCHEL-Related Homeobox Gene WOX11 Is Required to Activate Shoot-Borne Crown Root Development in Rice. Plant Cell 2009, 21, 736–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pont, C.; Leroy, T.; Seidel, M.; Tondelli, A.; Duchemin, W.; Armisen, D.; Lang, D.; Bustos-Korts, D.; Goue, N.; Balfourier, F.; et al. Tracing the ancestry of modern bread wheats. Nat. Genet. 2019, 51, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-Wide Analysis of WOX Gene Family in Rice, Sorghum, Maize, Arabidopsis and Poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL Is a Bifunctional Transcription Factor That Acts as a Repressor in Stem Cell Regulation and as an Activator in Floral Patterning. Plant Cell 2009, 21, 3493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL Related Homeobox Protein WOX7 Regulates the Sugar Response of Lateral Root Development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Jiang, Q.T.; Ma, J.; Zhang, X.W.; Zhao, Q.Z.; Wang, X.Y.; Wang, C.S.; Cao, X.; Lu, Z.X.; Zheng, Y.L.; et al. Characterization and expression analysis of WOX5 genes from wheat and its relatives. Gene 2014, 537, 63–69. [Google Scholar] [CrossRef]
- Liu, X.; Kim, Y.J.; Müller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS Terminates Floral Stem Cell Maintenance in Arabidopsis by Directly Repressing WUSCHEL through Recruitment of Polycomb Group Proteins. Plant Cell 2011, 23, 3654. [Google Scholar] [CrossRef] [Green Version]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 Gene of Arabidopsis thaliana Functions in Floral Organ and Meristem Identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.W.; Chang, K.Y.; Wu, K. Genome-Wide Analysis of Gene Regulatory Networks of the FVE-HDA6-FLD Complex in Arabidopsis. Front. Plant Sci. 2016, 7, 555. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.S.; Lu, Y.Q.; Meng, Y.Y.; Zhang, R.Z.; Zhang, H.; Sun, J.M.; Wang, M.M.; Li, L.H.; Li, R.Y. Identification of interacting proteins of the TaFVE protein involved in spike development in bread wheat. Proteomics 2017, 17, 1600331. [Google Scholar] [CrossRef]
- Lee, J.; Amasino, R.M. Two FLX family members are non-redundantly required to establish the vernalization requirement in Arabidopsis. Nat. Commun. 2013, 4, 2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Su, X.; Wang, Y.; Yang, W.; Pan, Y.; Su, C.; Zhang, X. Genome-wide identification and expression analysis of the BTB domain-containing protein gene family in tomato. Genes Genom. 2018, 40, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tian, X.; Zhao, Q.; Liu, Z.; Li, X.; Ren, Y.; Tang, J.; Fang, J.; Xu, Q.; Bu, Q. The E3 Ligase DROUGHT HYPERSENSITIVE Negatively Regulates Cuticular Wax Biosynthesis by Promoting the Degradation of Transcription Factor ROC4 in Rice. Plant Cell 2018, 30, 228–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, A.; Seo, M.; Kamiya, Y.; Sawa, S. Mystery in genetics: PUB4 gives a clue to the complex mechanism of CLV signaling pathway in the shoot apical meristem. Plant Signal. Behav. 2015, 10, e1028707. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, A.; ten Hove, C.A.; Tabata, R.; Yamada, M.; Shimizu, N.; Ishida, T.; Yamaguchi, K.; Shigenobu, S.; Takebayashi, Y.; Iuchi, S.; et al. A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 2015, 142, 444. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Lu, Y.; Jiang, T.; Berg, H.; Li, C.; Xia, Y. The Arabidopsis U–box/ARM repeat E3 ligase AtPUB4 influences growth and degeneration of tapetal cells, and its mutation leads to conditional male sterility. Plant J. 2013, 74, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. TeXshade: Shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, L.; Liu, Y.; Lv, Q.; Zhang, H.; Zhu, J.; Li, X. Influence of TaGW2-6A on seed development in wheat by negatively regulating gibberellin synthesis. Plant Sci. 2017, 263, 226–235. [Google Scholar] [CrossRef]
- Walter, M.; Chaban, C.; Schütze, K.; Batistic, O.; Weckermann, K.; Näke, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C.; et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004, 40, 428–438. [Google Scholar] [CrossRef]
- Bhat, R.A.; Lahaye, T.; Panstruga, R. The visible touch: In planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods 2006, 2, 12. [Google Scholar] [CrossRef] [Green Version]
- Chai, G.; Li, C.; Xu, F.; Li, Y.; Shi, X.; Wang, Y.; Wang, Z. Three endoplasmic reticulum-associated fatty acyl-coenzyme a reductases were involved in the production of primary alcohols in hexaploid wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Liu, D.; Xia, Y.; Li, Z.; Niu, N.; Ma, S.; Wang, J.; Song, Y.; Zhang, G. Identification and Functional Analysis of the CLAVATA3/EMBRYO SURROUNDING REGION (CLE) Gene Family in Wheat. Int. J. Mol. Sci. 2019, 20, 4319. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Liu, D.; Xia, Y.; Li, Z.; Jing, D.; Du, J.; Niu, N.; Ma, S.; Wang, J.; Song, Y.; et al. Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family, and Interaction and Functional Analysis of TaWOX9 and TaWUS in Wheat. Int. J. Mol. Sci. 2020, 21, 1581. https://doi.org/10.3390/ijms21051581
Li Z, Liu D, Xia Y, Li Z, Jing D, Du J, Niu N, Ma S, Wang J, Song Y, et al. Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family, and Interaction and Functional Analysis of TaWOX9 and TaWUS in Wheat. International Journal of Molecular Sciences. 2020; 21(5):1581. https://doi.org/10.3390/ijms21051581
Chicago/Turabian StyleLi, Zheng, Dan Liu, Yu Xia, Ziliang Li, Doudou Jing, Jingjing Du, Na Niu, Shoucai Ma, Junwei Wang, Yulong Song, and et al. 2020. "Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family, and Interaction and Functional Analysis of TaWOX9 and TaWUS in Wheat" International Journal of Molecular Sciences 21, no. 5: 1581. https://doi.org/10.3390/ijms21051581




